Elongating Effect of the Peptide AEDL on the Root of Nicotiana tabacum under Salinity
Abstract
1. Introduction
2. Results
2.1. Morphometric Data
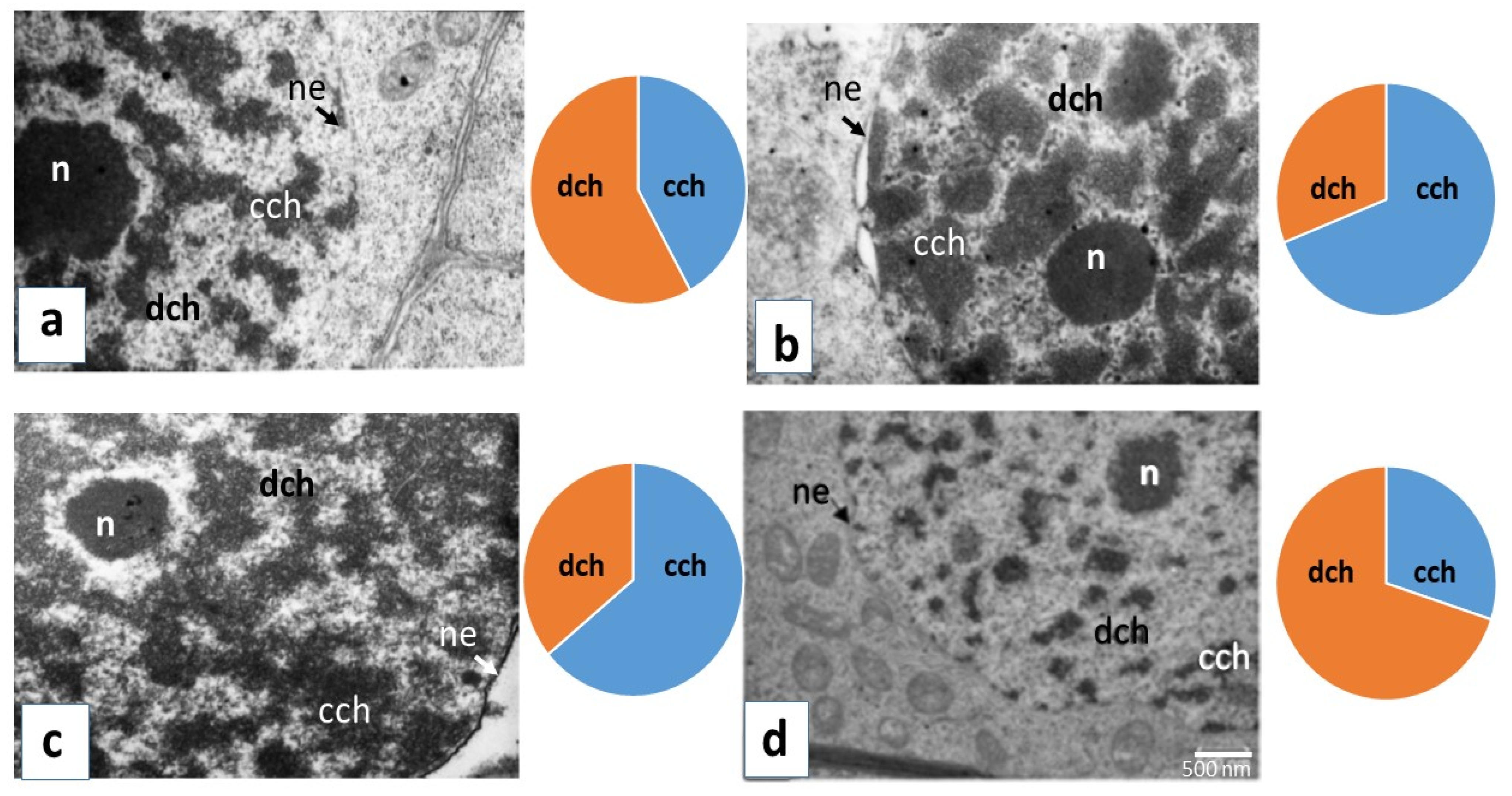
2.2. TEM Analysis
2.3. ROS Distribution
2.4. Expression of EXPA3 and EXPA5 Genes and Genes of the WOX Family
3. Discussion
4. Materials and Methods
4.1. Object of Study
4.2. Preparation of FITC-Labeled AEDL
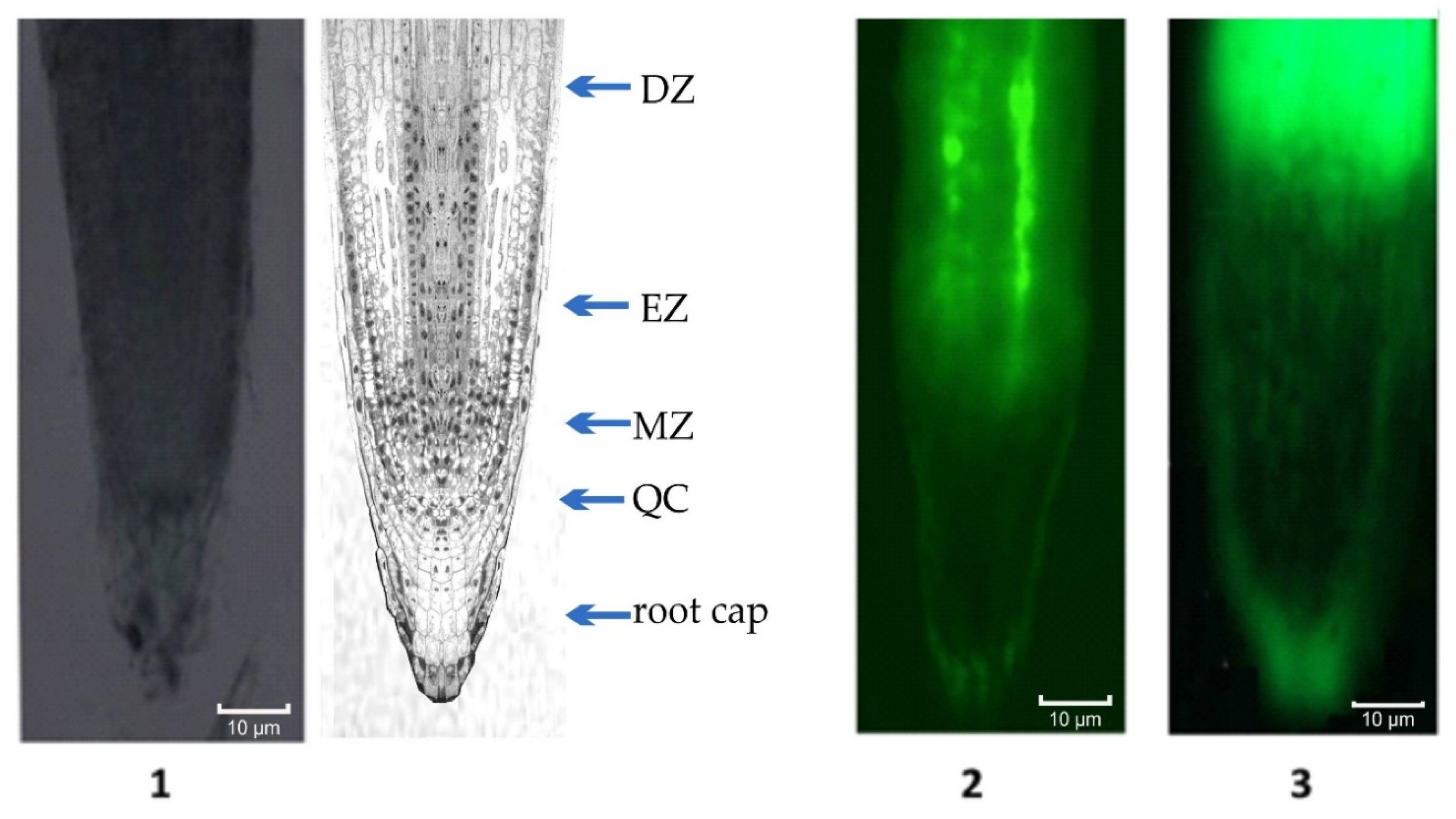
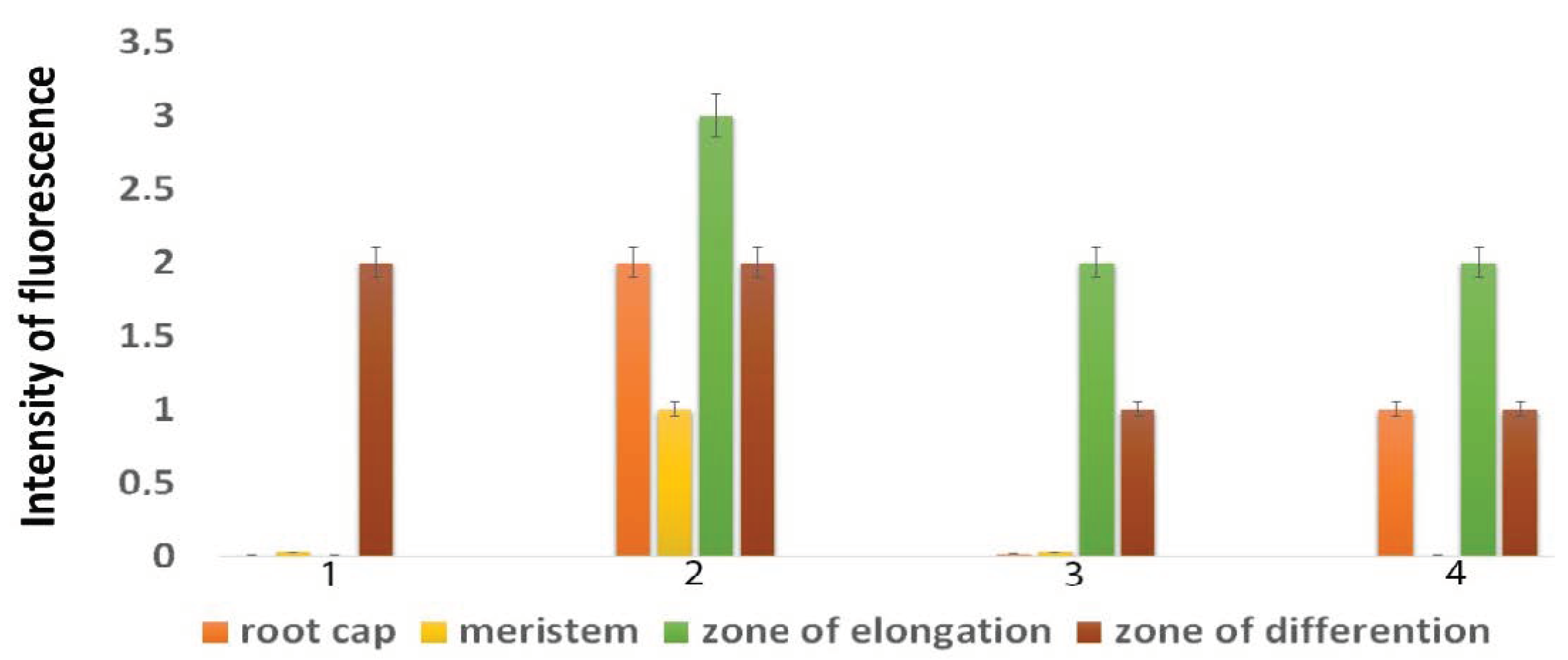
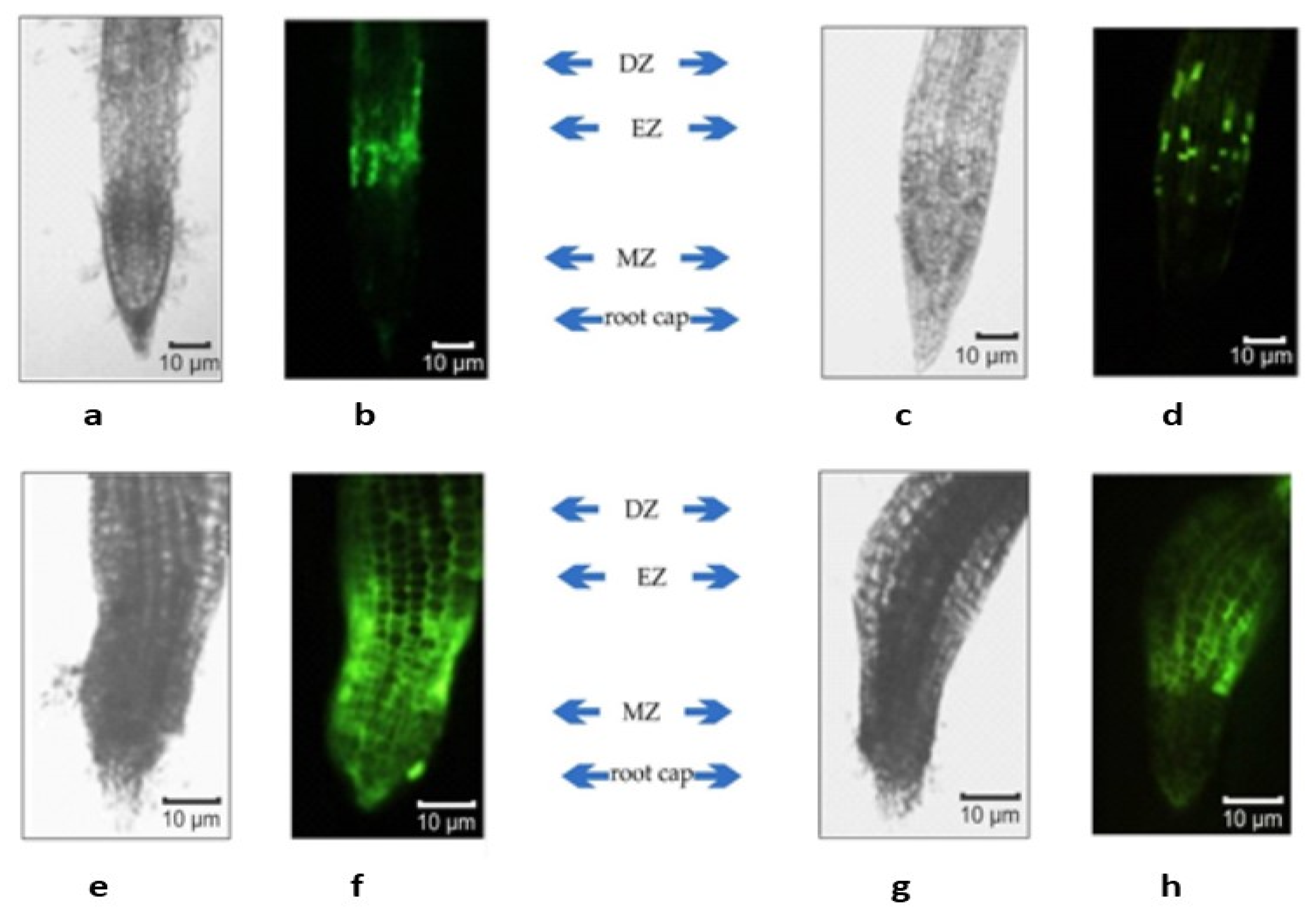
4.3. Fluorescence Microscopy
4.4. Transmission Electron Microscopy (TEM)
4.5. RNA Isolation
4.6. Real-Time PCR
4.7. Statistical Processing of Results
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis Root Development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef] [PubMed]
- Slovak, R.; Ogura, T.; Satbhai, S.B.; Ristova, D.; Busch, W. Genetic control of root growth: From genes to networks. Ann. Bot. 2016, 117, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Wachsman, G.; Sparks, E.E.; Benfey, P.N. Genes and networks regulating root anatomy and architecture. New Phytol. 2015, 208, 26–38. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, C.; Willemsen, V.; Hendriks, G.; Weisbeek, P.; Scheres, B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 1997, 390, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, K.; Shasha, D.E.; Wang, J.Y.; Jung, J.W.; Lambert, G.M.; Galbraith, D.W.; Benfey, P.N. A gene expression map of the Arabidopsis root. Science 2003, 302, 1956–1960. [Google Scholar] [CrossRef]
- Fizames, C.; Munos, S.; Cazettes, C.; Nacry, P.; Boucherez, J.; Gaymard, F.; Piquemal, D.; Delorme, V.; Commes, T.; Doumas, P.; et al. The Arabidopsis root transcriptome by serial analysis of gene expression. Gene identification using the genome sequence. Plant Physiol. 2004, 134, 67–80. [Google Scholar] [CrossRef]
- Gehring, W.J.; Affolter, M.; Burglin, T. Homeodomain proteins. Annu. Rev. Biochem. 1994, 63, 487–526. [Google Scholar] [CrossRef]
- Dolzblasz, A.; Nardmann, J.; Clerici, E.; Causier, B.; van der Graaff, E.; Chen, J.; Devies, B.; Werr, W.; Laux, N. Stem cell regulation by Arabidopsis WOX genes. Mol. Plant. 2016, 9, 1028–1039. [Google Scholar] [CrossRef]
- Breuninger, H.; Rikirsch, E.; Hermann, M.; Ueda, M.; Laux, T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 2008, 14, 867–876. [Google Scholar] [CrossRef]
- Costanzo, E.; Trehin, C.; Vandenbussche, M. The role of WOX genes in flower development. Ann. Bot. 2014, 114, 1545–1553. [Google Scholar] [CrossRef]
- van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, R.; Qin, G.; Chen, Z.; Gu, H.; Qu, L.J. Over-expression of WOX1 leads to defects in meristem development and polyamine homeostasis in Arabidopsis. J. Integr. Plant Biol. 2011, 53, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Haecker, A.; Gross-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Zhang, Z.; Laux, T. Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell. 2011, 20, 264–270. [Google Scholar] [CrossRef]
- Suer, S.; Agusti, J.; Sanchez, P.; Schwarz, M.; Greb, T. WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 2011, 23, 3247–3259. [Google Scholar] [CrossRef]
- Gonzali, S.; Novi, G.; Loreti, E.; Paolicchi, F.; Poggi, A.; Alpi, A.; Perata, P. A turanose-insensitive mutant suggests a role for WOX5 in auxin homeostasis in Arabidopsis thaliana. Plant J. 2005, 44, 633–645. [Google Scholar] [CrossRef]
- Kong, D.; Hao, Y.; Cui, H. The WUSCHEL related Homeobox protein WOX7 regulates the sugar response of lateral root development in Arabidopsis thaliana. Mol. Plant 2016, 9, 261–270. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef]
- Brand, U.; Fletcher, J.C.; Hobe, M.; Meyerowitz, E.M.; Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 2000, 289, 617–619. [Google Scholar] [CrossRef]
- Guo, Y.; Han, L.; Hymes, M.; Denver, R.; Clark, S.E. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010, 63, 889–900. [Google Scholar] [CrossRef]
- Stahl, Y.; Wink, R.H.; Ingram, G.C.; Simon, R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 2009, 19, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Ogawa-Ohnishi, M.; Mori, A.; Matsubayashi, Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 2010, 329, 1065–1067. [Google Scholar] [CrossRef] [PubMed]
- Geitmann, A.; Ortega, J. Mechanics and modeling of plant cell growth. Trends Plant Sci. 2009, 14, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Hamant, O.; Traas, J. The mechanics behind plant development. New Phytol. 2010, 185, 369–385. [Google Scholar] [CrossRef] [PubMed]
- ZhiMing, Y.; Bo, K.; XiaoWei, H.; ShaoLei, L.; YouHuang, B.; WoNa, D.; Ming, C.; Hyung-Taeg, C.; Ping, W. Root hair-specific expansins modulate root hair elongation in rice. Plant J. 2011, 66, 725–734. [Google Scholar] [CrossRef]
- Maris, A.; Suslov, D.; Fry, S.C.; Verbelen, J.P.; Vissenberg, K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 2009, 60, 3959–3972. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, S.; Alvarez, S.; Asirvatham, V.S.; Schachtman, D.P.; Wu, Y.; Sharp, R.E. Cell wall proteome in the maize primary root elongation zone. I. Extraction and identification of water-soluble and lightly ionically bound proteins. Plant Physiol. 2006, 140, 311–325. [Google Scholar] [CrossRef]
- Muller, K.; Linkies, A.; Vreeburg, R.A.; Fry, S.C.; Krieger-Liszkay, A.; Leubner-Metzger, G. In vivo cell wall loosening by hydroxyl radicals during cress seed germination and elongation growth. Plant Physiol. 2009, 150, 1855–1865. [Google Scholar] [CrossRef]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef]
- Cosgrove, D.J. 2005. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Lionetti, V.; Raiola, A.; Camardella, L.; Giovane, A.; Obel, N.; Pauly, M.; Favaron, F.; Cervone, F.; Bellincampi, D. Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol. 2007, 143, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Siedlecka, A.; Wiklund, S.; Peronne, M.A.; Micheli, F.; Lesniewska, J.; Sethson, I.; Edlund, U.; Richard, L.; Sundberg, B.; Mellerowicz, E.J. Pectin methyl esterase inhibits intrusive and symplastic cell growth in developing wood cells of Populus. Plant Physiol. 2008, 146, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhao, J.; Li, X.; Qin, L.; Yan, X.; Liao, H. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011, 66, 541–552. [Google Scholar] [CrossRef]
- Cho, H.T.; Kende, H. Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 1997, 9, 1661–1671. [Google Scholar]
- Benkova, E.; Hejatko, J. Hormone interactions at the root apical meristem. Plant Mol. Biol. 2009, 69, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Wolters, H.; Jürgens, G. Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009, 10, 305–317. [Google Scholar] [CrossRef]
- Tsukagoshi, H.; Busch, W.; Benfey, P.N. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 2010, 143, 606–616. [Google Scholar] [CrossRef]
- Kaya, C.; Tuna, A.L.; Yokaş, I. The role of plant hormones in plants under salinity stress. In Salinity and Water Stress; Ashraf, M., Ozturk, M., Athar, H., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 45–50. [Google Scholar]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.K.; Kim, S.T.; Gupta, R. Ethylene: A master regulator of salinity stress tolerance in plants. Biomolecules 2020, 10, 959. [Google Scholar] [CrossRef]
- Cao, W.H.; Liu, J.; He, X.J.; Mu, R.L.; Zhou, H.L.; Chen, S.Y.; Zhang, J.S. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007, 143, 707–719. [Google Scholar] [CrossRef]
- Kevin, L.; Wang, C.; Li, H.; Ecker, J.R. Ethylene biosynthesis and signaling networks. Plant Cell 2002, 14, 131–151. [Google Scholar]
- Guzman, P.; Ecker, J.R. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 1990, 2, 513–523. [Google Scholar] [PubMed]
- Daszkowska-Golec, A.; Szarejko, I. Open or close the gate–stomata action under the control of phytohormones in drought stress conditions. Front. Plant Sci. 2013, 4, 138. [Google Scholar] [CrossRef] [PubMed]
- Fedoreyeva, L.I.; Dilovarova, T.A.; Ashapkin, V.V.; Martirosyan, Y.T.; Khavinson, V.K.; Kharchenko, P.N.; Vanyushin, B.F. Short exogenous peptides regulate expression of CLE, KNOX1 and GRF family genes in Nicotiana tabacum. Biochemistry 2017, 82, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Van Breusegem, F.; Dat, J.F. Reactive oxygen species in plant cell death. Plant Physiol. 2006, 141, 384–390. [Google Scholar] [CrossRef]
- Beemster, G.T.S.; Baskin, T.I. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 1998, 116, 1515–1526. [Google Scholar] [CrossRef]
- Culligan, K.; Tissier, A.; Britt, A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 2004, 16, 1091–1104. [Google Scholar] [CrossRef]
- Cools, T.; Iantcheva, A.; Weimer, A.K. The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell 2011, 23, 1435–1448. [Google Scholar] [CrossRef]
- Spadafora, N.D.; Doonan, J.H.; Herbert, R.J.; Bitonti, M.B.; Wallace, E.; Rogers, H.J.; Francis, D. Arabidopsis T-DNA insertional lines for CDC25 are hypersensitive to hydroxyurea but not to zeocin or salt stress. Ann Bot. 2011, 107, 1183–1192. [Google Scholar] [CrossRef]
- Takatsuka, H.; Umeda, M. Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 2014, 65, 2633–2643. [Google Scholar] [CrossRef]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef]
- Laux, T. The stem cell concept in plants: A matter of debate. Cell 2003, 113, 281–283. [Google Scholar]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Gibson, S.W.; Todd, C.D. Arabidopsis AIR12 influences root development. Physiol. Mol. Biol. Plants 2015, 21, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Perales, M.; Gruel, J.; Girke, T.; Jönsson, H.; Reddy, G.V. WUSCHEL protein movement mediates stem cell homeostasis in the arabidopsis shoot apex. Genes Dev. 2011, 25, 2025–2030. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.E.; Williams, R.W.; Meyerowitz, E.M. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in arabidopsis. Cell 1997, 89, 575–585. [Google Scholar] [CrossRef]
- Lenhard, M.; Laux, T. Stem cell homeostasis in the arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 2003, 130, 3163–3173. [Google Scholar] [CrossRef]
- Fletcher, J.C.; Brand, U.; Running, M.P.; Simon, R.; Meyerowitz, E.M. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 1999, 283, 1911–1914. [Google Scholar] [CrossRef]
- Rojo, E.; Sharma, V.K.; Kovaleva, V.; Raikhel, N.V.; Fletcher, J.C. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 2002, 14, 969–977. [Google Scholar] [CrossRef]
- Hobe, M.; Müller, R.; Grünewald, M.; Brand, U.; Simon, R. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev. Genes Evol. 2003, 213, 371–381. [Google Scholar] [CrossRef]
- Faulker, C. Plasmodesmata and the symplast. Curr. Biol. 2018, 28, R1374–R1378. [Google Scholar] [CrossRef]
- Mongiovi, M.; Shran, R. Global alignment of protein-protein interaction networks. Methods Mol. Biol. 2013, 939, 21–34. [Google Scholar] [PubMed]
- Marga, F.; Grandbois, M.; Cosgrove, D.J.; Baskin, T.I. Cell wall extension results in the coordinate separation of parallel microfibrils: Evidence from scanning electron microscopy and atomic force microscopy. Plant J. 2005, 43, 181–190. [Google Scholar] [CrossRef] [PubMed]
- McQueen-Mason, S.J.; Fry, S.C.; Durachko, D.M.; Cosgrove, D.J. The relationship between xyloglucan endotransglycosylase and in vitro cell wall extension in cucumber hypocotyls. Planta 1993, 190, 327–331. [Google Scholar] [CrossRef] [PubMed]
- McQueen-Mason, S.; Cosgrove, D.J. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc. Natl. Acad. Sci. USA 1994, 91, 6574–6578. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef]
- Lee, Y.; Choi, D.; Kende, H. Expansins: Ever-expanding numbers and functions. Curr. Opin. Plant Biol. 2001, 4, 527–532. [Google Scholar] [CrossRef]
- Reinhard, D.; Wittwer, F.; Mandel, T.; Kuhlemeier, C. Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell 1998, 10, 1427–1437. [Google Scholar] [CrossRef]
- Cho, H.T.; Cosgrove, D.J. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 2002, 14, 3237–3253. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef][Green Version]
- Fedoreyeva, L.I.; Dilovarova, T.A.; Kononenko, N.V.; Baranova, E.N.; Smirnova, E.A.; Vanyushin, B.F. Influence of glycylglycine, glycine, and glycylaspartic acid on growth, development, and gene expression in a tobacco (Nicotiana tabacum) callus culture. Biol. Bull. 2018, 45, 351–358. [Google Scholar] [CrossRef]
- Kononenko, N.; Baranova, E.; Dilovarova, T.; Akanov, E.; Fedoreyeva, L. Oxidative Damage to Various Root and Shoot Tissues of Durum and Soft Wheat Seedlings during Salinity. Agriculture 2020, 10, 55. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Variant | Shoot Height, mm | Root Length, mm |
|---|---|---|
| Control | 7.2 ± 2.3 | 10.2 ± 2.1 |
| +AEDL | 9.1 ± 3.1 | 21.2 ± 3.1 |
| +150 mM NaCl | 7.3 ± 1.2 | 12.7 ± 2.4 |
| + AEDL + | ||
| 150 mM NaCl | 4.2 ± 1.5 | 7.1 ± 2.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedoreyeva, L.I.; Baranova, E.N.; Chaban, I.A.; Dilovarova, T.A.; Vanyushin, B.F.; Kononenko, N.V. Elongating Effect of the Peptide AEDL on the Root of Nicotiana tabacum under Salinity. Plants 2022, 11, 1352. https://doi.org/10.3390/plants11101352
Fedoreyeva LI, Baranova EN, Chaban IA, Dilovarova TA, Vanyushin BF, Kononenko NV. Elongating Effect of the Peptide AEDL on the Root of Nicotiana tabacum under Salinity. Plants. 2022; 11(10):1352. https://doi.org/10.3390/plants11101352
Chicago/Turabian StyleFedoreyeva, Larisa I., Ekaterina N. Baranova, Inn A. Chaban, Tatyana A. Dilovarova, Boris F. Vanyushin, and Neonila V. Kononenko. 2022. "Elongating Effect of the Peptide AEDL on the Root of Nicotiana tabacum under Salinity" Plants 11, no. 10: 1352. https://doi.org/10.3390/plants11101352
APA StyleFedoreyeva, L. I., Baranova, E. N., Chaban, I. A., Dilovarova, T. A., Vanyushin, B. F., & Kononenko, N. V. (2022). Elongating Effect of the Peptide AEDL on the Root of Nicotiana tabacum under Salinity. Plants, 11(10), 1352. https://doi.org/10.3390/plants11101352









