Enhanced Carbonylation of Photosynthetic and Glycolytic Proteins in Antibiotic Timentin-Treated Tobacco In Vitro Shoot Culture
Abstract
1. Introduction
2. Results
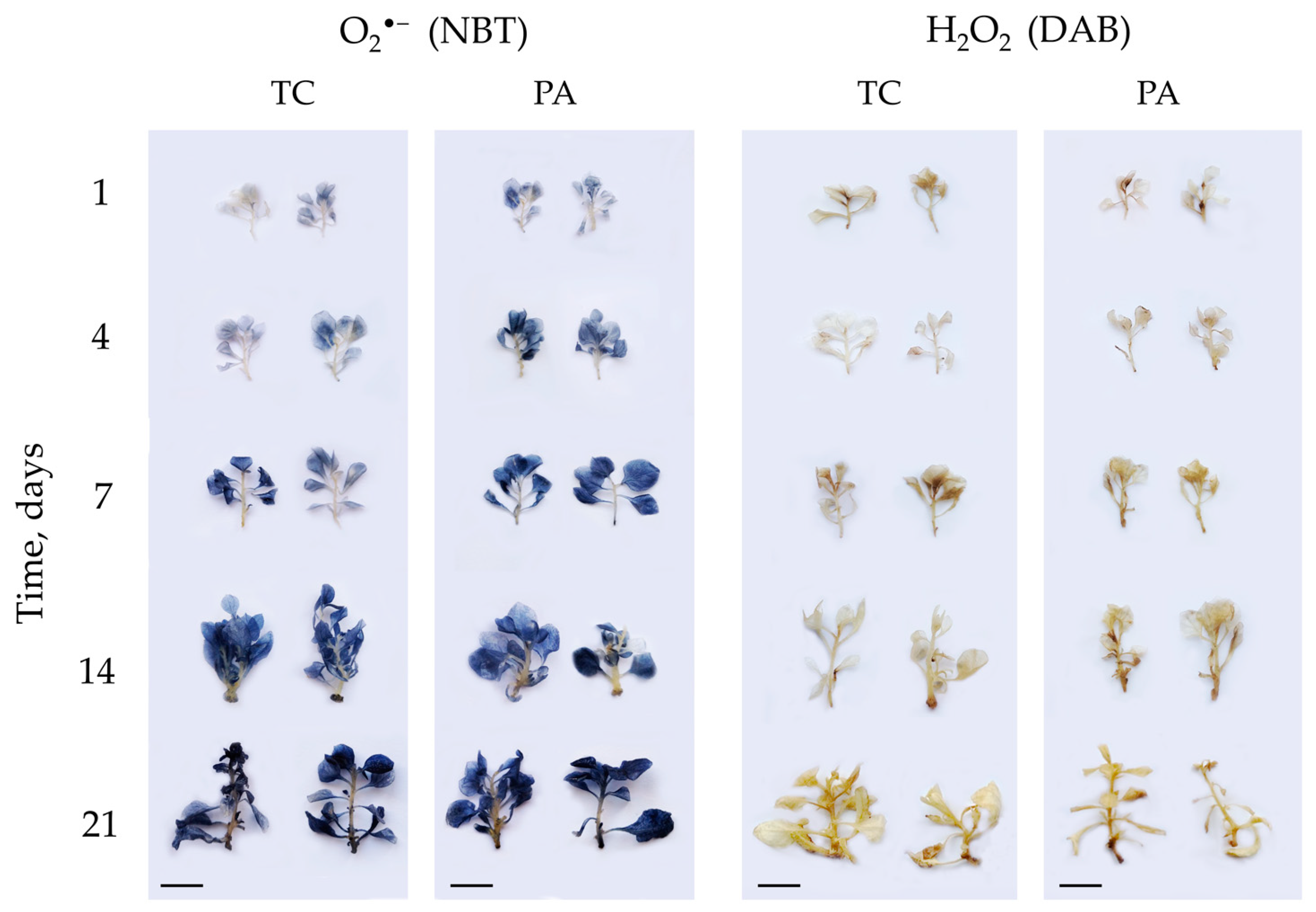
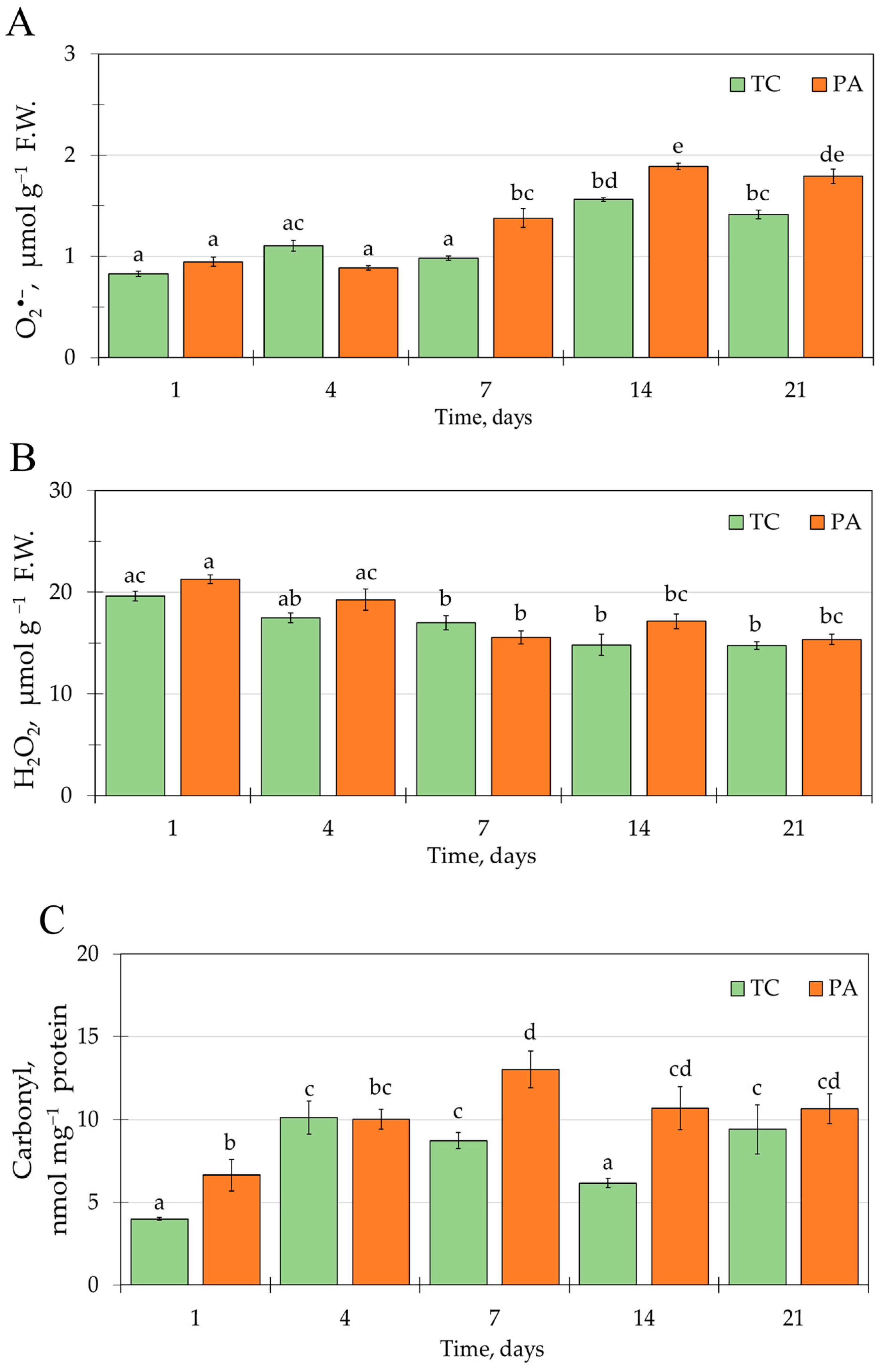
2.1. ROS Accumulation and Protein Carbonylation in Tobacco In Vitro Shoots
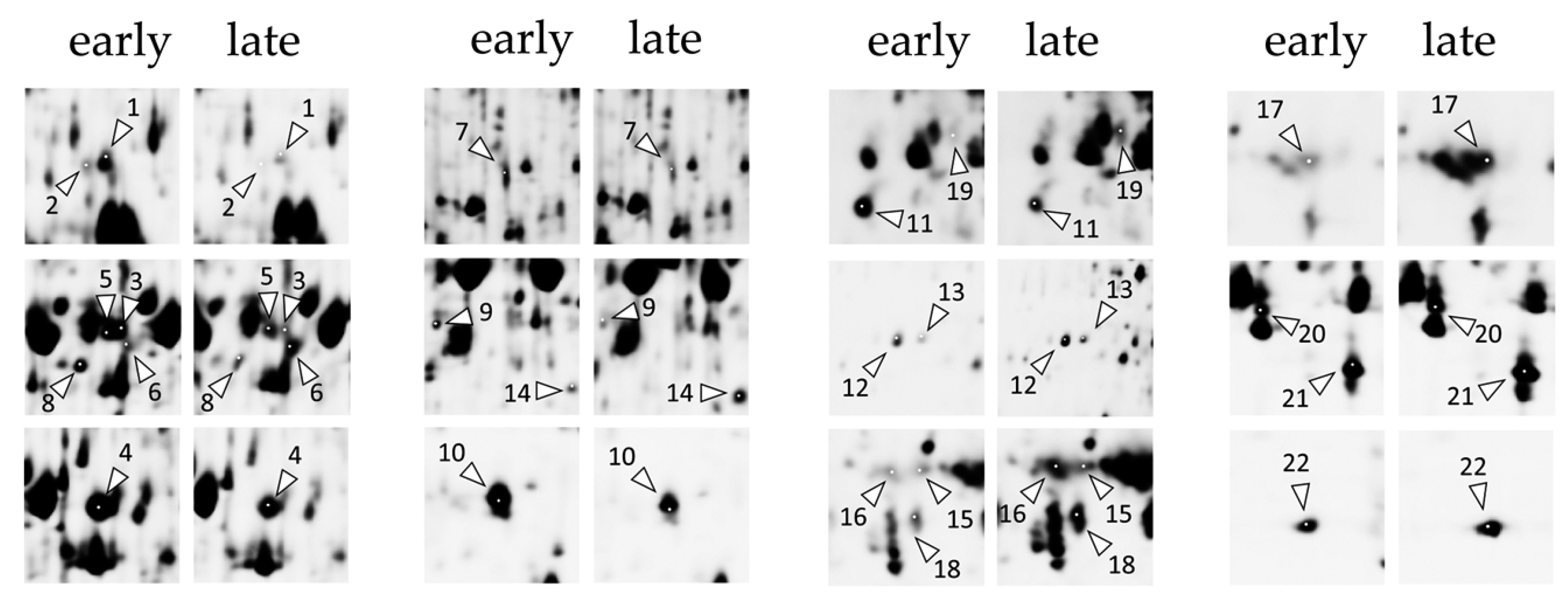
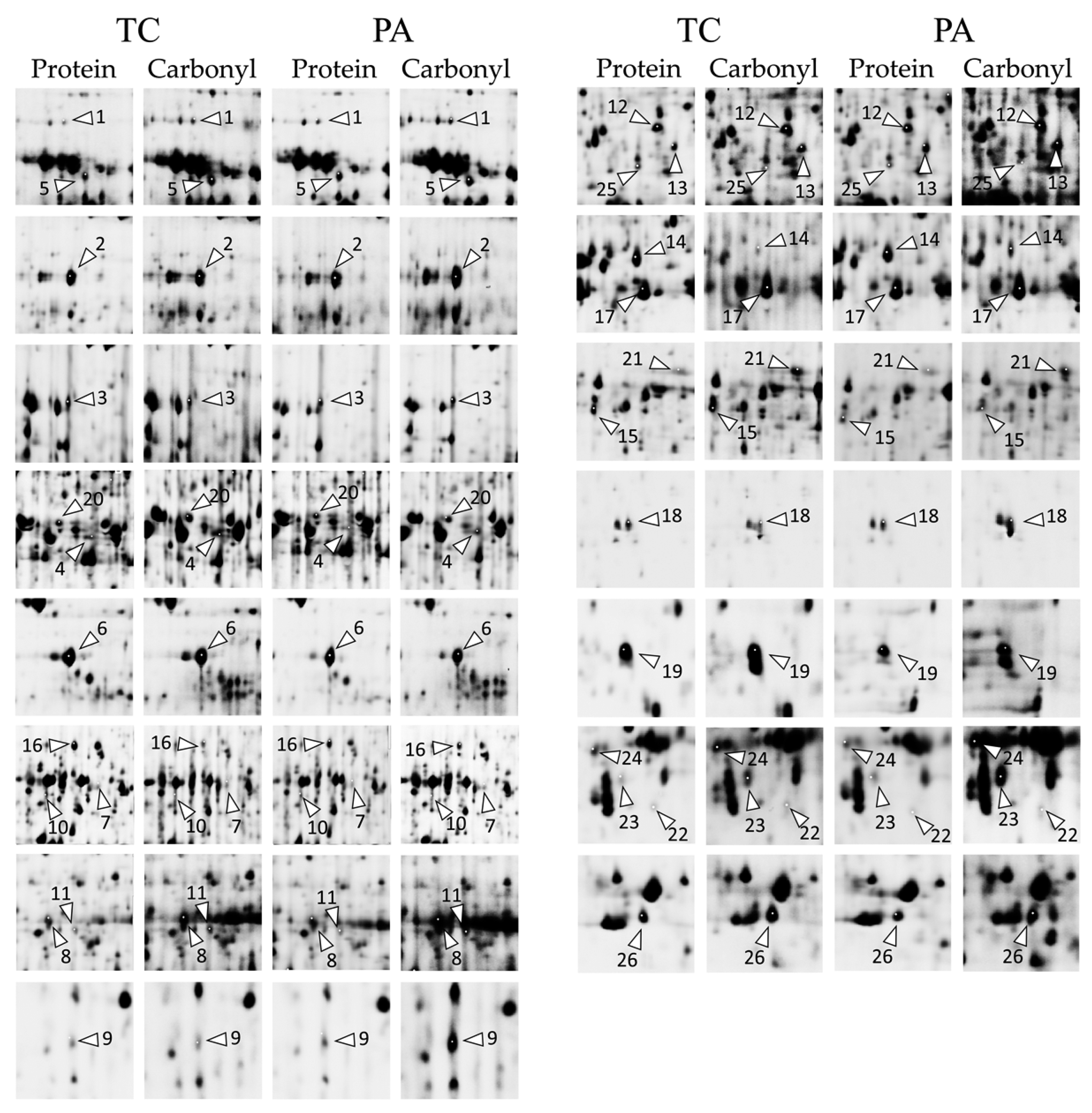
2.2. Differences in Protein Expression Associated with Tobacco Shoot Response to Timentin Treatment or Tissue Culture Senescence
2.3. Timentin-Induced Changes of Tobacco Shoot Protein Carbonylation
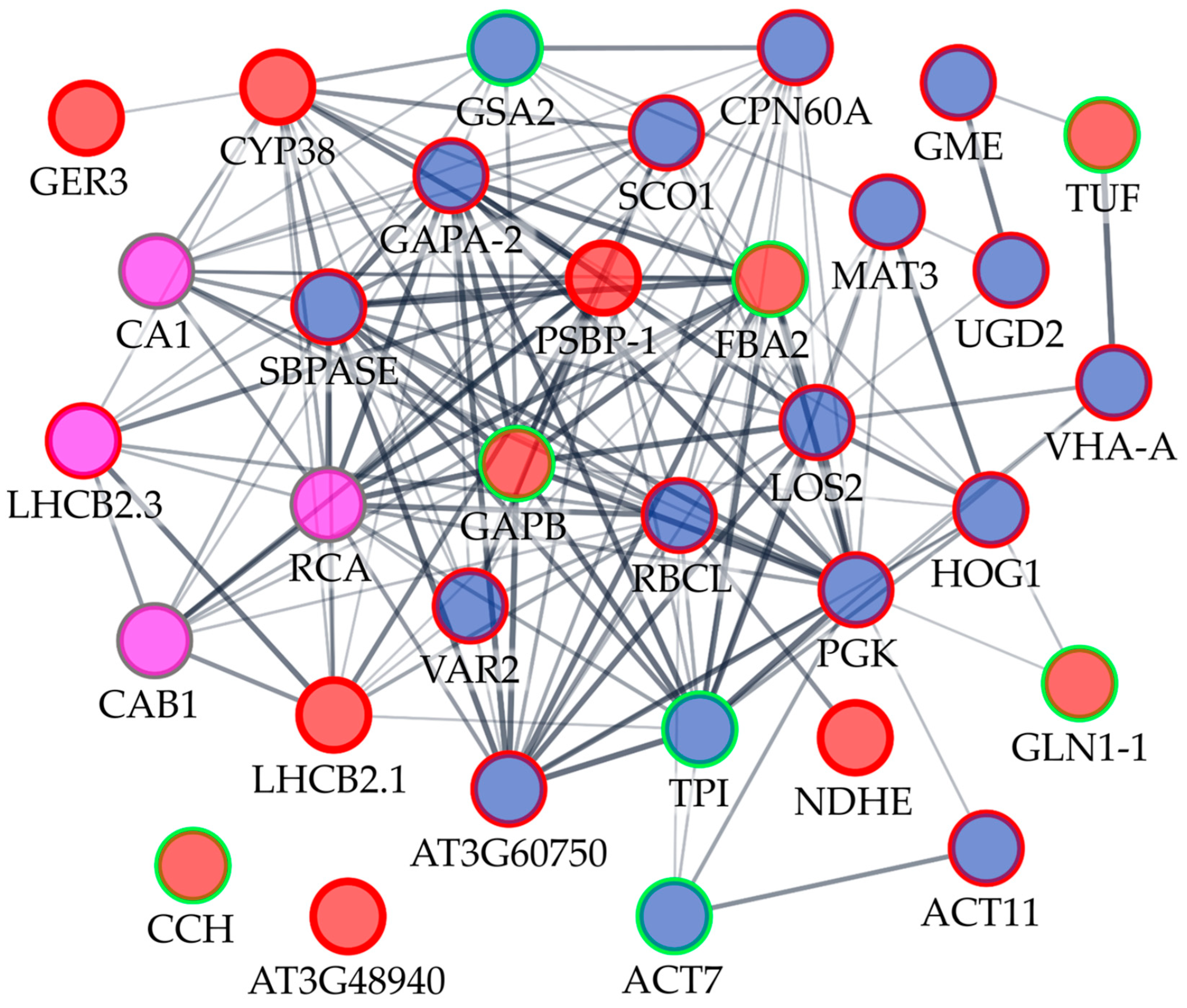
2.4. Functional Interactions of Differentially Expressed or Carbonylated Tobacco Shoot Proteins
3. Discussion
3.1. Accumulation of ROS and Protein Expression Patterns Related to Tobacco In Vitro Shoot Culture Senescence
3.2. Enhanced Protein Carbonylation in Timentin-Treated Tobacco In Vitro Shoots
4. Materials and Methods
4.1. Tobacco In Vitro Shoot Culture
4.2. Analysis of ROS Accumulation
4.3. Protein Extraction
4.4. Quantitative Analysis of Protein Carbonylation
4.5. Proteomics Analysis
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Withers, L.A.; Engelmann, F. In Vitro conservation of plant genetic resources. In Biotechnology in Agriculture; Altman, A., Ed.; Marcel Dekker Inc.: New York, NY, USA, 1998; Volume 13, pp. 57–88. [Google Scholar]
- George, E.F.; Hall, M.A.; Klerk, G.J.D. Micropropagation: Uses and methods. In Plant Propagation by Tissue Culture; George, E.F., Hall, M.A., Klerk, G.J.D., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 29–64. [Google Scholar]
- Colgan, R.; Atkinson, C.J.; Paul, M.; Hassan, S.; Drake, P.M.W.; Sexton, A.L.; Santa-Cruz, S.; James, D.; Hamp, K.; Gutteridge, C.; et al. Optimisation of contained Nicotiana tabacum cultivation for the production of recombinant protein pharmaceuticals. Transgenic Res. 2010, 19, 241–256. [Google Scholar] [CrossRef]
- Tremblay, R.; Wang, D.; Jevnikar, A.M.; Ma, S. Tobacco, a highly efficient green bioreactor for production of therapeutic proteins. Biotechnol. Adv. 2010, 28, 214–221. [Google Scholar] [CrossRef]
- Moustafa, K.; Makhzoum, A.; Tremouillaux-Guiller, J. Molecular farming on rescue of pharma industry for next generations. Crit. Rev. Biotechnol. 2016, 36, 840–850. [Google Scholar] [CrossRef]
- Ganapathi, T.R.; Suprasanna, P.; Rao, P.S.; Bapat, V.A. Tobacco (Nicotiana tabacum L.)—A model system for tissue culture interventions and genetic engineering. Indian J. Biotechnol. 2004, 3, 171–184. [Google Scholar]
- Sierro, N.; Battey, J.N.D.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef]
- Cassells, A.C. Problems in tissue culture: Culture contamination. In Micropropagation: Technology and Application; Debergh, P.C., Zimmerman, R.H., Eds.; Springer: Dordrecht, The Netherlands, 1991; pp. 31–44. [Google Scholar]
- Reed, B.M.; Buckley, P.M.; DeWilde, T.N. Detection and eradication of endophytic bacteria from micropropagated mint plants. In Vitro Cell. Dev. Biol. Plant 1995, 31, 53–57. [Google Scholar] [CrossRef]
- Cassells, A.C.; Tahmatsidou, V. The influence of local plant growth conditions on non-fastidious bacterial contamination of meristem-tips of Hydrangea cultured in vitro. Plant Cell Tiss. Org. Cult. 1996, 47, 15–26. [Google Scholar] [CrossRef]
- Mantell, S.H. Microbes intimately associated with tissue and cell cultures of tropical Dioscorea yams. Plant Cell Tiss. Org. Cult. 1998, 52, 47–52. [Google Scholar] [CrossRef]
- Anami, S.; Njuguna, E.; Coussens, G.; Aesaert, S.; Van Lijsebettens, M. Higher plant transformation: Principles and molecular tools. Int. J. Dev. Biol. 2013, 57, 483–494. [Google Scholar] [CrossRef]
- Hansen, G. Evidence for Agrobacterium-induced apoptosis in maize cells. Mol. Plant-Microbe Interact. 2000, 13, 649–657. [Google Scholar] [CrossRef]
- Ditt, R.F.; Kerr, K.F.; de Figueiredo, P.; Delrow, J.; Comai, L.; Nester, E.W. The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol. Plant-Microbe Interact. 2006, 19, 665–681. [Google Scholar] [CrossRef]
- Jiang, V.H.; Doerge, R.W.; Gelvin, S.B. Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J. 2003, 35, 219–236. [Google Scholar] [CrossRef]
- Deng, W.; Pu, X.A.; Goodman, R.N.; Gordon, M.P.; Nester, E.W. T-DNA genes responsible for inducing a necrotic response on grape vines. Mol. Plant-Microbe Interact. 1995, 8, 538–548. [Google Scholar] [CrossRef]
- Pu, X.-a.; Goodman, R.N. Induction of necrogenesis by Agrobacterium tumefaciens on grape explants. Physiol. Mol. Plant Pathol. 1992, 41, 241–254. [Google Scholar] [CrossRef]
- Ozawa, K. Establishment of a high efficiency Agrobacterium-mediated transformation system of rice (Oryza sativa L.). Plant Sci. 2009, 176, 522–527. [Google Scholar] [CrossRef]
- Thomas, P.; Prakash, G.S. Sanitizing long-term micropropagated grapes from covert and endophytic bacteria and preliminary field testing of plants after 8 years in vitro. In Vitro Cell. Dev. Biol. Plant 2004, 40, 603–607. [Google Scholar] [CrossRef]
- Kulkarni, A.A.; Kelkar, S.; Watve, M.; Krishnamurthy, K. Characterization and control of endophytic bacterial contaminants in in vitro cultures of Piper spp., Taxus baccata subsp. wallichiana, and Withania somnifera. Can. J. Microbiol. 2007, 53, 63–74. [Google Scholar] [CrossRef]
- Buckley, P.M.; Traci, N.D.; Barbara, M.R. Characterization and identification of bacteria isolated from micropropagated mint plants. In Vitro Cell. Dev. Biol. Plant 1995, 31, 58–64. [Google Scholar] [CrossRef]
- Leifert, C.; Li, H.; Chidburee, S.; Hampson, S.; Workman, S.; Sigee, D.; Epton, H.A.S.; Harbour, A. Antibiotic production and biocontrol activity by Bacillus subtilis CL27 and Bacillus pumilus CL45. J. Appl. Microbiol. 1995, 78, 97–108. [Google Scholar] [CrossRef]
- Pirttilä, A.M.; Podolich, O.; Koskimäki, J.J.; Hohtola, E.; Hohtola, A. Role of origin and endophyte infection in browning of bud-derived tissue cultures of Scots pine (Pinus sylvestris L.). Plant Cell Tiss. Org. Cult. 2008, 95, 47–55. [Google Scholar] [CrossRef]
- Thomas, P.; Swarna, G.K.; Patil, P.; Rawal, R.D. Ubiquitous presence of normally non-culturable endophytic bacteria in field shoot-tips of banana and their gradual activation to quiescent cultivable form in tissue cultures. Plant Cell Tiss. Org. Cult. 2008, 93, 39–54. [Google Scholar] [CrossRef]
- Ray, S.S.; Ali, M.N.; Mukherjee, S.; Chatterjee, G.; Banerjee, M. Elimination and molecular identification of endophytic bacterial contaminants during in vitro propagation of Bambusa balcooa. World J. Microbiol. Biotechnol. 2017, 33, 31. [Google Scholar] [CrossRef] [PubMed]
- Tewelde, S.; Patharajan, S.; Teka, Z.; Sbhatu, D.B. Assessing the efficacy of broad-spectrum antibiotics in controlling bacterial contamination in the in vitro micropropagation of ginger (Zingiber officinale Rosc). Sci. World J. 2020, 2020, 6431301. [Google Scholar] [CrossRef]
- Shehata, A.M.; Wannarat, W.; Skirvin, R.M.; Norton, M.A. The dual role of carbenicillin in shoot regeneration and somatic embryogenesis of horseradish (Armoracia rusticana) in vitro. Plant Cell Tissue Organ Cult. (PCTOC) 2010, 102, 397–402. [Google Scholar] [CrossRef]
- Tang, H.; Ren, Z.; Krczal, G. An evaluation of antibiotics for the elimination of Agrobacterium tumefaciens from walnut somatic embryos and for the effects on the proliferation of somatic embryos and regeneration of transgenic plants. Plant Cell Rep. 2000, 19, 881–887. [Google Scholar] [CrossRef]
- Izarra, M.L.; Panta, A.L.; Maza, C.R.; Zea, B.C.; Cruzado, J.; Gutarra, L.R.; Rivera, C.R.; Ellis, D.; Kreuze, J.F. Identification and control of latent bacteria in in vitro cultures of sweetpotato [Ipomoea batatas (L.) Lam]. Front. Plant Sci. 2020, 11, 903. [Google Scholar] [CrossRef]
- Nauerby, B.; Billing, K.; Wyndaele, R. Influence of the antibiotic timentin on plant regeneration compared to carbenicillin and cefotaxime in concentrations suitable for elimination of Agrobacterium tumefaciens. Plant Sci. 1997, 123, 169–177. [Google Scholar] [CrossRef]
- Ogawa, Y.; Mii, M. Screening for highly active β-lactam antibiotics against Agrobacterium tumefaciens. Arch. Microbiol. 2004, 181, 331–336. [Google Scholar] [CrossRef]
- Priya, A.M.; Pandian, S.K.; Manikandan, R. The effect of different antibiotics on the elimination of Agrobacterium and high frequency Agrobacterium-mediated transformation of indica rice (Oryza sativa L.). Czech J. Genet. Plant Breed. 2012, 48, 120–130. [Google Scholar] [CrossRef]
- Cheng, Z.M.; Schnurr, J.A.; Kapaun, J.A. Timentin as an alternative antibiotic for suppression of Agrobacterium tumefaciens in genetic transformation. Plant Cell Rep. 1998, 17, 646–649. [Google Scholar] [CrossRef]
- Leone, G.F.; Andrade, P.A.M.; de Almeida, C.V.; de Almeida, C.V.; Dini Andreote, F.; de Almeida, M. Use of antibiotics to control endophytic bacterial growth migration onto culture medium in Eucalyptus cloeziana F. Muell.: A micropropagation approach. In Vitro Cell. Dev. Biol. Plant 2019, 55, 421–432. [Google Scholar] [CrossRef]
- Tamošiūnė, I.; Andriūnaitė, E.; Vinskienė, J.; Stanys, V.; Rugienius, R.; Baniulis, D. Enduring effect of antibiotic timentin treatment on tobacco in vitro shoot growth and microbiome diversity. Plants 2022, 11, 832. [Google Scholar] [CrossRef]
- Andriūnaitė, E.; Tamošiūnė, I.; Gelvonauskienė, D.; Vinskienė, J.; Stanys, V.; Baniulis, D.; Rugienius, R. Effect of endophytic bacteria isolates on growth and oxidative stress injury of transgenic tobacco shoots in vitro. Zemdirbyste 2022, in press. [Google Scholar]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Su, L.-J.; Zhang, J.-H.; Gomez, H.; Murugan, R.; Hong, X.; Xu, D.; Jiang, F.; Peng, Z.-Y. Reactive oxygen species-induced lipid peroxidation in apoptosis, autophagy, and ferroptosis. Oxid. Med. Cell Longev. 2019, 2019, 5080843. [Google Scholar] [CrossRef]
- Shapiguzov, A.; Vainonen, J.P.; Wrzaczek, M.; Kangasjarvi, J. ROS-talk—How the apoplast, the chloroplast, and the nucleus get the message through. Front. Plant Sci. 2012, 3, 292. [Google Scholar] [CrossRef]
- Li, M.; Kim, C. Chloroplast ROS and stress signaling. Plant Commun. 2022, 3, 100264. [Google Scholar] [CrossRef]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef]
- Bauwe, H.; Hagemann, M.; Fernie, A.R. Photorespiration: Players, partners and origin. Trends Plant Sci. 2010, 15, 330–336. [Google Scholar] [CrossRef]
- Voss, I.; Sunil, B.; Scheibe, R.; Raghavendra, A.S. Emerging concept for the role of photorespiration as an important part of abiotic stress response. Plant Biol. 2013, 15, 713–722. [Google Scholar] [CrossRef]
- Miyake, C. Alternative electron flows (water–water cycle and cyclic electron flow around psi) in photosynthesis: Molecular mechanisms and physiological functions. Plant Cell Physiol. 2010, 51, 1951–1963. [Google Scholar] [CrossRef]
- Frohnert, B.I.; Bernlohr, D.A. Protein carbonylation, mitochondrial dysfunction, and insulin resistance. Adv. Nutr. 2013, 4, 157–163. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Toxicity of iron and hydrogen peroxide: The Fenton reaction. Toxicol. Lett. 1995, 82–83, 969–974. [Google Scholar] [CrossRef]
- Kehrer, J.P. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology 2000, 149, 43–50. [Google Scholar] [CrossRef]
- Stadtman, E.R.; Levine, R.L. Protein Oxidation. Ann. N. Y. Acad. Sci. 2000, 899, 191–208. [Google Scholar] [CrossRef]
- Møller, I.M.; Rogowska-Wrzesinska, A.; Rao, R.S. Protein carbonylation and metal-catalyzed protein oxidation in a cellular perspective. J. Proteom. 2011, 74, 2228–2242. [Google Scholar] [CrossRef]
- Stadtman, E.R.; Levine, R.L. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids 2003, 25, 207–218. [Google Scholar] [CrossRef]
- Ciacka, K.; Tymiński, M.; Gniazdowska, A.; Krasuska, U. Carbonylation of proteins—An element of plant ageing. Planta 2020, 252, 12. [Google Scholar] [CrossRef]
- Levine, R.L.; Garland, D.; Oliver, C.N.; Amici, A.; Climent, I.; Lenz, A.-G.; Ahn, B.-W.; Shaltiel, S.; Stadtman, E.R. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990, 186, 464–478. [Google Scholar] [CrossRef]
- Fedorova, M.; Bollineni, R.C.; Hoffmann, R. Protein carbonylation as a major hallmark of oxidative damage: Update of analytical strategies. Mass Spectrom. Rev. 2014, 33, 79–97. [Google Scholar] [CrossRef]
- Georgiou, C.D.; Zisimopoulos, D.; Argyropoulou, V.; Kalaitzopoulou, E.; Ioannou, P.V.; Salachas, G.; Grune, T. Protein carbonyl determination by a rhodamine B hydrazide-based fluorometric assay. Redox Biol. 2018, 17, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Cassells, A.C.; Curry, R.F. Oxidative stress and physiological, epigenetic and genetic variability in plant tissue culture: Implications for micropropagators and genetic engineers. Plant Cell Tiss. Org. Cult. 2001, 64, 145–157. [Google Scholar] [CrossRef]
- Saher, S.; Piqueras, A.; Hellin, E.; Olmos, E. Hyperhydricity in micropropagated carnation shoots: The role of oxidative stress. Physiol. Plant. 2004, 120, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Bairu, M.W.; Kane, M.E. Physiological and developmental problems encountered by in vitro cultured plants. Plant Growth Regul. 2011, 63, 101–103. [Google Scholar] [CrossRef]
- Jajic, I.; Sarna, T.; Strzalka, K. Senescence, stress, and reactive oxygen species. Plants 2015, 4, 393–411. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.D.; Fernandez-Pozo, N.; Drake-Stowe, K.; Humphry, M.; Evans, A.D.; Bombarely, A.; Allen, F.; Hurst, R.; White, B.; Kernodle, S.P.; et al. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genom. 2017, 18, 448. [Google Scholar] [CrossRef] [PubMed]
- Minibayeva, F.; Kolesnikov, O.; Chasov, A.; Beckett, R.P.; Luthje, S.; Vylegzhanina, N.; Buck, F.; Bottger, M. Wound-induced apoplastic peroxidase activities: Their roles in the production and detoxification of reactive oxygen species. Plant Cell Environ. 2009, 32, 497–508. [Google Scholar] [CrossRef]
- Monshausen, G.B.; Bibikova, T.N.; Weisenseel, M.H.; Gilroy, S. Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 2009, 21, 2341–2356. [Google Scholar] [CrossRef]
- Prasad, A.; Sedlářová, M.; Balukova, A.; Rác, M.; Pospíšil, P. Reactive oxygen species as a response to wounding: In vivo imaging in Arabidopsis thaliana. Front. Plant Sci. 2020, 10, 1660. [Google Scholar] [CrossRef]
- Benson, E.E. In vitro plant recalcitrance: An introduction. In Vitro Cell. Dev. Biol. Plant 2000, 36, 141–148. [Google Scholar] [CrossRef]
- Benson, E.E. Special symposium: In vitro plant recalcitrance do free radicals have a role in plant tissue culture recalcitrance? In Vitro Cell. Dev. Biol. Plant 2000, 36, 163–170. [Google Scholar] [CrossRef]
- Schafer, C.; Simper, H.; Hofmann, B. Glucose feeding results in coordinated changes of chlorophyll content, ribulose-1,5-bisphosphate carboxylase-oxygenase activity and photosynthetic potential in photoautrophic suspension cultured cells of Chenopodium rubrum. Plant Cell Environ. 1992, 15, 343–350. [Google Scholar] [CrossRef]
- Hdider, C.; Desjardins, Y. Reduction of ribulose-1,5-bisphosphate carboxylase/oxygenase efficiency by the presence of sucrose during the tissue culture of strawberry plantlets. In Vitro Cell. Dev. Biol. Plant 1995, 31, 165–170. [Google Scholar] [CrossRef]
- Van Huylenbroeck, J.M.; Debergh, P.C. Impact of sugar concentration in vitro on photosynthesis and carbon metabolism during ex vitro acclimatization of Spathiphyllum plantlets. Physiol. Plant. 1996, 96, 298–304. [Google Scholar] [CrossRef]
- Santamaría, J.M.; Talavera, C.; Lavergne, D.; Trabelsi, S.; Verdeil, J.L.; Huet, C.; Rival, A.; Hamon, S.; Nato, A. Effect of medium sucrose on the photosynthetic capacity of coconut vitroplants formed from zygotic embryos. In Current Advances in Coconut Biotechnology; Oropeza, C., Verdeil, J.L., Ashburner, G.R., Cardeña, R., Santamaría, J.M., Eds.; Springer: Dordrecht, The Netherlands, 1999; pp. 371–381. [Google Scholar]
- Laetsch, W.M.; Stetler, D.A. Chloroplast structure and function in cultured tobacco tissue. Am. J. Bot. 1965, 52, 798–804. [Google Scholar] [CrossRef]
- Jo, E.A.; Tewari, R.K.; Hahn, E.J.; Paek, K.Y. In vitro sucrose concentration affects growth and acclimatization of Alocasia amazonica plantlets. Plant Cell Tiss. Org. Cult. 2008, 96, 307. [Google Scholar] [CrossRef]
- Fu, A.; He, Z.; Cho, H.S.; Lima, A.; Buchanan, B.B.; Luan, S. A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 15947–15952. [Google Scholar] [CrossRef]
- Sirpiö, S.; Khrouchtchova, A.; Allahverdiyeva, Y.; Hansson, M.; Fristedt, R.; Vener, A.V.; Scheller, H.V.; Jensen, P.E.; Haldrup, A.; Aro, E.M. AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 2008, 55, 639–651. [Google Scholar] [CrossRef]
- Parry, M.A.; Andralojc, P.J.; Mitchell, R.A.; Madgwick, P.J.; Keys, A.J. Manipulation of Rubisco: The amount, activity, function and regulation. J. Exp. Bot. 2003, 54, 1321–1333. [Google Scholar] [CrossRef]
- Salvucci, M.E. Association of Rubisco activase with chaperonin-60β: A possible mechanism for protecting photosynthesis during heat stress. J. Exp. Bot. 2008, 59, 1923–1933. [Google Scholar] [CrossRef]
- Bracher, A.; Whitney, S.M.; Hartl, F.U.; Hayer-Hartl, M. Biogenesis and metabolic maintenance of Rubisco. Annu. Rev. Plant Biol. 2017, 68, 29–60. [Google Scholar] [CrossRef] [PubMed]
- Lawson, T.; Bryant, B.; Lefebvre, S.; Lloyd, J.C.; Raines, C.A. Decreased SBPase activity alters growth and development in transgenic tobacco plants. Plant Cell Environ. 2006, 29, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Ölçer, H.l.; Lloyd, J.C.; Raines, C.A. Photosynthetic capacity is differentially affected by reductions in sedoheptulose-1,7-bisphosphatase activity during leaf development in transgenic tobacco plants. Plant Physiol. 2001, 125, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Raines, C.A. The Calvin cycle revisited. Photosynthesis Res. 2003, 75, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Henkes, S.; Sonnewald, U.; Badur, R.; Flachmann, R.; Stitt, M. A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 2001, 13, 535–551. [Google Scholar] [CrossRef]
- Hines, K.M.; Chaudhari, V.; Edgeworth, K.N.; Owens, T.G.; Hanson, M.R. Absence of carbonic anhydrase in chloroplasts affects C3 plant development but not photosynthesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2107425118. [Google Scholar] [CrossRef]
- Chen, M.; Choi, Y.; Voytas, D.F.; Rodermel, S. Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J. 2000, 22, 303–313. [Google Scholar] [CrossRef]
- Jansson, S. The light-harvesting chlorophyll A/B-binding proteins. Biochim. Biophys. Acta 1994, 1184, 1–19. [Google Scholar] [CrossRef]
- Rochaix, J.-D.; Bassi, R. LHC-like proteins involved in stress responses and biogenesis/repair of the photosynthetic apparatus. Biochem. J. 2019, 476, 581–593. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y.; Hua, W.; Last, R.L. A new light on photosystem II maintenance in oxygenic photosynthesis. Front. Plant Sci. 2019, 10, 975. [Google Scholar] [CrossRef]
- Johansson, E.; Olsson, O.; Nyström, T. Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 22204–22208. [Google Scholar] [CrossRef] [PubMed]
- Job, C.; Rajjou, L.C.; Lovigny, Y.; Belghazi, M.; Job, D. Patterns of protein oxidation in arabidopsis seeds and during germination. Plant Physiol. 2005, 138, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Reverter-Branchat, G.; Cabiscol, E.; Tamarit, J.; Ros, J. Oxidative damage to specific proteins in replicative and chronological-aged Saccharomyces cerevisiae: Common targets and prevention by calorie restriction. J. Biol. Chem. 2004, 279, 31983–31989. [Google Scholar] [CrossRef]
- Shanmuganathan, A.; Avery, S.V.; Willetts, S.A.; Houghton, J.E. Copper-induced oxidative stress in Saccharomyces cerevisiae targets enzymes of the glycolytic pathway. FEBS Lett. 2004, 556, 253–259. [Google Scholar] [CrossRef]
- Holmgren, A. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 1989, 264, 13963–13966. [Google Scholar] [CrossRef]
- Tamarit, J.; Cabiscol, E.; Ros, J. Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J. Biol. Chem. 1998, 273, 3027–3032. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Rossi, R.; Giustarini, D.; Gagliano, N.; Lusini, L.; Milzani, A.; Di Simplicio, P.; Colombo, R. Actin carbonylation: From a simple marker of protein oxidation to relevant signs of severe functional impairment. Free Radic. Biol. Med. 2001, 31, 1075–1083. [Google Scholar] [CrossRef]
- Boone, C.H.; Grove, R.A.; Adamcova, D.; Braga, C.P.; Adamec, J. Revealing oxidative damage to enzymes of carbohydrate metabolism in yeast: An integration of 2D DIGE, quantitative proteomics, and bioinformatics. Proteomics 2016, 16, 1889–1903. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Bournonville, C.F.; Díaz-Ricci, J.C. Quantitative determination of superoxide in plant leaves using a modified NBT staining method. Phytochem. Anal. 2011, 22, 268–271. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Srivastava, S.; Lokhande, V.H.; D’Souza, S.F.; Suprasanna, P. Salt stress reveals differential antioxidant and energetics responses in glycophyte (Brassica juncea L.) and halophyte (Sesuvium portulacastrum L.). Front. Environ. Sci. 2015, 3, 1–9. [Google Scholar] [CrossRef]
- Daudi, A.; O’Brien, J.A. Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio Protoc. 2012, 2, e263. [Google Scholar] [CrossRef] [PubMed]
- Kotchoni, S.O.; Kuhns, C.; Ditzer, A.; Kirch, H.-H.; Bartels, D. Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ. 2006, 29, 1033–1048. [Google Scholar] [CrossRef] [PubMed]
- Isaacson, T.; Damasceno, C.M.B.; Saravanan, R.S.; He, Y.; Catalá, C.; Saladié, M.; Rose, J.K.C. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 2006, 1, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Wehr, N.; Williams, J.A.; Stadtman, E.R.; Shacter, E. Determination of carbonyl groups in oxidized proteins. In Stress Response: Methods and Protocols; Walker, J.M., Keyse, S.M., Eds.; Humana Press: Totowa, NJ, USA, 2000; pp. 15–24. [Google Scholar]
- Xia, Q.; El-Maarouf-Bouteau, H.; Bailly, C.; Meimoun, P. Determination of protein carbonylation and proteasome activity in seeds. Methods Mol. Biol. 2016, 1450, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Tamošiūnė, I.; Stanienė, G.; Haimi, P.; Stanys, V.; Rugienius, R.; Baniulis, D. Endophytic Bacillus and Pseudomonas spp. modulate apple shoot growth, cellular redox balance, and protein expression under in vitro conditions. Front. Plant Sci. 2018, 9, 889. [Google Scholar] [CrossRef]
- Schwartz, R.; Ting, C.S.; King, J. Whole proteome pI values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res. 2001, 11, 703–709. [Google Scholar] [CrossRef]
- Wu, S.; Wan, P.; Li, J.; Li, D.; Zhu, Y.; He, F. Multi-modality of pI distribution in whole proteome. Proteomics 2006, 6, 449–455. [Google Scholar] [CrossRef]
- Nikolic, A.; Perc, M.; Ladouce, R.; Mikecin, A.-M.; Martin, F.A.; Kralj, M.; Kriako, A.; Radman, M. Death by UVC light correlates with protein damage in isogenic human tumor cells: Primary tumor SW480 versus its metastasis SW620. J. Proteom. Comput. Biol. 2016, 2, 12. [Google Scholar] [CrossRef][Green Version]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Toronen, P.; Medlar, A.; Holm, L. PANNZER2: A rapid functional annotation web server. Nucleic Acids Res. 2018, 46, W84–W88. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Lin, D. An Information-theoretic definition of dimilarity. In Proceedings of the 15th International Conference on Machine Learning, San Francisco, CA, USA, 24–27 July 1998; pp. 296–304. [Google Scholar]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
| No. | Peptide ID 1 | Protein Name 1 | TAIR ID | Protein Symbol | Score/P.N./S.C. | M.W./pI | R.A./p-Value |
|---|---|---|---|---|---|---|---|
| 1. | 0009646g0060.1 | Glyceraldehyde 3-phosphate dehydrogenase | AT1G42970.1 | GAPB | 359/7/25 | 45.5/8.3 | −1.72/0.002 |
| 2. | 0006485g0040.1 | Rubisco activase | AT2G39730.1 | RCA | 898/18/47 | 47.9/7.6 | −1.75/0.007 |
| 3. | 0000722g0100.1 | 466/9/35 | 49.4/7.5 | −1.67/0.002 | |||
| 4. | 0000722g0100.1 | 1303/18/45 | 49.4/7.5 | −2.03/0.001 | |||
| 5. | 0023724g0010.1 | 1136/27/58 | 47.4/8.2 | −1.79/0.001 | |||
| 6. | 0000527g0230.1 | 421/11/36 | 45.1/7.6 | 1.74/0.008 | |||
| 7. | 0002564g0010.1 | Vacuolar ATP synthase subunit E1 | AT4G11150.1 | TUF | 86/2/9 | 28.9/6.8 | −1.81/0.001 |
| 8. | 0009806g0020.1 | Glutamine synthetase | AT5G37600.1 | GLN1-1 | 423/8/20 | 38.9/5.4 | −1.75/0.006 |
| 9. | 0001317g0050.1 | Fructose-bisphosphate aldolase | AT4G38970.1 | FBA2 | 797/12/39 | 43.7/6.5 | −1.56/0.006 |
| 10. | 0003337g0010.1 | Carbonic anhydrase | AT3G01500.2 | CA1 | 617/9/48 | 32.7/6.7 | −1.87/0.001 |
| 11. | 0007257g0020.1 | Heavy metal-associated protein 31 | AT3G56240.1 | CCH | 278/5/34 | 12.3/4.8 | −1.55/0.006 |
| 12. | 0003337g0100.1 | Cyclophilin-like protein | AT3G01480.1 | CYP38 | 549/7/30 | 42.8/4.7 | 1.65/0.003 |
| 13 | 0003337g0100.1 | 646/11/41 | 42.8/4.7 | 1.86/0.003 | |||
| 14. | 0004193g0010.1 | Remorin | AT3G48940.1 | AT3G48940 | 111/5/17 | 23.3/5.5 | 2.27/0.004 |
| 15. | 0002814g0040.1 | Chlorophyll A/B binding protein | AT1G29930.1 | CAB1 | 709/13/40 | 54.1/5.5 | 1.89/0.001 |
| 16. | 0002814g0040.1 | 469/10/37 | 54.1/5.5 | 2.12/0.007 | |||
| 17. | 0002814g0040.1 | 576/10/39 | 54.1/5.5 | 2.17/0.003 | |||
| 18. | 0005511g0010.1 | AT2G05100.1 | LHCB2.1 | 677/11/64 | 28.6/5.5 | 2.22/0.003 | |
| 19. | 0000441g0070.1 | AT3G27690.1 | LHCB2.3 | 187/3/18 | 28.6/5.5 | 2.20/0.003 | |
| 20. | 0008321g0040.1 | Photosystem II PsbP | AT1G06680.1 | PSBP-1 | 396/8/40 | 27.1/8.6 | 2.17/0.005 |
| 21. | 0004422g0010.1 | Germin | AT5G20630.1 | GER3 | 381/5/44 | 21.5/5.8 | 2.25/0.001 |
| 22. | 0001313g0070.1 | NADH-ubiquinone/plastoquinone oxidoreductase chain 4L | ATCG01070.1 | NDHE | 209/3/15 | 27.3/8.6 | 1.81/0.005 |
| No. | Peptide ID 1 | Protein Name 1 | TAIR ID | Protein Symbol | Score/P.N./S.C. | M.W./pI | R.A./p-Value |
|---|---|---|---|---|---|---|---|
| 1. | 0003983g0050.1 | Translation elongation factor EFG related | AT1G62750.1 | SCO1 | 108.4/4/7.6 | 85.9/5.4 | 1.5/0.008 |
| 2. | 0001329g0110.1 | Transketolase | AT3G60750.1 | AT3G60750 | 104/3/5 | 79.8/6.2 | 1.4/0.035 |
| 3. | 0002354g0050.1 | V-type ATP synthase catalytic subunit alpha | AT1G78900.2 | VHA-A | 262/8/16 | 68.6/5.2 | 1.4/0.009 |
| 4. | 0023724g0010.1 | Rubisco activase | AT2G39730.1 | RCA | 190/5/17 | 47.4/8.2 | 1.8/0.009 |
| 5. | 0000249g0010.1 | Peptidase M41 | AT2G30950.1 | VAR2 | 436/12/24 | 75.6/5.9 | 1.5/0.024 |
| 6. | 0004126g0010.1 | Chaperonin Cpn60 | AT2G28000.1 | CPN60A | 130/4/8 | 62.1/5.3 | 1.5/0.035 |
| 7. | 0000697g0210.1 | S-adenosyl-L-homocysteine hydrolase | AT4G13940.1 | HOG1 | 76/3/6 | 52.9/6.4 | 1.5/0.023 |
| 8. | 0002183g0050.1 | Ribulose bisphosphate carboxylase, large subunit | ATCG00490.1 | RBCL | 43/2/2 | 50/5.8 | 1.3/0.034 |
| 9. | 0002183g0050.1 | 107/2/4 | 50/5.8 | 1.4/0.011 | |||
| 10. | 0010922g0010.1 | Enolase | AT2G36530.1 | LOS2 | 1003/21/64 | 47.6/6.5 | 1.4/0.005 |
| 11. | 0000565g0160.1 | UDP-glucose/GDP- mannose dehydrogenase | AT3G29360.1 | UGD2 | 113/2/6 | 51.8/6.6 | 1.2/0.005 |
| 12. | 0000578g0090.1 | S-adenosylmethionine synthetase | AT2G36880.2 | MAT3 | 39/2/3 | 51.4/6.5 | 1.4/0.003 |
| 13. | 0001592g0070.1 | NAD-dependent epimerase/dehydratase | AT5G28840.2 | GME | 206/6/21 | 42.5/5.9 | 1.2/0.035 |
| 14. | 0000029g0330.1 | Phosphoglycerate kinase | AT1G79550.2 | PGK | 49/2/4 | 42.3/5.7 | 1.4/0.001 |
| 15. | 0000369g0010.1 | Sedoheptulose-1,7- bisphosphatase | AT3G55800.1 | SBPase | 609/14/40 | 44.4/6.1 | 1.4/0.040 |
| 16. | 0012115g0030.1 | Fructose-bisphosphate aldolase 2 | AT4G38970.1 | FBA2 | 410/9/26 | 42.4/6.1 | 1.4/0.016 |
| 17. | 0012714g0020.1 | 259/7/23 | 43.4/6.8 | 1.4/0.004 | |||
| 18. | 0010299g0040.1 | Glyceraldehyde-3- phosphate dehydrogenase | AT1G12900.1 | GAPA-2 | 1041/23/65 | 40.8/8.5 | 1.2/0.020 |
| 19. | 0003337g0010.1 | Carbonic anhydrase | AT3G01500.2 | CA1 | 352/10/41 | 32.7/6.7 | 1.7/0.007 |
| 20. | 0003600g0030.1 | Actin-related protein | AT3G12110.1 | ACT11 | 451/9/32 | 41.7/5.4 | 1.7/0.039 |
| 21. | 0002064g0070.1 | AT5G09810.1 | ACT7 | 308/9/36 | 38.9/5.2 | −1.5/0.009 | |
| 22. | 0000441g0070.1 | Chlorophyll A/B binding protein | AT3G27690.1 | LHCB2.3 | 318/8/45 | 28.6/5.5 | 1.42/0.001 |
| 23. | 0000441g0070.1 | 219/4/19 | 28.6/5.5 | 1.42/0.002 | |||
| 24. | 0001434g0050.1 | AT1G29930.1 | CAB1 | 41/2/3 | 30.7/6.4 | −1.7/0.026 | |
| 25. | 0011777g0030.1 | Glutamate-1-semialdehyde aminotransferase | AT3G48730.1 | GSA2 | 362/8/32 | 47.7/7.6 | −1.4/0.001 |
| 26. | 0004607g0010.1 | Triosephosphate isomerase | AT3G55440.1 | TPI | 128/4/21 | 27.2/5.7 | −1.7/0.044 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andriūnaitė, E.; Rugienius, R.; Tamošiūnė, I.; Haimi, P.; Vinskienė, J.; Baniulis, D. Enhanced Carbonylation of Photosynthetic and Glycolytic Proteins in Antibiotic Timentin-Treated Tobacco In Vitro Shoot Culture. Plants 2022, 11, 1572. https://doi.org/10.3390/plants11121572
Andriūnaitė E, Rugienius R, Tamošiūnė I, Haimi P, Vinskienė J, Baniulis D. Enhanced Carbonylation of Photosynthetic and Glycolytic Proteins in Antibiotic Timentin-Treated Tobacco In Vitro Shoot Culture. Plants. 2022; 11(12):1572. https://doi.org/10.3390/plants11121572
Chicago/Turabian StyleAndriūnaitė, Elena, Rytis Rugienius, Inga Tamošiūnė, Perttu Haimi, Jurgita Vinskienė, and Danas Baniulis. 2022. "Enhanced Carbonylation of Photosynthetic and Glycolytic Proteins in Antibiotic Timentin-Treated Tobacco In Vitro Shoot Culture" Plants 11, no. 12: 1572. https://doi.org/10.3390/plants11121572
APA StyleAndriūnaitė, E., Rugienius, R., Tamošiūnė, I., Haimi, P., Vinskienė, J., & Baniulis, D. (2022). Enhanced Carbonylation of Photosynthetic and Glycolytic Proteins in Antibiotic Timentin-Treated Tobacco In Vitro Shoot Culture. Plants, 11(12), 1572. https://doi.org/10.3390/plants11121572







