Root Breeding in the Post-Genomics Era: From Concept to Practice in Apple
Abstract
:1. Introduction
2. Rootstock Genetic and Molecular Regulation
2.1. Abundance of Rootstock Resources and Genomic Diversity
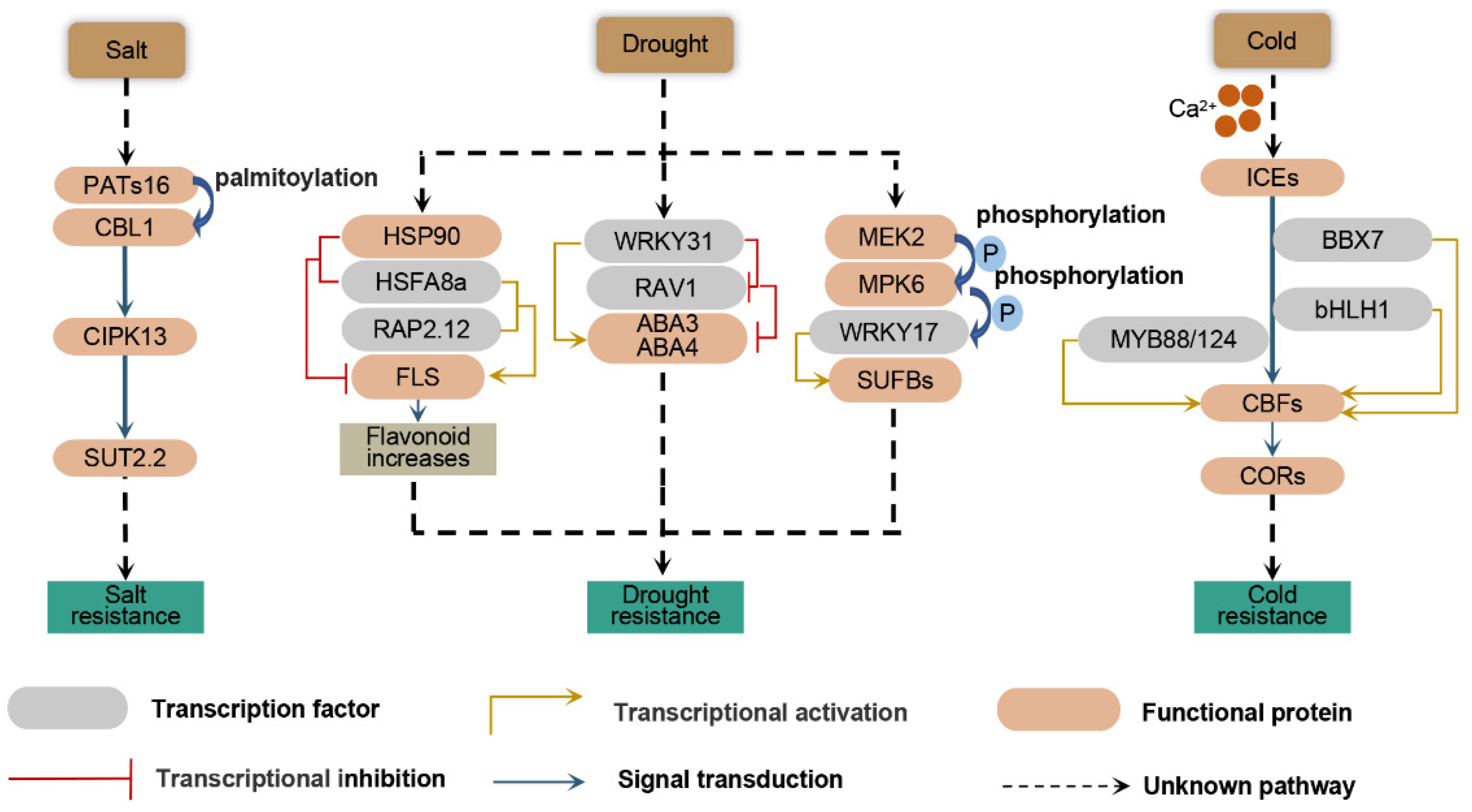
2.2. Biotic and Abiotic Stress Modulation in Apple
2.3. Mineral Nutrient Regulation of the Apple Root System and Design of the Ideal Root System Architecture
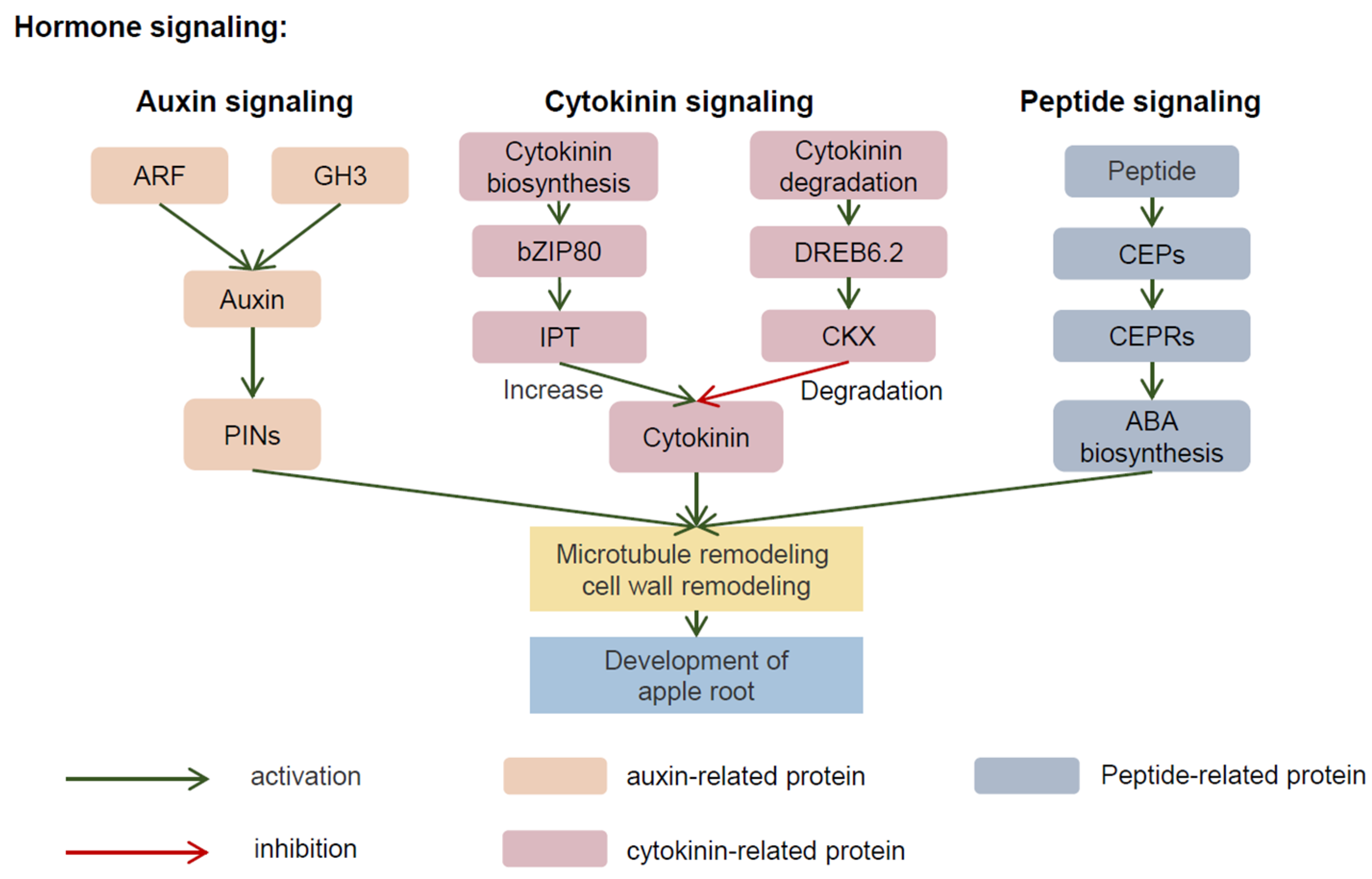
2.4. Root Architecture and Hormone Regulation
3. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Igarashi, M.; Hatsuyama, Y.; Harada, T.; Fukasawa-Akada, T. Biotechnology and apple breeding in Japan. Breed. Sci. 2016, 66, 18–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonso, S.; Ribeiro, C.; Bacelar, E.; Ferreira, H.; Oliveira, I. Influence of training system on physiological performance, biochemical composition and antioxidant parameters in apple tree (Malus domestica Borkh.). Sci. Hortic. 2017, 225, 394–398. [Google Scholar] [CrossRef]
- Warschefsky, E.J.; Klein, L.L.; Frank, M.H.; Chitwood, D.H.; Londo, J.P.; von Wettberg, E.J.B.; Miller, A.J. Rootstocks: Diversity, Domestication, and Impacts on Shoot Phenotypes. Trends Plant Sci. 2016, 21, 418–437. [Google Scholar] [CrossRef] [PubMed]
- Bohra, A. Emerging paradigms in genomics-based crop improvement. Sci. World J. 2013, 2013, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Chen, J.; Schwaninger, H.; Chao, C.T.; Fei, Z. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 2020, 52, 1423–1432. [Google Scholar] [CrossRef]
- Lin, L.; Luo, X.C.; Ge, H.J.; Guo, C.; Sha, G.L.; Jiang, Z.S.; Zhang, S.Z.; Shu, H.R. Apple, from omics to systemic function. Plant Growth Regul. 2017, 83, 1–11. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y. Drought Resistance in Rice from Conventional to Molecular Breeding: A Review. Int. J. Mol. Sci. 2019, 20, 3519. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Liu, F.; Gao, Y.; Wang, D.; Gong, X.; Liu, L. The natural distribution, diversity and utilization of wild in China. J. Plant Genet. Resour. 2013, 6, 14. [Google Scholar]
- Wang, D.; Xiao, Y.; Gao, Y.; Sun, S.; Li, L.; Zhang, F.; Wang, K. Current status of research on collection, conservation and utilization of wild Malus resources in China. China Fruit Tree 2021, 10, 6–11. [Google Scholar] [CrossRef]
- Li, Y. A primarily modern systematics of genus malus mill. In the world. J. Fruit Sci. 1996, 13, 82–92. [Google Scholar]
- Chu, A.; Tang, G. Research progress on the classification of ornamental begonia varieties. Biol. Bull. 2008, 43, 4. [Google Scholar]
- Chen, X.; Li, S.; Zhang, D.; Han, M.; Jin, X.; Zhao, C.; Wang, S.; Xing, L.; Ma, J.; Ji, J.; et al. Sequencing of a wild apple (Malus baccata) genome unravels the differences between cultivated and wild apple species regarding disease resistance and cold tolerance. G3 2019, 9, 2051–2060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.; Liu, F.; Wang, Y.; Fu, C.; Yan, Y.; Sha, G.; Shu, H. Evaluation of zinc deficiency tolerance in different kinds of apple rootstocks. J. Appl. Ecol. 2015, 26, 3045–3052. [Google Scholar]
- Yang, F.; Liu, Z.; He, M.; Wang, D.; Yang, Z.; Lv, T.; Yi, K. ‘LZ106’, A new apomixis and semi-dwarfing rootstock for apple. North Fruit Trees 2018, 2, 8–10. [Google Scholar] [CrossRef]
- Wada, M.; Nishitani, C.; Komori, S. Stable and efficient transformation of apple. Plant Biotechnol. (Tokyo) 2020, 25, 163–170. [Google Scholar] [CrossRef]
- Johnson, W.; Aldwinckle, H.; Cummins, J.; Forsline, P.; Holleran, H.; Norelli, J.; Robinson, T. The new usda-ars/cornell university apple rootstock breeding and evaluation program. Acta Hortic. 1999, 557, 35–40. [Google Scholar] [CrossRef]
- Rufato, L.; Silva, P.S.; Kretzschmar, A.A.; Bogo, A.; Macedo, T.A.; Welter, J.F.; Fazio, G.; Petry, D. Geneva® series rootstocks for apple trees under extreme replanting conditions in southern brazil. Front Plant Sci. 2021, 30, 712162. [Google Scholar] [CrossRef]
- Yu, D.; Yan, Z.; Zhang, P. Chinese fruit tree rootstock resources. China Fruit Tree 1979, 1, 1–7. [Google Scholar] [CrossRef]
- Chen, E.; Huang, X.; Tian, Z.; Wing, R.A.; Han, B. The genomics of oryza species provides insights into rice domestication and heterosis. Annu. Rev. Plant Biol. 2019, 70, 639–665. [Google Scholar] [CrossRef]
- Großkinsky, D.K.; Svensgaard, J.; Christensen, S.; Roitsch, T. Plant phenomics and the need for physiological phenotyping across scales to narrow the genotype-to-phenotype knowledge gap. J. Exp. Bot. 2015, 66, 5429–5440. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.; Lee, T.; Lee, J.; Shim, S.; Jeong, H.; Satyawan, D.; Kim, M.; Lee, S. Translational genomics for plant breeding with the genome sequence explosion. Plant Biotechol. J. 2016, 14, 1057–1069. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Hilario, E.; Deng, C. Turbocharging introgression breeding of perennial fruit crops: A case study on apple. Hortic. Res. 2020, 7, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soundararajan, P.; Won, S.; Kim, J. Insight on rosaceae family with genome sequencing and functional genomics perspective. Biomed. Res. Int. 2019, 2019, 7519687. [Google Scholar] [CrossRef]
- Giovannoni, J. Harvesting the apple genome. Nat. Genet. 2010, 42, 822–823. [Google Scholar] [CrossRef]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Nishio, S.; Hayashi, T.; Shirasawa, K.; Saito, T.; Terakami, S.; Takada, N.; Takeuchi, Y.; Moriya, S.; Itai, A. Genome-wide association study of individual sugar content in fruit of Japanese pear (Pyrus spp.). BMC Plant Biol. 2021, 21, 378. [Google Scholar] [CrossRef] [PubMed]
- Peace, C.; Bianco, L.; Troggio, M.; Van de Weg, E. Apple whole genome sequences: Recent advances and new prospects. Hortic. Res. 2019, 6, 59. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Hu, J.; Han, X.; Li, J.; Gao, Y.; Richards, C.M.; Zhang, C.; Tian, Y.; Liu, G.; Gul, H.; et al. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 2019, 10, 1494. [Google Scholar] [CrossRef] [Green Version]
- Duan, N.; Bai, Y.; Sun, H.; Wang, N.; Ma, Y.; Li, M.; Wang, X.; Jiao, C.; Legall, N.; Mao, L.; et al. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat. Commun. 2017, 15, 249. [Google Scholar] [CrossRef]
- Chen, P.; Li, Z. Insights into the effect of human civilization on Malus evolution and domestication. Plant Biotechnol. J. 2021, 11, 2206–2220. [Google Scholar] [CrossRef]
- Xu, Y.; Li, P.; Zou, C.; Lu, Y.; Xie, C.; Zhang, X.; Prasanna, B.M.; Olsen, M.S. Enhancing genetic gain in the era of molecular breeding. J. Exp. Bot. 2017, 68, 2641–2666. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Cózar, I.; Perez-Garcia, C.; Benzekri, H.; Sánchez, J.; Seoane, P.; Cruz, F.; Gut, M.; Zamorano, M.; Claros, M.; Manchado, M. Development of whole-genome multiplex assays and construction of an integrated genetic map using SSR markers in Senegalese sole. Sci. Rep. 2020, 10, 21905. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, X.; Wei, J.; Li, Y.; Tigabu, M.; Zhao, X. Development and Transferability of EST-SSR Markers for Pinus koraiensis from Cold-Stressed Transcriptome through Illumina Sequencing. Genes 2020, 11, 500. [Google Scholar] [CrossRef]
- Taheri, S.; Lee Abdullah, T.; Yusop, M. Mining and development of novel ssr markers using next generation sequencing (ngs) data in plants. Molecules 2018, 23, 399. [Google Scholar] [CrossRef] [Green Version]
- Reim, S.; Lochschmidt, F.; Proft, A.; Höfer, M. Genetic integrity is still maintained in natural populations of the indigenous wild apple species Malus sylvestris (Mill.) in Saxony as demonstrated with nuclear SSR and chloroplast DNA markers. Ecol. Evol. 2020, 10, 11798–11809. [Google Scholar] [CrossRef] [PubMed]
- Garkava-Gustavsson, L.; Kolodinska Brantestam, A.; Sehic, J.; Nybom, H. Molecular characterisation of indigenous Swedish apple cultivars based on SSR and S-allele analysis. Hereditas 2008, 145, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Jamann, T.; Balint-Kurti, P.; Holland, J. QTL mapping using high-throughput sequencing. Methods Mol. Biol. 2015, 1284, 257–285. [Google Scholar] [CrossRef]
- Powder, K. Quantitative Trait Loci (QTL) Mapping. Methods Mol. Biol. 2020, 2082, 211–229. [Google Scholar] [CrossRef]
- Calenge, F.; Drouet, D.; Denancé, C.; Van de Weg, W.; Brisset, M.; Paulin, J.; Durel, C. Identification of a major QTL together with several minor additive or epistatic QTLs for resistance to fire blight in apple in two related progenies. Theor. Appl. Genet. 2005, 111, 128–135. [Google Scholar] [CrossRef]
- Gardiner, S.; Norelli, J.; De Silva, N.; Fazio, G.; Peil, A.; Malnoy, M.; Horner, M.; Bowatte, D.; Carlisle, C.; Wiedow, C.; et al. Putative resistance gene markers associated with quantitative trait loci for fire blight resistance in Malus ‘Robusta 5’ accessions. BMC Genet. 2012, 13, 25. [Google Scholar] [CrossRef]
- Khan, M.; Zhao, Y.; Korban, S. Identification of genetic loci associated with fire blight resistance in Malus through combined use of QTL and association mapping. Physiol. Plant 2013, 148, 344–353. [Google Scholar] [CrossRef]
- Kostick, S.; Teh, S.; Norelli, J.; Vanderzande, S.; Peace, C.; Evans, K. Fire blight QTL analysis in a multi-family apple population identifies a reduced-susceptibility allele in ‘Honeycrisp’. Hortic. Res. 2021, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Le Roux, P.M.; Khan, M.A.; Broggini, G.; Duffy, B.; Gessler, C.; Patocchi, A. Mapping of quantitative trait loci for fire blight resistance in the apple cultivars ‘Florina’ and ‘Nova Easygro’. Genome 2010, 53, 710–722. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Sha, G.; Wei, T.; Ma, B.; Ma, F. Linkage map and qtl mapping of red flesh locus in apple using a r1r1 × r6r6 population. Hortic. Plant J. 2020, 7, 8. [Google Scholar] [CrossRef]
- Zheng, W.; Shen, F.; Wang, W.; Wu, B.; Wang, X.; Xiao, C.; Tian, Z.; Yang, X.; Yang, J.; Wang, Y.; et al. Quantitative trait loci-based genomics-assisted prediction for the degree of apple fruit cover color. Plant Genome 2020, 13, e20047. [Google Scholar] [CrossRef]
- Ngou, B.M.; Ding, P.; Jones, J. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 2022, 10, 1494. [Google Scholar] [CrossRef] [PubMed]
- Bjornson, M.; Pimprikar, P.; Nürnberger, T.; Zipfel, C. The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat. Plants 2021, 7, 579–586. [Google Scholar] [CrossRef]
- Dangl, J.; Horvath, D.; Staskawicz, B. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Dong, C.; Sun, X.; Zhang, Y.; Dai, H.; Bai, S. Overexpression of an apple LysM-containing protein gene, MdCERK1-2, confers improved resistance to the pathogenic fungus, Alternaria alternata, in Nicotiana benthamiana. BMC Plant Biol. 2020, 20, 146. [Google Scholar] [CrossRef]
- Zhou, Z.; Tian, Y.; Cong, P.; Zhu, Y. Functional characterization of an apple (Malus × domestica) LysM domain receptor encoding gene for its role in defense response. Plant Sci. 2018, 269, 56–65. [Google Scholar] [CrossRef]
- Chen, Q.; Li, Q.; Qiao, X.; Yin, H.; Zhang, S. Genome-wide identification of lysin motif containing protein family genes in eight rosaceae species, and expression analysis in response to pathogenic fungus Botryosphaeria dothidea in Chinese white pear. BMC Genom. 2020, 21, 612. [Google Scholar] [CrossRef] [PubMed]
- Piazza, S.; Campa, M.; Pompili, V.; Costa, L.D.; Malnoy, M. The Arabidopsis pattern recognition receptor EFR enhances fire blight resistance in apple. Hortic. Res. 2021, 8, 204. [Google Scholar] [CrossRef] [PubMed]
- Min, C.; Ya-Ning, C.; Wei-Hong, L.; Ya-Peng, C.; Bin, H.E.; Jiang-Hui, W. Drought Resistance Mechanism of Three Plant Species in the Middle Reaches of Tarim River, China. Acta Bot. Boreali-Occident. Sin. 2007, 27, 747–754. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Wang, N.; Xue, Y.; Guan, Q.; Van Nocker, S.; Liu, C.; Ma, F. Overexpression of the RNA binding protein MhYTP1 in transgenic apple enhances drought tolerance and WUE by improving ABA level under drought condition. Plant Sci. 2019, 280, 397–407. [Google Scholar] [CrossRef]
- Lian, X.; Wang, X.; Gao, H.; Jiang, H.; Mao, K.; You, C.; Li, Y.; Hao, Y. Genome wide analysis and functional identification of MdKCS genes in apple. Plant Physiol. Biochem. 2020, 151, 299–312. [Google Scholar] [CrossRef]
- Ren, Y.; Yang, Y.; Zhang, R.; You, C.; Zhao, Q.; Hao, Y. MdGRF11, an apple 14-3-3 protein, acts as a positive regulator of drought and salt tolerance. Plant Sci. 2019, 288, 110219. [Google Scholar] [CrossRef]
- Tan, Y.; Li, M.; Yang, Y.; Sun, X.; Wang, N.; Liang, B.; Ma, F. Overexpression of MpCYS4, A Phytocystatin Gene from Malus prunifolia (Willd.) Borkh., Enhances Stomatal Closure to Confer Drought Tolerance in Transgenic Arabidopsis and Apple. Front. Plant Sci. 2017, 8, 33. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Guo, T.; Sun, X.; Jia, X.; Wang, P.; Shao, Y.; Liang, B.; Gong, X.; Ma, F. Functions of two Malus hupehensis (Pamp.) Rehd. YTPs (MhYTP1 and MhYTP2) in biotic- and abiotic-stress responses. Plant Sci. 2017, 261, 18–27. [Google Scholar] [CrossRef]
- Wang, N.; Guo, T.; Wang, P.; Sun, X.; Shao, Y.; Jia, X.; Liang, B.; Gong, X.; Ma, F. MhYTP1 and MhYTP2 from apple confer tolerance to multiple abiotic stresses in Arabidopsis thaliana. Front Plant Sci. 2017, 8, 1367. [Google Scholar] [CrossRef] [Green Version]
- Zhong, M.; Jiang, H.; Cao, Y.; Wang, Y.; You, C.; Li, Y.; Hao, Y. MdCER2 conferred to wax accumulation and increased drought tolerance in plants. Plant Physiol. Biochem. 2020, 149, 277–285. [Google Scholar] [CrossRef]
- Li, R.; Ge, H.; Dai, Y.; Yuan, L.; Liu, X.; Sun, Q.; Wang, X. Genomewide analysis of homeobox gene family in apple (Malus domestica Borkh.) and their response to abiotic stress. J. Genet. 2019, 98, 13. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, P.; Xie, Y.; Yan, Y.; Wang, L.; Dang, H.; Zhang, J.; Xu, L.; Ma, F.; Guan, Q. Apple SERRATE negatively mediates drought resistance by regulating MdMYB88 and MdMYB124 and microRNA biogenesis. Hortic. Res. 2020, 7, 98. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, R.; Dai, Y.; Yuan, L.; Sun, Q.; Zhang, S.; Wang, X. A B-box zinc finger protein, MdBBX10, enhanced salt and drought stresses tolerance in Arabidopsis. Planta 2019, 99, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zheng, P.; Ren, Y.; Yao, Y.; You, C.; Wang, X.; Hao, Y. Apple MdSAT1 encodes a bHLHm1 transcription factor involved in salinity and drought responses. Planta 2021, 253, 46. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, C.; Wang, G.; Wang, Y.; Qi, C.; You, C.; Li, Y.; Hao, Y. Apple AP2/EREBP transcription factor MdSHINE2 confers drought resistance by regulating wax biosynthesis. Planta 2019, 249, 1627–1643. [Google Scholar] [CrossRef]
- Zhao, Q.; Fan, Z.; Qiu, L.; Che, Q.; Wang, T.; Li, Y.; Wang, Y. MdbHLH130, an Apple bHLH Transcription Factor, Confers Water Stress Resistance by Regulating Stomatal Closure and ROS Homeostasis in Transgenic Tobacco. Plant Mol. Biol. 2020, 11, 543696. [Google Scholar] [CrossRef]
- Zhao, X.; Qi, C.; Jiang, H.; You, C.; Guan, Q.; Ma, F.; Li, Y.; Hao, Y. The MdWRKY31 transcription factor binds to the MdRAV1 promoter to mediate ABA sensitivity. Hortic. Res. 2019, 6, 66. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Xu, Y.; Huang, J.; Jiang, Z.; Shu, H.; Wang, H.; Zhang, S. Global identification, classification, and expression analysis of MAPKKK genes: Functional characterization of MdRaf5 reveals evolution and drought-responsive profile in apple. Sci. Rep. 2017, 7, 13511. [Google Scholar] [CrossRef] [Green Version]
- Shan, D.; Wang, C.; Song, H.; Bai, Y.; Zhang, H.; Hu, Z.; Wang, L.; Shi, K.; Zheng, X.; Yan, T.; et al. The MdMEK2-MdMPK6-MdWRKY17 pathway stabilizes chlorophyll levels by directly regulating MdSUFB in apple under drought stress. Plant J. 2021, 108, 814–828. [Google Scholar] [CrossRef]
- Wang, N.; Liu, W.; Yu, L.; Guo, Z.; Chen, Z.; Jiang, S.; Xu, H.; Fang, H.; Wang, Y.; Zhang, Z.; et al. HEAT SHOCK FACTOR A8a modulates flavonoid synthesis and drought tolerance. Plant Physiol. 2020, 184, 1273–1290. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Zhang, X.; Bi, S.; You, C.; Wang, X.; Hao, Y. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Deng, Z.; Liang, C.; Sun, H.; Li, D.; Song, J.; Zhang, S.; Wang, R. Genome-wide analysis of rav transcription factors and functional characterization of anthocyanin-biosynthesis-related RAV genes in pear. Plant Cell Environ. 2021, 22, 5567. [Google Scholar] [CrossRef]
- Sun, X.; Jia, X.; Huo, L.; Che, R.; Gong, X.; Wang, P.; Ma, F. MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 2018, 41, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.; Guo, Z.; Jia, X.; Sun, X.; Wang, P.; Gong, X.; Ma, F. Increased autophagic activity in roots caused by overexpression of the autophagy-related gene MdATG10 in apple enhances salt tolerance. Plant Sci. 2020, 294, 110444. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Zhu, Y.; Zhang, R.; Zhu, Z.; Zhao, T.; Cheng, L.; Gao, L.; Liu, B.; Zhang, X.; Wang, Y. Ionomic and metabolomic analyses reveal the resistance response mechanism to saline-alkali stress in Malus halliana seedlings. Plant Physiol. Bioch. 2020, 147, 77–90. [Google Scholar] [CrossRef]
- Liu, C.; Li, C.; Liang, D.; Wei, Z.; Zhou, S.; Wang, R.; Ma, F. Differential expression of ion transporters and aquaporins in leaves may contribute to different salt tolerance in Malus species. Plant Physiol. Bioch. 2012, 58, 159–165. [Google Scholar] [CrossRef]
- Sun, T.; Pei, T.; Yang, L.; Zhang, Z.; Li, M.; Liu, Y.; Ma, F.; Liu, C. Exogenous application of xanthine and uric acid and nucleobase-ascorbate transporter MdNAT7 expression regulate salinity tolerance in apple. BMC Plant Biol. 2021, 21, 52. [Google Scholar] [CrossRef]
- Yao, Y.; Dong, Q.; You, C.; Zhai, H.; Hao, Y. Expression analysis and functional characterization of apple MdVHP1 gene reveals its involvement in Na (+), malate and soluble sugar accumulation. BMC Plant Biol. 2011, 49, 1201–1208. [Google Scholar] [CrossRef]
- Zhu, J. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [Green Version]
- Hu, D.; Li, M.; Luo, H.; Dong, Q.; Yao, Y.; You, C.; Hao, Y. Molecular cloning and functional characterization of MdSOS2 reveals its involvement in salt tolerance in apple callus and Arabidopsis. Plant Cell Rep. 2012, 31, 713–722. [Google Scholar] [CrossRef]
- Wang, R.; Li, L.; Cao, Z.; Zhao, Q.; Li, M.; Zhang, L.; Hao, Y. Molecular cloning and functional characterization of a novel apple MdCIPK6L gene reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Plant Mol. Biol. 2012, 79, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Wu, C.; Luo, C.; Wei, M.; Qu, S.; Wang, S. Overexpression of MdCPK1a gene, a calcium dependent protein kinase in apple, increase tobacco cold tolerance via scavenging ROS accumulation. PLoS ONE 2020, 15, e0242139. [Google Scholar] [CrossRef]
- Jiang, H.; Ma, Q.; Zhong, M.; Gao, H.; Li, Y. The apple palmitoyltransferase MdPAT16 influences sugar content and salt tolerance via an MdCBL1-MdCIPK13-MdSUT2.2 pathway. Plant J. 2021, 106, 689–705. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, R.; Luo, X.; Jiang, Z.; Shu, H. Genome-wide identification and expression analysis of MAPK and MAPKK gene family in Malus domestica. Gene 2013, 531, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Teige, M.; Scheikl, E.; Eulgem, T.; Dóczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Dong, C.; Zhu, J. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 2007, 10, 290–295. [Google Scholar] [CrossRef]
- Agurla, S.; Gahir, S.; Munemasa, S.; Murata, Y.; Raghavendra, A.; Davies, J.; Knight, H.; Knight, M.; Gong, Z.; Guo, Y.; et al. Mechanism of stomatal closure in plants exposed to drought and cold stress. Adv. Exp. Med. Biol. 2018, 1081, 215–232. [Google Scholar] [CrossRef]
- Liu, Q.; Ding, Y.; Shi, Y.; Ma, L.; Wang, Y.; Song, C.; Wilkins, K. The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. EMBO J. 2021, 40, e104559. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.; Sintaha, M.; Cheung, M.; Lam, H. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, V.; Ravindran, P.; Kumar, P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomashow, M. So what’s new in the field of plant cold acclimation? Lots! Plant Physiol. 2001, 125, 89–93. [Google Scholar] [CrossRef] [Green Version]
- Maity, S.; de Crombrugghe, B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 1998, 23, 174–178. [Google Scholar] [CrossRef]
- Xie, Z.; Lin, W.; Yu, G.; Cheng, Q.; Xu, B.; Huang, B. Improved cold tolerance in switchgrass by a novel CCCH-type zinc finger transcription factor gene, PvC3H72, associated with ICE1-CBF-COR regulon and ABA-responsive genes. Biotechnol. Biofuels 2019, 12, 224. [Google Scholar] [CrossRef]
- Feng, X.; Zhao, Q.; Zhao, L.; Qiao, Y.; Xie, X.; Li, H.; Yao, Y.; You, C.; Hao, Y. The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol. 2012, 12, 22. [Google Scholar] [CrossRef] [Green Version]
- An, J.; Wang, X.; Zhang, X.; You, C.; Hao, Y. Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 2021, 229, 2707–2729. [Google Scholar] [CrossRef]
- An, J.; Yao, J.; Wang, X.; You, C.; Wang, X.; Hao, Y. MdHY5 positively regulates cold tolerance via CBF-dependent and CBF-independent pathways in apple. J. Plant Physiol. 2017, 218, 275–281. [Google Scholar] [CrossRef]
- Han, D.; Du, M.; Zhou, Z.; Wang, S.; Li, T.; Han, J.; Xu, T.; Yang, G. Overexpression of a Malus baccata NAC Transcription Factor Gene MbNAC25 Increases Cold and Salinity Tolerance in Arabidopsis. New Phytol. 2020, 21, 1198. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Mao, Z.; Jiang, H.; Zhang, Z.; Chen, X. A feedback loop involving MdMYB108L and MdHY5 controls apple cold tolerance. Biochem. Biophys. Res. Commun. 2019, 512, 381–386. [Google Scholar] [CrossRef]
- Xie, Y.; Chen, P.; Yan, Y.; Bao, C.; Li, X.; Wang, L.; Shen, X.; Li, H.; Liu, X.; Niu, C.; et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 2018, 218, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Wang, X.; Tong, Y.; Chen, X.; Liao, H. Bioengineering and management for efficient phosphorus utilization in crops and pastures. Curr. Opin. Biotech. 2012, 23, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yan, X.; Liao, H. Genetic improvement for phosphorus efficiency in soybean: A radical approach. Ann. Bot. 2010, 106, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Corratgé-Faillie, C.; Lacombe, B. Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 2017, 68, 3107–3113. [Google Scholar] [CrossRef]
- Léran, S.; Varala, K.; Boyer, J.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B.; et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef]
- Chai, S.; Li, E.; Zhang, Y.; Li, S. NRT1.1-Mediated nitrate suppression of root coiling relies on PIN2- and AUX1-mediated auxin transport. Front. Plant Sci. 2020, 11, 671. [Google Scholar] [CrossRef]
- Jian, S.; Luo, J.; Liao, Q.; Liu, Q.; Guan, C.; Zhang, Z. NRT1.1 regulates nitrate allocation and cadmium tolerance in arabidopsis. Front. Plant Sci. 2019, 10, 384. [Google Scholar] [CrossRef]
- Maghiaoui, A.; Bouguyon, E.; Cuesta, C.; Perrine-Walker, F.; Alcon, C.; Krouk, G.; Benková, E.; Nacry, P.; Gojon, A.; Bach, L. The Arabidopsis NRT1.1 transceptor coordinately controls auxin biosynthesis and transport to regulate root branching in response to nitrate. J. Exp. Bot. 2020, 71, 4480–4494. [Google Scholar] [CrossRef]
- Saito, S.; Uozumi, N. Calcium-regulated phosphorylation systems controlling uptake and balance of plant nutrients. Front. Plant Sci. 2020, 11, 44. [Google Scholar] [CrossRef] [Green Version]
- Camañes, G.; Bellmunt, E.; García-Andrade, J.; García-Agustín, P.; Cerezo, M. Reciprocal regulation between AtNRT2.1 and AtAMT1.1 expression and the kinetics of NH4+ and NO3- influxes. J. Plant Physiol. 2012, 169, 268–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chopin, F.; Wirth, J.; Dorbe, M.; Lejay, L.; Krapp, A.; Gojon, A.; Daniel-Vedele, F. The Arabidopsis nitrate transporter AtNRT2.1 is targeted to the root plasma membrane. Plant Physiol. Bioch. 2007, 45, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, Y.; Okamoto, M.; Crawford, N.; Siddiqi, M.; Glass, A. Dissection of the AtNRT2.1: AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 2007, 143, 425–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazoa, P.; Vidmar, J.; Tranbarger, T.; Mouline, K.; Damiani, I.; Tillard, P.; Zhuo, D.; Glass, A.; Touraine, B. Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: Responses to nitrate, amino acids and developmental stage. Plant Mol. Biol. 2003, 52, 689–703. [Google Scholar] [CrossRef]
- Orsel, M.; Eulenburg, K.; Krapp, A.; Daniel-Vedele, F. Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 2004, 219, 714–721. [Google Scholar] [CrossRef]
- Hu, H.; Wang, Y.; Tsay, Y. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 2009, 57, 264–278. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, W.; Li, Z.; Li, Z.; Bi, Y.; Crawford, N.M.; Wang, Y. FIP1 plays an important role in nitrate signaling and regulates CIPK8 and CIPK23 expression in Arabidopsis. Front. Plant Sci. 2018, 9, 593. [Google Scholar] [CrossRef]
- Yin, X.; Xia, Y.; Xie, Q.; Cao, Y.; Wang, Z.; Hao, G.; Song, J.; Zhou, Y.; Jiang, X. The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance. J. Exp. Bot. 2020, 71, 1801–1814. [Google Scholar] [CrossRef]
- Alvarez, J.M.; Schinke, A.L.; Brooks, M.D.; Pasquino, A. Transient genome-wide interactions of the master transcription factor NLP7 initiate a rapid nitrogen-response cascade. Nat. Commun. 2020, 11, 1157. [Google Scholar] [CrossRef] [Green Version]
- Coruzzi, G.; Yu, L.; Wu, J.; Tang, H.; Yuan, Y.; Wang, S.; Wang, Y.; Zhu, Q.; Li, S.; Xiang, C. Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Nat. Commun. 2016, 6, 27795. [Google Scholar] [CrossRef]
- Liu, K.H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Sun Chung, H.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchive, C.; Roudier, F.; Castaings, L.; Bréhaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013, 4, 1713. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, W.; Yang, Y.; Li, Z.; Li, N.; Qi, S.; Crawford, N.M.; Wang, Y. The Arabidopsis NLP7 gene regulates nitrate signaling via NRT1.1-dependent pathway in the presence of ammonium. Sci. Rep. 2018, 8, 1487. [Google Scholar] [CrossRef] [Green Version]
- Kiba, T.; Feria-Bourrellier, A.B.; Lafouge, F.; Lezhneva, L.; Boutet-Mercey, S.; Orsel, M.; Bréhaut, V.; Miller, A.; Daniel-Vedele, F.; Sakakibara, H.; et al. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 2012, 24, 245–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Angeli, A.; Monachello, D.; Ephritikhine, G.; Frachisse, J.M.; Thomine, S.; Gambale, F.; Barbier-Brygoo, H. The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 2006, 442, 939–942. [Google Scholar] [CrossRef] [PubMed]
- Demes, E.; Besse, L.; Cubero-Font, P. Dynamic measurement of cytosolic pH and [NO3−] uncovers the role of the vacuolar transporter AtCLCa in cytosolic pH homeostasis. Proc. Natl. Acad. Sci. USA 2020, 117, 15343–15353. [Google Scholar] [CrossRef]
- Nedelyaeva, O.; Shuvalov, A.; Karpichev, I.; Beliaev, D.; Myasoedov, N.; Khalilova, L.; Khramov, D.; Popova, L.G.; Balnokin, Y.V. Molecular cloning and characterisation of SaCLCa1, a novel protein of the chloride channel (CLC) family from the halophyte Suaeda altissima (L.) Pall. Proc. Natl. Acad. Sci. USA 2019, 240, 152995. [Google Scholar] [CrossRef]
- Zifarelli, G.; Pusch, M. CLC transport proteins in plants. FEBS Lett. 2010, 584, 2122–2127. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Yang, Q.; Liu, W.; Lin, J.; Chang, Y. The AMT1 family genes from Malus robusta display differential transcription features and ammonium transport abilities. Mol. Biol. Rep. 2017, 44, 379–390. [Google Scholar] [CrossRef]
- Venkatachalam, P.; Jain, A.; Sahi, S.; Raghothama, K. Molecular cloning and characterization of phosphate (Pi) responsive genes in Gulf ryegrass (Lolium multiflorum L.): A Pi hyperaccumulator. Plant Mol. Biol. 2009, 69, 1–21. [Google Scholar] [CrossRef]
- Sun, T.; Li, M.; Shao, Y.; Yu, L.; Ma, F. Comprehensive Genomic Identification and Expression Analysis of the Phosphate Transporter (PHT) Gene Family in Apple. Front. Plant Sci. 2017, 8, 426. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Ren, Y.; Zheng, P.; Qu, F.; Song, L.; You, C.; Wang, X.; Hao, Y. Functional identification of apple MdMYB2 gene in phosphate-starvation response. J. Plant Physiol. 2020, 244, 153089. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhou, L.; Li, Y.; You, C.; Sha, G.; Hao, Y. Apple SUMO E3 ligase MdSIZ1 is involved in the response to phosphate deficiency. J. Plant Physiol. 2019, 232, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Barberon, M.; Dubeaux, G.; Kolb, C.; Isono, E.; Zelazny, E.; Vert, G. Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, 8293–8298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Liu, H.; Cao, H.; Qi, R.; Yang, K.; Zhao, R.; Lv, W.; Zhang, Y. Genome-wide analysis of zinc- and iron-regulated transporter-like protein family members in apple and functional validation of ZIP10. Biometals 2019, 32, 657–669. [Google Scholar] [CrossRef]
- Tinoco, A.; Saxena, M.; Sharma, S.; Noinaj, N.; Delgado, Y.; Quiñones González, E.; Conklin, S.; Zambrana, N.; Loza-Rosas, S.; Parks, T. Unusual synergism of transferrin and citrate in the regulation of ti(iv) speciation, transport, and toxicity. J. Am. Chem. Soc. 2016, 138, 5659–5665. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Sun, W.; Wang, X.; Tong, X.; Ji, X.; An, J.; Zhao, Q.; You, C.; Hao, Y. Phosphate regulates malate/citrate-mediated iron uptake and transport in apple. Int. J. Mol. Sci. 2020, 297, 110526. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Sun, W.; Wang, T. The adaptive mechanism of plants to iron deficiency via iron uptake, transport, and homeostasis. Int. J. Mol. Sci. 2019, 20, 2424. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Wang, X.; Zhang, J.; Ma, F.; Hao, Y. MdMYB58 modulates fe homeostasis by directly binding to the MdMATE43 promoter in plants. Plant Cell Physiol. 2018, 59, 2476–2489. [Google Scholar] [CrossRef]
- Zhao, Q.; Ren, Y.; Wang, Q.; Yao, Y.; You, C.; Hao, Y. Overexpression of MdbHLH104 gene enhances the tolerance to iron deficiency in apple. Plant Cell Physiol. 2016, 14, 1633–1645. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Zhang, C.; Zhang, R.; Wang, G.; Li, Y.; Hao, Y. The SUMO E3 ligase MdSIZ1 targets MdbHLH104 to regulate plasma membrane H(+)-atpase activity and iron homeostasis. Plant Physiol. 2019, 179, 88–106. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Xu, J.; Wang, Y.; Hu, C.; Shi, K. The phyB-dependent induction of HY5 promotes iron uptake by systemically activating FER expression. EMBO Rep. 2021, 22, e51944. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.; Botto, J. The multifaceted roles of hy5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Xie, Y.; Wang, H.; Ma, X.; Yao, W.; Wang, H. Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell 2017, 29, 2269–2284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, C.; Kobayashi, K.; Fujii, S.; Fukaki, H.; Mitsuda, N.; Ohme-Takagi, M. Blue light regulates phosphate deficiency-dependent primary root growth inhibition in Arabidopsis. Plant Cell 2019, 10, 1803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, X.; Sun, M.; Xu, R.; Shu, H.; Wang, J.; Zhang, S. Genomewide identification and expression analysis of the ARF gene family in apple. J. Genet. 2014, 93, 785–797. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, N.; Xu, H.; Jiang, S.; Fang, H.; Su, M.; Zhang, Z.; Zhang, T.; Chen, X. Auxin regulates anthocyanin biosynthesis through the Aux/IAA-ARF signaling pathway in apple. Hortic. Res. 2018, 5, 59. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Yu, Z.; Xu, Y.; Guo, C.; Zhang, L.; Wu, C.; Yang, G.; Huang, J.; Yan, K.; Shu, H.; et al. Function identification of MdTIR1 in apple root growth benefited from the predicted Md PPI network. J. Inter. Plant Biol. 2021, 63, 723–736. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, Y.; Feng, C.; Wei, Y.; Peng, X.; Guo, X.; Guo, X.; Zhai, Z.; Li, J.; Shen, X.; et al. Overexpression of MsGH3.5 inhibits shoot and root development through the auxin and cytokinin pathways in apple plants. Plant J. 2020, 103, 166–183. [Google Scholar] [CrossRef]
- Devoghalaere, F.; Doucen, T.; Guitton, B.; Keeling, J.; Payne, W.; Ling, T.; Ross, J.; Hallett, I.; Gunaseelan, K.; Dayatilake, G.; et al. A genomics approach to understanding the role of auxin in apple (Malus × domestica) fruit size control. BMC Plant Biol. 2012, 12, 7. [Google Scholar] [CrossRef] [Green Version]
- Gan, Z.; Wang, Y.; Wu, T.; Xu, X.; Zhang, X.; Han, Z. MdPIN1b encodes a putative auxin efflux carrier and has different expression patterns in BC and M9 apple rootstocks. Plant Mol. Biol. 2018, 96, 353–365. [Google Scholar] [CrossRef]
- Wang, Z.; Li, J.; Mao, Y.; Zhang, M.; Wang, R.; Hu, Y.; Mao, Z.; Shen, X. Transcriptional regulation of MdPIN3 and MdPIN10 by MdFLP during apple self-rooted stock adventitious root gravitropism. BMC Plant Biol. 2019, 19, 229. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Han, M. Genomewide analysis of ABCBs with a focus on ABCB1 and ABCB19 in Malus domestica. J. Genet. 2016, 95, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Li, G.; Qi, S.; Liu, X.; Chen, X.; Ma, J.; Zhang, D.; Han, M. Identification and expression analysis of the IPT and CKX gene families during axillary bud outgrowth in apple (Malus domestica Borkh.). Gene 2018, 651, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Liu, J.; Zhai, L.; Gan, Z.; Zhang, G.; Yang, S.; Wang, Y.; Wu, T.; Zhang, X.; Xu, X.; et al. Natural variation in cytokinin maintenance improves salt tolerance in apple rootstocks. Plant Cell Environ. 2019, 42, 424–436. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, X.; Wu, T.; Xu, X.; Han, Z.; Wang, Y. Methylation effect on IPT5b gene expression determines cytokinin biosynthesis in apple rootstock. Biochem. Biophys. Res. Commun. 2017, 482, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Guo, X.; Wang, Q.; Wang, Y.; Zhao, D.; Yao, L.; Wang, S.; Liu, G.; Li, T. Overexpression of MsDREB6.2 results in cytokinin-deficient developmental phenotypes and enhances drought tolerance in transgenic apple plants. Plant J. 2017, 89, 510–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Xu, Y.; Liu, L.; Guo, Y.; Yuan, X.; Man, X.; Liu, C.; Yang, G.; Huang, J.; Yan, K.; et al. The importance of conserved serine for c-terminally encoded peptides function exertion in apple. Int. J. Mol. Sci. 2019, 20, 775. [Google Scholar] [CrossRef] [Green Version]
- Gregory, E.; Dao, T. The signaling peptide-encoding genes CLE16, CLE17 and CLE27 are dispensable for Arabidopsis shoot apical meristem activity. Nat. Plants 2018, 13, e0202595. [Google Scholar] [CrossRef] [Green Version]
- Holzwart, E.; Huerta, A.; Glöckner, N.; Garnelo Gómez, B.; Wanke, F.; Augustin, S.; Askani, J.; Schürholz, A.; Harter, K.; Wolf, S. BRI1 controls vascular cell fate in the Arabidopsis root through RLP44 and phytosulfokine signaling. Proc. Natl. Acad. Sci. USA 2018, 115, 11838–11843. [Google Scholar] [CrossRef] [Green Version]
- Dickinson, A.; Zhang, J.; Luciano, M. A plant lipocalin promotes retinal-mediated oscillatory lateral root initiation. Science 2021, 373, 1532–1536. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Lin, R.; Liang, J.; King, G.; Wu, J.; Wang, X. Transposable element insertion: A hidden major source of domesticated phenotypic variation in Brassica rapa. Plant Biotechnol. J. 2022, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Guo, Y. Less Is More. Natural Loss-of-Function Mutation Is a Strategy for Adaptation. Plant Commun. 2020, 13, 100103. [Google Scholar] [CrossRef] [PubMed]
- Reusch, T.B.H.; Baums, I.B.; Werner, B. Evolution via somatic genetic variation in modular species. Trends Ecol. Evol. 2021, 36, 1083–1092. [Google Scholar] [CrossRef]
- Gupta, D.; Bhattacharjee, O.; Mandal, D.; Sen, M.K.; Dey, D.; Dasgupta, A.; Kazi, T.A.; Gupta, R.; Sinharoy, S.; Acharya, K.; et al. CRISPR-Cas9 system: A new-fangled dawn in gene editing. Life Sci. 2019, 232, 116636. [Google Scholar] [CrossRef]
- Janik, E.; Niemcewicz, M.; Ceremuga, M.; Krzowski, L.; Saluk-Bijak, J.; Bijak, M. Various aspects of a gene editing system-CRISPR-Cas9. Int. J. Mol. Sci. 2020, 21, 9604. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Z.; Zhang, L.; Shu, J.; Wang, M.; Li, H.; Shu, H.; Wang, X.; Sun, Q.; Zhang, S. Root Breeding in the Post-Genomics Era: From Concept to Practice in Apple. Plants 2022, 11, 1408. https://doi.org/10.3390/plants11111408
Zhou Z, Zhang L, Shu J, Wang M, Li H, Shu H, Wang X, Sun Q, Zhang S. Root Breeding in the Post-Genomics Era: From Concept to Practice in Apple. Plants. 2022; 11(11):1408. https://doi.org/10.3390/plants11111408
Chicago/Turabian StyleZhou, Zhou, Lei Zhang, Jing Shu, Mengyu Wang, Han Li, Huairui Shu, Xiaoyun Wang, Qinghua Sun, and Shizhong Zhang. 2022. "Root Breeding in the Post-Genomics Era: From Concept to Practice in Apple" Plants 11, no. 11: 1408. https://doi.org/10.3390/plants11111408
APA StyleZhou, Z., Zhang, L., Shu, J., Wang, M., Li, H., Shu, H., Wang, X., Sun, Q., & Zhang, S. (2022). Root Breeding in the Post-Genomics Era: From Concept to Practice in Apple. Plants, 11(11), 1408. https://doi.org/10.3390/plants11111408





