The Dynamics of
Abstract
1. Introduction
2. Results
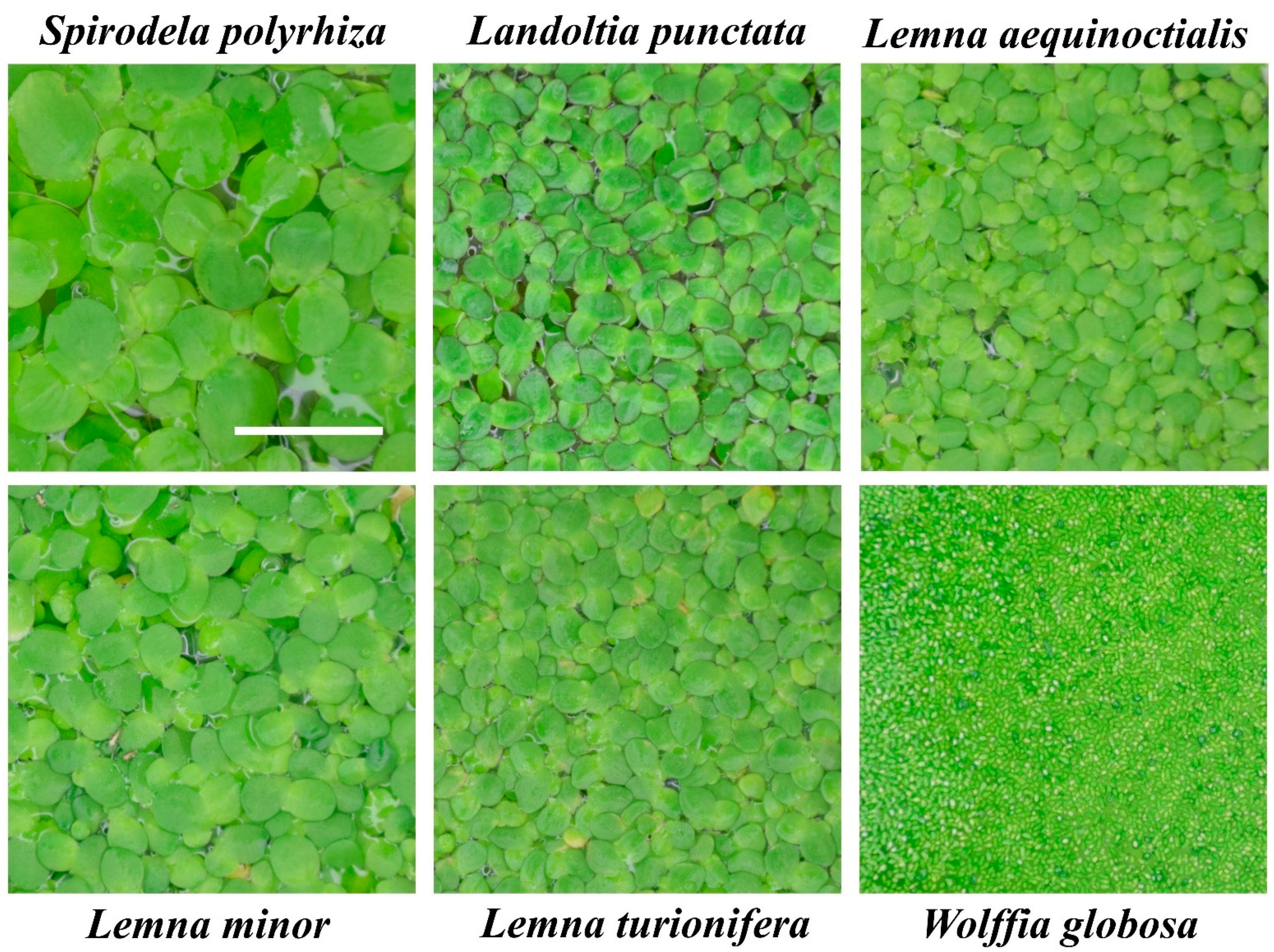
2.1. Identity of the Analyzed Species
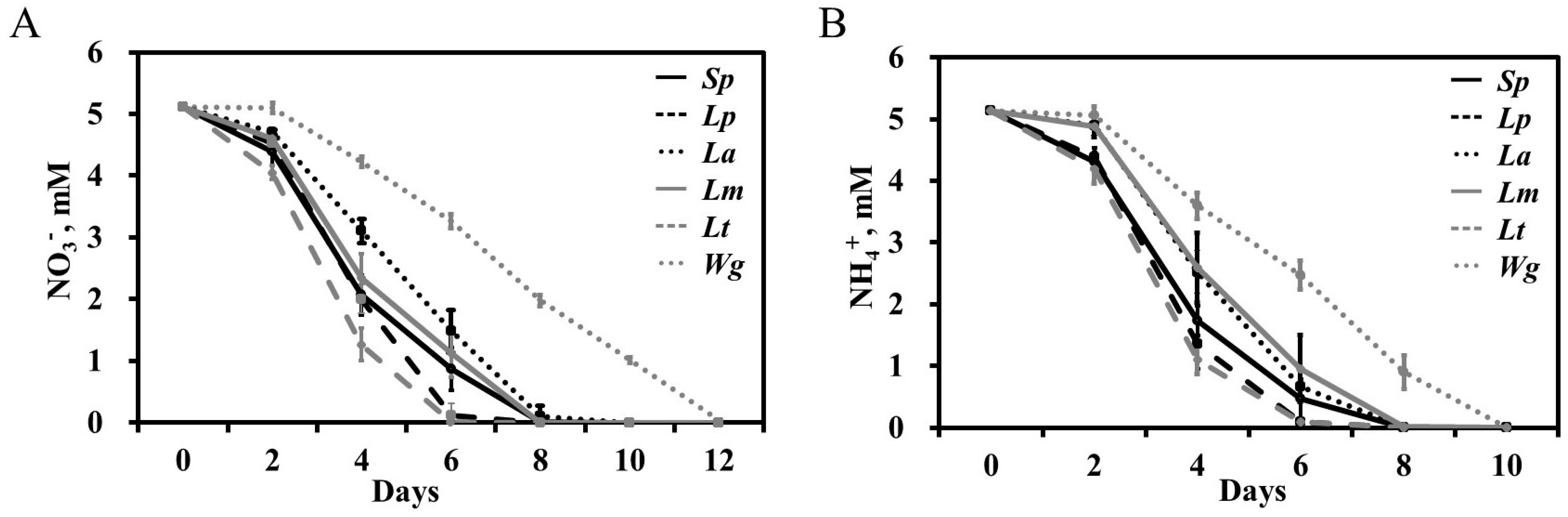
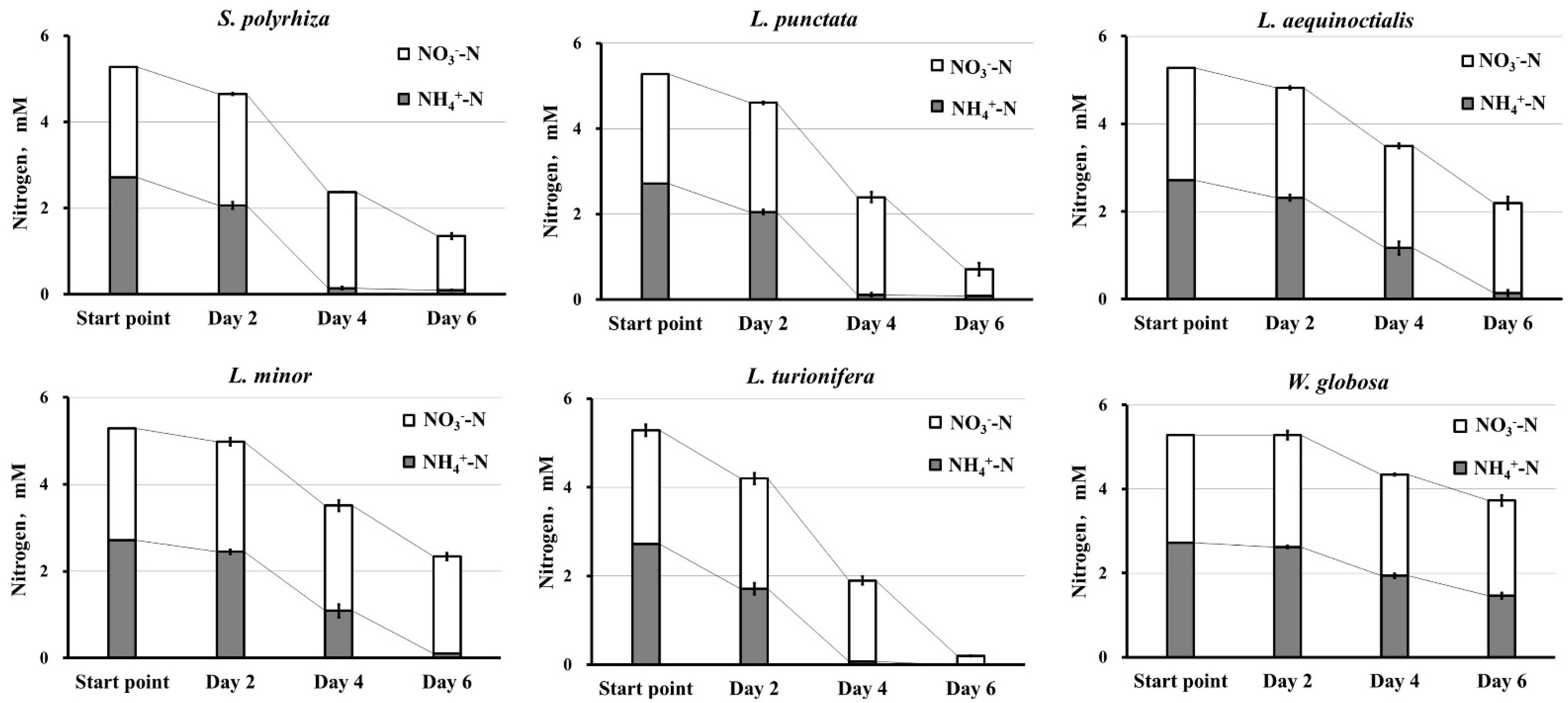
2.2. All Six Duckweed Species Demonstrate a Preference for over NO3
2.3. Key Genes for N Assimilation in the Genome of S. polyrhiza
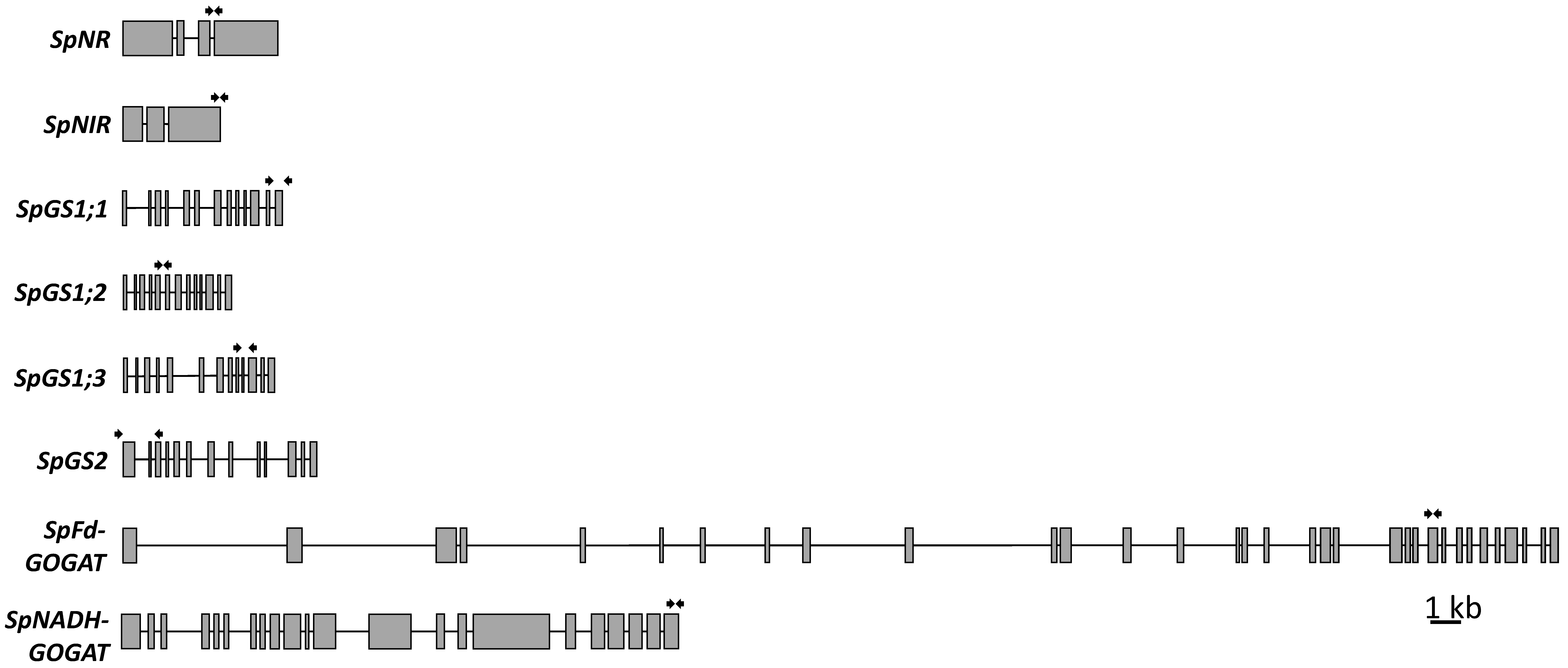
2.3.1. Nitrate and Nitrite Reductases
2.3.2. Glutamine Synthetases
2.3.3. Fd-GOGAT and NADH-GOGAT
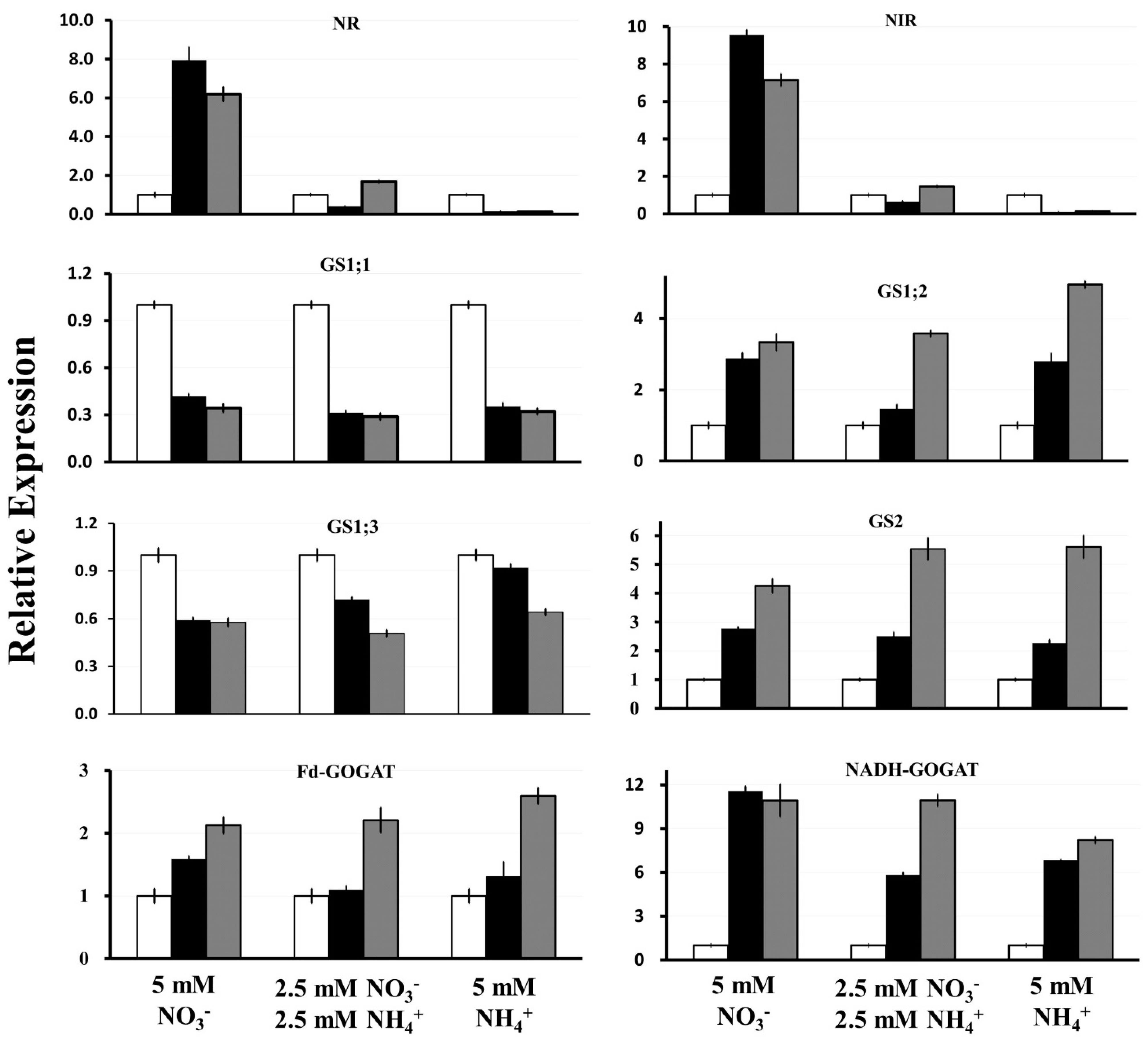
2.4. Expression of Key S. polyrhiza Genes Involved in N Assimilation
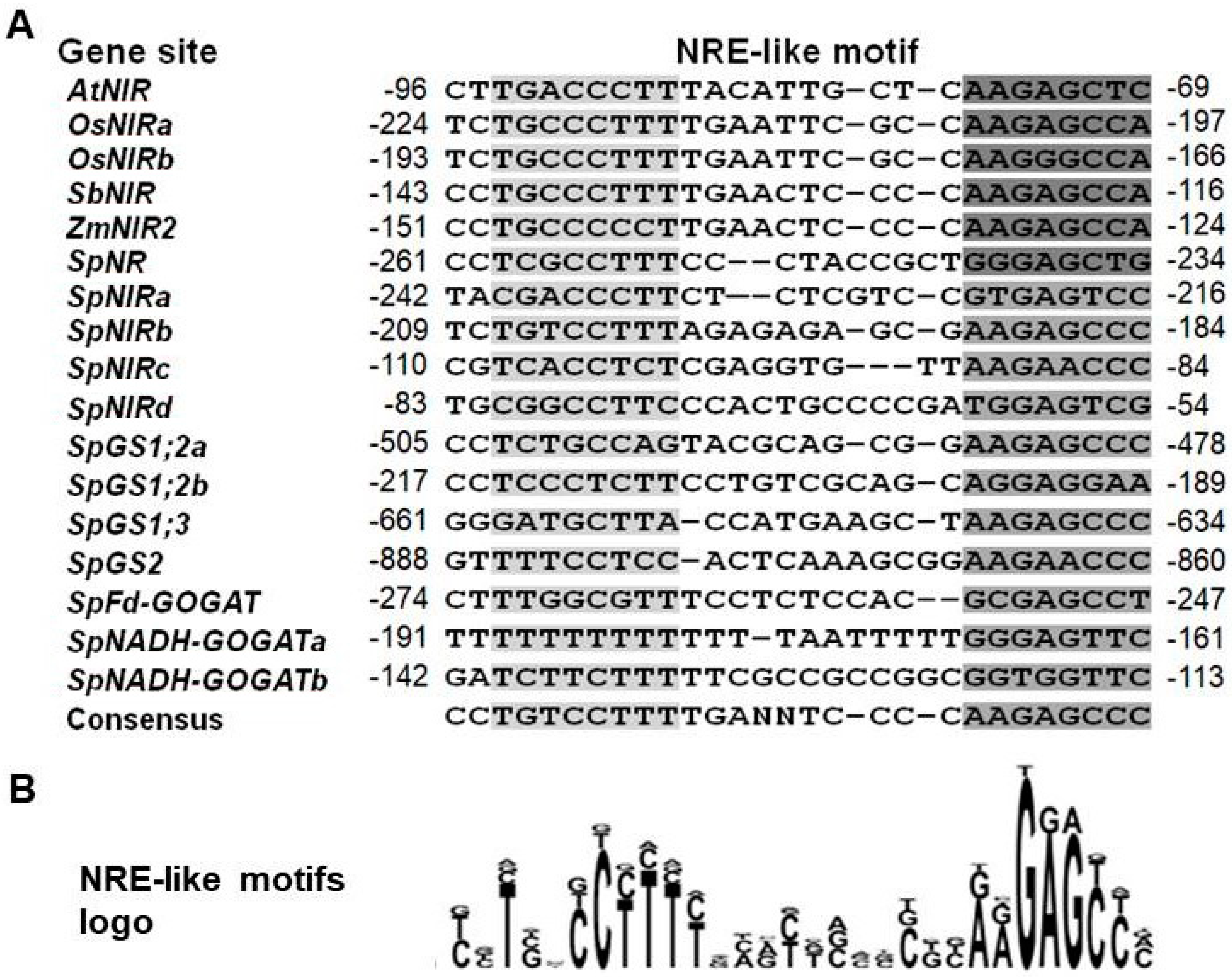
2.5. Survey for Possible N-Responsive Promoter Cis-Elements in the N Assimilation Genes
3. Discussion
3.1. Duckweeds’ Preference for as a Source of N
3.2. NR and NiR Are Co-Ordinately Expressed, Stimulated by and Suppressed by
3.3. SpGS1;2 and SpGS2 Are Regulated in a Very Similar Manner, Which Is Different from SpGS1;1 and SpGS1;3
3.4. Fd-GOGAT and NADH-GOGAT Have a Complex Exon-Intron Structure, and Are Upregulated by and
3.5. Distinctive Promoter Elements in the N Assimilation Genes in S. polyrhiza
4. Materials and Methods
4.1. Plant Materials
4.2. Duckweed Cultivation Parameters and Determination of N Uptake
4.3. Characterization of Major Duckweed Genes Related to N Assimilation
4.4. Phylogenetic Analysis
4.5. Gene Expression Analysis by RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Xu, H.; Yu, C.; Zhou, G. Multifaceted Roles of Duckweed in Aquatic Phytoremediation and Bioproducts Synthesis. GCB Bioenergy 2021, 13, 70–82. [Google Scholar] [CrossRef]
- Coskun, D.; Britto, D.T.; Shi, W.; Kronzucker, H.J. Nitrogen Transformations in Modern Agriculture and the Role of Biological Nitrification Inhibition. Nat. Plants 2017, 3, 17074. [Google Scholar] [CrossRef] [PubMed]
- Camargo, J.A.; Alonso, Á. Ecological and Toxicological Effects of Inorganic Nitrogen Pollution in Aquatic Ecosystems: A Global Assessment. Environ. Int. 2006, 32, 831–849. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Huang, X.; Chen, H.; Godfray, H.C.J.; Wright, J.S.; Hall, J.W.; Gong, P.; Ni, S.; Qiao, S.; Huang, G.; et al. Managing Nitrogen to Restore Water Quality in China. Nature 2019, 567, 516–520. [Google Scholar] [CrossRef]
- Smith, V.H. Eutrophication of Freshwater and Coastal Marine Ecosystems a Global Problem. Environ. Sci. Pollut. Res. 2003, 10, 126–139. [Google Scholar] [CrossRef]
- Food and Agriculture Organization (FAO). World Fertilizer Trends and Outlook to 2022. 2019. Available online: https://www.fao.org/3/ca6746en/CA6746EN.pdf?eloutlink=imf2fao (accessed on 5 October 2021).
- Rai, P.K. Heavy Metal Pollution in Aquatic Ecosystems and Its Phytoremediation Using Wetland Plants: An Ecosustainable Approach. Int. J. Phytoremediation 2008, 10, 131–158. [Google Scholar] [CrossRef]
- Queiroz, R.C.S.; Lôbo, I.P.; Ribeiro, V.S.; Rodrigues, L.B.; Almeida Neto, J.A. Assessment of Autochthonous Aquatic Macrophytes with Phytoremediation Potential for Dairy Wastewater Treatment in Floating Constructed Wetlands. Int. J. Phytoremediation 2020, 22, 518–528. [Google Scholar] [CrossRef]
- Kanwar, V.S.; Sharma, A.; Srivastav, A.L.; Rani, L. Phytoremediation of Toxic Metals Present in Soil and Water Environment: A Critical Review. Environ. Sci. Pollut. Res. Int. 2020, 27, 44835–44860. [Google Scholar] [CrossRef]
- Hammer, D.A. Constructed Wetlands for Wastewater Treatment: Municipal, Industrial, and Agricultural, 1st ed.; Hammer, D.A., Ed.; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar] [CrossRef]
- Koottatep, T.; Polprasert, C. Role of Plant Uptake on Nitrogen Removal in Constructed Wetlands Located in the Tropics. Water Sci. Technol. 1997, 36, 1–8. [Google Scholar] [CrossRef]
- Landolt, E. Biosystematic Investigations in the Family of Duckweeds (Lemnaceae) (Vol. 2). The family of Lemnaceae-a Monographic Study. vol. 1.; Veroeffentlichungen des Geobotanischen Instituts der ETH, Stiftung Ruebel: Zurich, Switzerland, 1986. [Google Scholar]
- Tippery, N.P.; Les, D.H. Tiny Plants with Enormous Potential: Phylogeny and Evolution of Duckweeds. In The Duckweed Genomes; Cao, X.H., Fourounjian, P., Wang, W., Eds.; Springer: Cham, Germany, 2020; pp. 19–38. [Google Scholar] [CrossRef]
- Acosta, K.; Appenroth, K.J.; Borisjuk, L.; Edelman, M.; Heinig, U.; Jansen, M.A.K.; Oyama, T.; Pasaribu, B.; Schubert, I.; Sorrels, S.; et al. Return of the Lemnaceae: Duckweed as a Model Plant System in the Genomics and Post-Genomics Era. Plant Cell 2021, 33, 3207–3234. [Google Scholar] [CrossRef]
- De Vasconcelos, V.M.; de Morais, E.R.C.; Faustino, S.J.B.; Hernandez, M.C.R.; Gaudêncio, H.R.S.C.; de Melo, R.R.; Bessa Junior, A.P. Floating Aquatic Macrophytes for the Treatment of Aquaculture Effluents. Environ. Sci. Pollut. Res. 2021, 28, 2600–2607. [Google Scholar] [CrossRef]
- Zhou, Y.; Borisjuk, N. Small Aquatic Duckweed Plants with Big Potential for the Production of Valuable Biomass and Wastewater Remediation. Int. J. Environ. Sci. Nat. Resour. 2019, 16, 555942. [Google Scholar] [CrossRef]
- Mohedano, R.A.; Costa, R.H.R.; Tavares, F.A.; Belli Filho, P. High Nutrient Removal Rate from Swine Wastes and Protein Biomass Production by Full-Scale Duckweed Ponds. Bioresour. Technol. 2012, 112, 98–104. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, G.; Peterson, A.; Zha, X.; Cheng, J.; Li, S.; Cui, D.; Zhu, H.; Kishchenko, O.; Borisjuk, N. Biodiversity of Duckweeds in Eastern China and Their Potential for Bioremediation of Municipal and Industrial Wastewater. J. Geosci. Environ. Protect. 2018, 6, 108–116. [Google Scholar] [CrossRef][Green Version]
- Xu, S.; Stapley, J.; Gablenz, S.; Boyer, J.; Appenroth, K.J.; Sree, K.S.; Gershenzon, J.; Widmer, A.; Huber, M. Low Genetic Variation Is Associated with Low Mutation Rate in the Giant Duckweed. Nat. Commun. 2019, 10, 1243. [Google Scholar] [CrossRef]
- Appenroth, K.-J.; Sree, K.S.; Böhm, V.; Hammann, S.; Vetter, W.; Leiterer, M.; Jahreis, G. Nutritional Value of Duckweeds (Lemnaceae) as Human Food. Food Chem. 2017, 217, 266–273. [Google Scholar] [CrossRef]
- Guo, L.; Jin, Y.; Xiao, Y.; Tan, L.; Tian, X.; Ding, Y.; He, K.; Du, A.; Li, J.; Yi, Z.; et al. Energy-Efficient and Environmentally Friendly Production of Starch-Rich Duckweed Biomass Using Nitrogen-Limited Cultivation. J. Clean. Produc. 2020, 251, 119726. [Google Scholar] [CrossRef]
- Ziegler, P.; Sree, K.S.; Appenroth, K.-J. Duckweeds for Water Remediation and Toxicity Testing. Toxicol. Environ. Chem. 2016, 98, 1127–1154. [Google Scholar] [CrossRef]
- Ekperusi, A.O.; Sikoki, F.D.; Nwachukwu, E.O. Application of Common Duckweed (Lemna minor) in Phytoremediation of Chemicals in the Environment: State and Future Perspective. Chemosphere 2019, 223, 285–309. [Google Scholar] [CrossRef]
- Walsh, É.; Paolacci, S.; Burnell, G.; Jansen, M.A.K. The Importance of the Calcium-to-Magnesium Ratio for Phytoremediation of Dairy Industry Wastewater Using the Aquatic Plant Lemna minor L. Int. J. Phytoremediat. 2020, 22, 694–702. [Google Scholar] [CrossRef]
- Petersen, F.; Demann, J.; Restemeyer, D.; Ulbrich, A.; Olfs, H.-W.; Westendarp, H.; Appenroth, K.-J. Influence of the Nitrate-N to Ammonium-N Ratio on Relative Growth Rate and Crude Protein Content in the Duckweeds Lemna minor and Wolffiella hyalina. Plants 2021, 10, 1741. [Google Scholar] [CrossRef]
- Wang, W.; Li, R.; Zhu, Q.; Tang, X.; Zhao, Q. Transcriptomic and physiological analysis of common duckweed Lemna minor responses to toxicity. BMC Plant Biol. 2016, 16, 92. [Google Scholar] [CrossRef]
- Tian, X.; Fang, Y.; Jin, Y.; Yi, Z.; Li, J.; Du, A.; He, K.; Huang, Y.; Zhao, H. Ammonium Detoxification Mechanism of Ammonium-Tolerant Duckweed (Landoltia punctata) Revealed by Carbon and Nitrogen Metabolism under Ammonium Stress. Environ. Pollut. 2021, 277, 116834. [Google Scholar] [CrossRef]
- Li, H.; Hu, B.; Chu, C. Nitrogen Use Efficiency in Crops: Lessons from Arabidopsis and Rice. J. Exp. Bot. 2017, 68, 2477–2488. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen Uptake, Assimilation and Remobilization in Plants: Challenges for Sustainable and Productive Agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef]
- Krapp, A. Plant Nitrogen Assimilation and Its Regulation: A Complex Puzzle with Missing Pieces. Curr. Opin. Plant Biol. 2015, 25, 115–122. [Google Scholar] [CrossRef]
- Bernard, S.M.; Møller, A.L.B.; Dionisio, G.; Kichey, T.; Jahn, T.P.; Dubois, F.; Baudo, M.; Lopes, M.S.; Tercé-Laforgue, T.; Foyer, C.H.; et al. Gene Expression, Cellular Localisation and Function of Glutamine Synthetase Isozymes in Wheat (Triticum aestivum L.). Plant Molec. Biol. 2008, 67, 89–105. [Google Scholar] [CrossRef]
- Bloom, A.J. The Increasing Importance of Distinguishing among Plant Nitrogen Sources. Curr. Opin. Plant Biol. 2015, 25, 10–16. [Google Scholar] [CrossRef]
- Britto, D.T.; Kronzucker, H.J. Ecological Significance and Complexity of N-Source Preference in Plants. Ann. Bot. 2013, 112, 957–963. [Google Scholar] [CrossRef]
- Britto, D.T.; Kronzucker, H.J. NH4+ Toxicity in Higher Plants: A Critical Review. J. Plant Physiol. 2002, 159, 567–584. [Google Scholar] [CrossRef]
- Esteban, R.; Ariz, I.; Cruz, C.; Moran, J.F. Review: Mechanisms of Ammonium Toxicity and the Quest for Tolerance. Plant Sci. 2016, 248, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.Y.; Babourina, O.; Rengel, Z.; Yang, X.E.; Pu, P.M. Ammonium and Nitrate Uptake by the Floating Plant Landoltia punctata. Ann. Bot. 2007, 99, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Michael, T.P.; Bryant, D.; Gutierrez, R.; Borisjuk, N.; Chu, P.; Zhang, H.; Xia, J.; Zhou, J.; Peng, H.; El Baidouri, M.; et al. Comprehensive Definition of Genome Features in Spirodela polyrhiza by High-Depth Physical Mapping and Short-Read DNA Sequencing Strategies. Plant J. 2017, 89, 617–635. [Google Scholar] [CrossRef] [PubMed]
- Borisjuk, N.; Chu, P.; Gutierrez, R.; Zhang, H.; Acosta, K.; Friesen, N.; Sree, K.S.; Garcia, C.; Appenroth, K.J.; Lam, E. Assessment, Validation and Deployment Strategy of a Two-Barcode Protocol for Facile Genotyping of Duckweed Species. Plant Biol. 2015, 17 (Suppl. S1), 42–49. [Google Scholar] [CrossRef]
- BLAST: Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 5 October 2021).
- Wang, W.; Haberer, G.; Gundlach, H.; Glasser, C.; Nussbaumer, T.; Luo, M.C.; Lomsadze, A.; Borodovsky, M.; Kerstetter, R.A.; Shanklin, J.; et al. The Spirodela polyrhiza Genome Reveals Insights into Its Neotenous Reduction Fast Growth and Aquatic Lifestyle. Nat Commun. 2014, 5, 3311. [Google Scholar] [CrossRef]
- An, D.; Zhou, Y.; Li, C.; Xiao, Q.; Wang, T.; Zhang, Y.; Wu, Y.; Li, Y.; Chao, D.-Y.; Messing, J.; et al. Plant evolution and environmental adaptation unveiled by long-read whole-genome sequencing of Spirodela. Proc. Natl. Acad. Sci. USA 2019, 116, 18893–18899. [Google Scholar] [CrossRef]
- Harkess, A.; McLoughlin, F.; Bilkey, N.; Elliott, K.; Emenecker, R.; Mattoon, E.; Miller, K.; Czymmek, K.; Vierstra, R.D.; Meyers, B.C.; et al. Improved Spirodela polyrhiza Genome and Proteomic Analyses Reveal a Conserved Chromosomal Structure with High Abundance of Chloroplastic Proteins Favoring Energy Production. J. Exp. Bot. 2021, 72, 2491–2500. [Google Scholar] [CrossRef]
- James, D.; Borphukan, B.; Fartyal, D.; Achary, V.M.M.; Reddy, M.K. Transgenic manipulation of glutamine synthetase: A target with untapped potential in various aspects of crop improvement. In Biotechnologies of Crop Improvement, Volume 2; Gosal, S.S., Wani, S.H., Eds.; Springer: Cham, Germany, 2018; pp. 367–416. [Google Scholar] [CrossRef]
- Bernard, S.M.; Habash, D.Z. The Importance of Cytosolic Glutamine Synthetase in Nitrogen Assimilation and Recycling. New Phytol. 2009, 182, 608–620. [Google Scholar] [CrossRef]
- Ferreira, M.J.; Vale, D.; Cunha, L.; Melo, P. Role of the C-Terminal Extension Peptide of Plastid Located Glutamine Synthetase from Medicago truncatula: Crucial for Enzyme Activity and Needless for Protein Import into the Plastids. Plant Physiol. Biochem. 2017, 111, 226–233. [Google Scholar] [CrossRef]
- Biesiadka, J.; Legocki, A.B. Evolution of the Glutamine Synthetase Gene in Plants. Plant Sci. 1997, 128, 51–58. [Google Scholar] [CrossRef]
- Thomsen, H.C.; Eriksson, D.; Møller, I.S.; Schjoerring, J.K. Cytosolic Glutamine Synthetase: A Target for Improvement of Crop Nitrogen Use Efficiency? Trends Plant Sci. 2014, 19, 656–663. [Google Scholar] [CrossRef]
- Pesole, G.; Bozzetti, M.P.; Lanave, C.; Preparata, G.; Saccone, C. Glutamine Synthetase Gene Evolution: A Good Molecular Clock. Proc. Natl. Acad. Sci. USA 1991, 88, 522–526. [Google Scholar] [CrossRef]
- Nigro, D.; Gu, Y.Q.; Huo, N.; Marcotuli, I.; Blanco, A.; Gadaleta, A.; Anderson, O.D. Structural Analysis of the Wheat Genes Encoding NADH-Dependent Glutamine-2-Oxoglutarate Amidotransferases and Correlation with Grain Protein Content. PLoS ONE 2013, 8, e73751. [Google Scholar] [CrossRef]
- Yamaya, T.; Kusano, M. Evidence Supporting Distinct Functions of Three Cytosolic Glutamine Synthetases and Two NADH-Glutamate Synthases in Rice. J. Exp. Bot. 2014, 65, 5519–5525. [Google Scholar] [CrossRef]
- García-Gutiérrez, Á.; Cánovas, F.M.; Ávila, C. Glutamate Synthases from Conifers: Gene Structure and Phylogenetic Studies. BMC Genomics 2018, 19, 65. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. Identification of a Nitrate-Responsive Cis-Element in the Arabidopsis NIR1 Promoter Defines the Presence of Multiple Cis-Regulatory Elements for Nitrogen Response: Nitrate-Responsive Element of Arabidopsis NIR1. Plant J. 2010, 63, 269–282. [Google Scholar] [CrossRef]
- Feng, H.; Yan, M.; Fan, X.; Li, B.; Shen, Q.; Miller, A.J.; Xu, G. Spatial Expression and Regulation of Rice High-Affinity Nitrate Transporters by Nitrogen and Carbon Status. J. Exp. Bot. 2011, 62, 2319–2332. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. Emergence of a New Step towards Understanding the Molecular Mechanisms Underlying Nitrate-Regulated Gene Expression. J. Exp. Bot. 2014, 65, 5589–5600. [Google Scholar] [CrossRef]
- Liu, X.; Feng, H.; Huang, D.; Song, M.; Fan, X.; Xu, G. Two Short Sequences in OsNAR2.1 Promoter Are Necessary for Fully Activating the Nitrate Induced Gene Expression in Rice Roots. Sci. Rep. 2015, 5, 11950. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. Roles of the Transcriptional Regulation Mediated by the Nitrate-Responsive Cis-Element in Higher Plants. Biochem. Biophy. Res. Commun. 2011, 411, 708–713. [Google Scholar] [CrossRef]
- Yu, J.; Xuan, W.; Tian, Y.; Fan, L.; Sun, J.; Tang, W.; Chen, G.; Wang, B.; Liu, Y.; Wu, W.; et al. Enhanced OsNLP4-OsNiR Cascade Confers Nitrogen Use Efficiency by Promoting Tiller Number in Rice. Plant Biotechnol. J. 2021, 19, 167–176. [Google Scholar] [CrossRef]
- Sawaki, N.; Tsujimoto, R.; Shigyo, M.; Konishi, M.; Toki, S.; Fujiwara, T.; Yanagisawa, S. A Nitrate-Inducible GARP Family Gene Encodes an Auto-Repressible Transcriptional Repressor in Rice. Plant Cell Physiol. 2013, 54, 506–517. [Google Scholar] [CrossRef]
- Medici, A.; Marshall-Colon, A.; Ronzier, E.; Szponarski, W.; Wang, R.; Gojon, A.; Crawford, N.M.; Ruffel, S.; Coruzzi, G.M.; Krouk, G. AtNIGT1/HRS1 Integrates Nitrate and Phosphate Signals at the Arabidopsis Root Tip. Nat. Commun. 2015, 6, 6274. [Google Scholar] [CrossRef]
- Ueda, Y.; Nosaki, S.; Sakuraba, Y.; Miyakawa, T.; Kiba, T.; Tanokura, M.; Yanagisawa, S. NIGT1 Family Proteins Exhibit Dual Mode DNA Recognition to Regulate Nutrient Response-Associated Genes in Arabidopsis. PLoS Genet. 2020, 16, e1009197. [Google Scholar] [CrossRef]
- Yanagisawa, S. Characterization of a Nitrate-Inducible Transcriptional Repressor NIGT1 Provides New Insights into DNA Recognition by the GARP Family Proteins. Plant Signal. Behav. 2013, 8, e24447. [Google Scholar] [CrossRef]
- Berger, N.; Dubreucq, B. Evolution Goes GAGA: GAGA Binding Proteins across Kingdoms. Biochim. Biophys. Acta 2012, 1819, 863–868. [Google Scholar] [CrossRef]
- Gong, R.; Cao, H.; Zhang, J.; Xie, K.; Wang, D.; Yu, S. Divergent Functions of the GAGA-binding Transcription Factor Family in Rice. Plant J. 2018, 94, 32–47. [Google Scholar] [CrossRef]
- Davison, B.L.; Egly, J.-M.; Mulvihill, E.R.; Chambon, P. Formation of Stable Preinitiation Complexes between Eukaryotic Class B Transcription Factors and Promoter Sequences. Nature 1983, 301, 680–686. [Google Scholar] [CrossRef]
- Brázda, V.; Bartas, M.; Bowater, R.P. Evolution of Diverse Strategies for Promoter Regulation. Trends Genet. 2021, 37, 730–744. [Google Scholar] [CrossRef]
- Komarnytsky, S.; Borisjuk, N. Functional Analysis of Promoter Elements in Plants. Genet. Eng. 2003, 25, 113–141. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-Quadruplexes and Their Regulatory Roles in Biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhao, Y.; Li, N. Genome-Wide Colonization of Gene Regulatory Elements by G4 DNA Motifs. Nucleic Acids Res. 2009, 37, 6784–6798. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Kim, N.; Tuteja, N.; Yadav, P. G Quadruplex in Plants: A Ubiquitous Regulatory Element and Its Biological Relevance. Front. Plant Sci. 2017, 8, 1163. [Google Scholar] [CrossRef] [PubMed]
- Hon, J.; Martínek, T.; Zendulka, J.; Lexa, M. Pqsfinder: An Exhaustive and Imperfection-Tolerant Search Tool for Potential Quadruplex-Forming Sequences in R. Bioinformatics 2017, 33, 3373–3379. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Budak, H.; Sen, T.Z. Genome-Wide Discovery of G-Quadruplexes in Barley. Sci. Rep. 2021, 11, 7876. [Google Scholar] [CrossRef]
- Andorf, C.M.; Kopylov, M.; Dobbs, D.; Koch, K.E.; Stroupe, M.E.; Lawrence, C.J.; Bass, H.W. G-Quadruplex (G4) Motifs in the Maize (Zea mays L.) Genome Are Enriched at Specific Locations in Thousands of Genes Coupled to Energy Status, Hypoxia, Low Sugar, and Nutrient Deprivation. J. Genet. Genom. 2014, 41, 627–647. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, M.; Zhang, Q.; Zhu, G.-F.; Li, F.-F.; Du, L.-F. Genomic Distribution and Possible Functional Roles of Putative G-Quadruplex Motifs in Two Subspecies of Oryza sativa. Comput. Biol. Chem. 2015, 56, 122–130. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Sen, T.Z. Genome-Wide Discovery of G-Quadruplexes in Wheat: Distribution and Putative Functional Roles. Sci. Rep. 2020, 10, 2021–2032. [Google Scholar] [CrossRef]
- Sekomo, C.B.; Rousseau, D.P.L.; Saleh, S.A.; Lens, P.N.L. Heavy Metal Removal in Duckweed and Algae Ponds as a Polishing Step for Textile Wastewater Treatment. Ecol. Eng. 2012, 44, 102–110. [Google Scholar] [CrossRef]
- Porath, D.; Pollock, J. Ammonia Stripping by Duckweed and Its Feasibility in Circulating Aquaculture. Aquat. Bot. 1982, 13, 125–131. [Google Scholar] [CrossRef]
- Huang, L.; Lu, Y.; Gao, X.; Du, G.; Ma, X.; Liu, M.; Guo, J.; Chen, Y. Ammonium-Induced Oxidative Stress on Plant Growth and Antioxidative Response of Duckweed (Lemna minor L.). Ecol. Engineer. 2013, 58, 355–362. [Google Scholar] [CrossRef]
- Gowri, G.; Kenis, J.D.; Ingemarsson, B.; Redinbaugh, M.G.; Campbell, W.H. Nitrate Reductase Transcript Is Expressed in the Primary Response of Maize to Environmental Nitrate. Plant Mol. Biol. 1992, 18, 55–64. [Google Scholar] [CrossRef]
- Ingemarsson, B.; Oscarson, P.; Af Ugglas, M.; Larsson, C.M. Nitrogen Utilization in Lemna: III. Short-Term Effects of Ammonium on Nitrate Uptake and Nitrate Reduction. Plant Physiol. 1987, 85, 865–867. [Google Scholar] [CrossRef]
- Lee, R.B.; Drew, M.C. Rapid, Reversible Inhibition of Nitrate Influx in Barley by Ammonium. J. Exp. Bot. 1989, 40, 741–752. [Google Scholar] [CrossRef]
- Kronzucker, H.J.; Glass, A.D.M.; Siddiqi, M.Y. Inhibition of Nitrate Uptake by Ammonium in Barley. Analysis of Component Fluxes. Plant Physiol. 1999, 120, 283–292. [Google Scholar] [CrossRef]
- Kronzucker, H.J.; Siddiqi, M.Y.; Glass, A.D.M.; Kirk, G.J.D. Nitrate-Ammonium Synergism in Rice. a Subcellular Flux Analysis. Plant Physiol. 1999, 119, 1041–1046. [Google Scholar] [CrossRef]
- Li, J.; Xue, L.; Yan, H.; Wang, L.; Liu, L.; Lu, Y.; Xie, H. The Nitrate Reductase Gene-Switch: A System for Regulated Expression in Transformed Cells of Dunaliella salina. Gene 2007, 403, 132–142. [Google Scholar] [CrossRef]
- Von der Heyde, E.; Klein, B.; Abram, L.; Hallmann, A. The Inducible NitA Promoter Provides a Powerful Molecular Switch for Transgene Expression in Volvox carteri. BMC Biotechnol. 2015, 15, 5. [Google Scholar] [CrossRef]
- Jackson, H.O.; Berepiki, A.; Baylay, A.J.; Terry, M.J.; Moore, C.M.; Bibby, T.S. An Inducible Expression System in the Alga Nannochloropsis gaditana Controlled by the Nitrate Reductase Promoter. J. Appl. Phycol. 2019, 31, 269–279. [Google Scholar] [CrossRef]
- Ji, Y.; Li, Q.; Liu, G.; Selvaraj, G.; Zheng, Z.; Zou, J.; Wei, Y. Roles of Cytosolic Glutamine Synthetases in Arabidopsis Development and Stress Responses. Plant Cell Physiol. 2019, 60, 657–671. [Google Scholar] [CrossRef]
- Lothier, J.; Gaufichon, L.; Sormani, R.; Lemaitre, T.; Azzopardi, M.; Morin, H.; Chardon, F.; Reisdorf-Cren, M.; Avice, J.-C.; Masclaux-Daubresse, C. The Cytosolic Glutamine Synthetase GLN1;2 Plays a Role in the Control of Plant Growth and Ammonium Homeostasis in Arabidopsis Rosettes When Nitrate Supply Is Not Limiting. J. Exp. Bot. 2011, 62, 1375–1390. [Google Scholar] [CrossRef]
- Guan, M.; de Bang, T.C.; Pedersen, C.; Schjoerring, J.K. Cytosolic Glutamine Synthetase Gln1;2 Is the Main Isozyme Contributing to GS1 Activity and Can Be Up-Regulated to Relieve Ammonium Toxicity. Plant Physiol. 2016, 171, 1921–1933. [Google Scholar] [CrossRef]
- Goodall, A.J.; Kumar, P.; Tobin, A.K. Identification and Expression Analyses of Cytosolic Glutamine Synthetase Genes in Barley (Hordeum vulgare L.). Plant Cell Physiol. 2013, 54, 492–505. [Google Scholar] [CrossRef]
- Ferreira, S.; Moreira, E.; Amorim, I.; Santos, C.; Melo, P. Arabidopsis thaliana Mutants Devoid of Chloroplast Glutamine Synthetase (GS2) Have Non-Lethal Phenotype under Photorespiratory Conditions. Plant Physiol. Biochem. 2019, 144, 365–374. [Google Scholar] [CrossRef]
- Tang, D.; Liu, M.-Y.; Zhang, Q.; Fan, K.; Ruan, J. Isolation and Characterization of Chloroplastic Glutamine Synthetase Gene (CsGS2) in Tea Plant Camellia sinensis. Plant Physiol. Biochem. 2020, 155, 321–329. [Google Scholar] [CrossRef]
- Németh, E.; Nagy, Z.; Pécsváradi, A. Chloroplast Glutamine Synthetase, the Key Regulator of Nitrogen Metabolism in Wheat, Performs Its Role by Fine Regulation of Enzyme Activity via Negative Cooperativity of Its Subunits. Front. Plant Sci. 2018, 9, 191. [Google Scholar] [CrossRef]
- Masclaux, C.; Valadier, M.-H.; Brugière, N.; Morot-Gaudry, J.-F.; Hirel, B. Characterization of the Sink/Source Transition in Tobacco (Nicotiana tabacum L.) Shoots in Relation to Nitrogen Management and Leaf Senescence. Planta 2000, 211, 510–518. [Google Scholar] [CrossRef]
- Hörtensteiner, S.; Feller, U. Nitrogen Metabolism and Remobilization during Senescence. J. Exp. Bot. 2002, 53, 927–937. [Google Scholar] [CrossRef]
- Orsel, M.; Moison, M.; Clouet, V.; Thomas, J.; Leprince, F.; Canoy, A.-S.; Just, J.; Chalhoub, B.; Masclaux-Daubresse, C. Sixteen Cytosolic Glutamine Synthetase Genes Identified in the Brassica napus L. Genome Are Differentially Regulated Depending on Nitrogen Regimes and Leaf Senescence. J. Exp. Bot. 2014, 65, 3927–3947. [Google Scholar] [CrossRef]
- Tabuchi, M.; Sugiyama, K.; Ishiyama, K.; Inoue, E.; Sato, T.; Takahashi, H.; Yamaya, T. Severe Reduction in Growth Rate and Grain Filling of Rice Mutants Lacking OsGS1;1, a Cytosolic Glutamine Synthetase1;1: GS1;1-Knockout Mutant in Rice. Plant J. 2005, 42, 641–651. [Google Scholar] [CrossRef]
- Gadaleta, A.; Nigro, D.; Marcotuli, I.; Giancaspro, A.; Giove, S.L.; Blanco, A. Isolation and Characterisation of Cytosolic Glutamine Synthetase (GSe) Genes and Association with Grain Protein Content in Durum Wheat. Crop Pasture Sci. 2014, 65, 38. [Google Scholar] [CrossRef]
- Wang, Y.; Teng, W.; Wang, Y.; Ouyang, X.; Xue, H.; Zhao, X.; Gao, C.; Tong, Y. The Wheat Cytosolic Glutamine Synthetase GS1.1 Modulates N Assimilation and Spike Development by Characterizing CRISPR-Edited Mutants. bioRxiv 2020. preprint. [Google Scholar] [CrossRef]
- Gao, Y.; Bang, T.C.; Schjoerring, J.K. Cisgenic Overexpression of Cytosolic Glutamine Synthetase Improves Nitrogen Utilization Efficiency in Barley and Prevents Grain Protein Decline under Elevated CO2. J. Plant Biotechnol. 2019, 17, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Lee, J.; Kichey, T.; Gerentes, D.; Zivy, M.; Tatout, C.; Dubois, F.; Balliau, T.; Valot, B.; Davanture, M.; et al. Two Cytosolic Glutamine Synthetase Isoforms of Maize Are Specifically Involved in the Control of Grain Production. Plant Cell 2006, 18, 3252–3274. [Google Scholar] [CrossRef]
- Konishi, N.; Ishiyama, K.; Beier, M.P.; Inoue, E.; Kanno, K.; Yamaya, T.; Takahashi, H.; Kojima, S. Contributions of Two Cytosolic Glutamine Synthetase Isozymes to Ammonium Assimilation in Arabidopsis Roots. J. Exp. Bot. 2017, 68, 613–625. [Google Scholar] [CrossRef]
- Wang, W.; Wu, Y.; Messing, J. RNA-Seq Transcriptome Analysis of Spirodela Dormancy without Reproduction. BMC Genom. 2014, 15, 60. [Google Scholar] [CrossRef]
- Lea, P.J.; Miflin, B.J. Alternative Route for Nitrogen Assimilation in Higher Plants. Nature 1974, 251, 614–616. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Reisdorf-Cren, M.; Pageau, K.; Lelandais, M.; Grandjean, O.; Kronenberger, J.; Valadier, M.-H.; Feraud, M.; Jouglet, T.; Suzuki, A. Glutamine Synthetase-Glutamate Synthase Pathway and Glutamate Dehydrogenase Play Distinct Roles in the Sink-Source Nitrogen Cycle in Tobacco. Plant Physiol. 2006, 140, 444–456. [Google Scholar] [CrossRef]
- Lea, P.J.; Miflin, B.J. Glutamate Synthase and the Synthesis of Glutamate in Plants. Plant Physiol. Biochem. 2003, 41, 555–564. [Google Scholar] [CrossRef]
- Suzuki, A.; Knaff, D.B. Glutamate Synthase: Structural, Mechanistic and Regulatory Properties, and Role in the Amino Acid Metabolism. Photosynth. Res. 2005, 83, 191–217. [Google Scholar] [CrossRef]
- Zeng, D.-D.; Qin, R.; Li, M.; Alamin, M.; Jin, X.-L.; Liu, Y.; Shi, C.-H. The Ferredoxin-Dependent Glutamate Synthase (OsFd-GOGAT) Participates in Leaf Senescence and the Nitrogen Remobilization in Rice. Mol. Genet. Genomics 2017, 292, 385–395. [Google Scholar] [CrossRef]
- Tamura, W.; Kojima, S.; Toyokawa, A.; Watanabe, H.; Tabuchi-Kobayashi, M.; Hayakawa, T.; Yamaya, T. Disruption of a Novel NADH-Glutamate Synthase 2 Gene Caused Marked Reduction in Spikelet Number of Rice. Front. Plant Sci. 2011, 2, 57. [Google Scholar] [CrossRef]
- Tsukiyama, T.; Becker, P.B.; Wu, C. ATP-Dependent Nucleosome Disruption at a Heat-Shock Promoter Mediated by Binding of GAGA Transcription Factor. Nature 1994, 367, 525–532. [Google Scholar] [CrossRef]
- Chetverina, D.; Erokhin, M.; Schedl, P. GAGA Factor: A Multifunctional Pioneering Chromatin Protein. Cell. Mol. Life Sci. 2021, 78, 4125–4141. [Google Scholar] [CrossRef]
- Hoang, P.T.N.; Fiebig, A.; Novák, P.; Macas, J.; Cao, H.X.; Stepanenko, A.; Chen, G.; Borisjuk, N.; Scholz, U.; Schubert, I. Chromosome-Scale Genome Assembly for the Duckweed Spirodela intermedia, Integrating Cytogenetic Maps, PacBio and Oxford Nanopore Libraries. Sci. Rep. 2020, 10, 19230. [Google Scholar] [CrossRef]
- Santi, L.; Wang, Y.; Stile, M.R.; Berendzen, K.; Wanke, D.; Roig, C.; Pozzi, C.; Müller, K.; Müller, J.; Rohde, W.; et al. The GA Octodinucleotide Repeat Binding Factor BBR Participates in the Transcriptional Regulation of the Homeobox Gene Bkn3. Plant J. 2003, 34, 813–826. [Google Scholar] [CrossRef]
- Schenk, R.U.; Hildebrandt, A.C. Medium and Techniques for Induction and Growth of Monocotyledonous and Dicotyledonous Plant Cell Cultures. Can. J. Bot. 1972, 50, 199–204. [Google Scholar] [CrossRef]
- Cedergreen, N.; Madsen, T.V. Nitrogen Uptake by the Floating Macrophyte Lemna minor. New Phytol. 2002, 155, 285–292. [Google Scholar] [CrossRef]
- American Water Works Association (AWWA). Standard Methods for the Examination of Water and Wastewater, 21st ed.; AWWA: Washington, DC, USA, 2005. [Google Scholar]
- National Center for Biotechnology Information (NCBI). Available online: https://www.ncbi.nlm.nih.gov (accessed on 5 October 2021).
- Movassat, M.; Forouzmand, E.; Reese, F.; Hertel, K.J. Exon Size and Sequence Conservation Improves Identification of Splice-Altering Nucleotides. RNA 2019, 25, 1793–1805. [Google Scholar] [CrossRef]
- Rice Genome Annotation Project. Available online: http://rice.plantbiology.msu.edu (accessed on 5 October 2021).
- LEMNA.ORG. Available online: https://www.lemna.org/ (accessed on 5 October 2021).
- The Arabidopsis Information Resource (TAIR). Available online: https://www.arabidopsis.org (accessed on 5 October 2021).
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Molecul. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Box, M.S.; Coustham, V.; Dean, C.; Mylne, J.S. Protocol: A Simple Phenol-Based Method for 96-Well Extraction of High Quality RNA from Arabidopsis. Plant Methods 2011, 7, 7. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- López-Arredondo, D.L.; Leyva-González, M.A.; Alatorre-Cobos, F.; Herrera-Estrella, L. Biotechnology of Nutrient Uptake and Assimilation in Plants. Int. J. Dev. Biol. 2013, 57, 595–610. [Google Scholar] [CrossRef]
- Borisjuk, N.; Borisjuk, L.; Komarnytsky, S.; Timeva, S.; Hemleben, V.; Gleba, Y.; Raskin, I. Tobacco Ribosomal DNA Spacer Element Stimulates Amplification and Expression of Heterologous Genes. Nat. Biotechnol. 2000, 18, 1303–1306. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Xu, S.; Tang, X.; Zhao, J.; Yu, C.; He, G.; Xu, H.; Wang, S.; Tang, Y.; et al. Efficient Genetic Transformation and CRISPR/Cas9-mediated Genome Editing in Lemna aequinoctialis. Plant Biotechnol. J. 2019, 17, 2143–2152. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Kishchenko, O.; Stepanenko, A.; Chen, G.; Wang, W.; Zhou, J.; Pan, C.; Borisjuk, N.
The Dynamics of
Zhou Y, Kishchenko O, Stepanenko A, Chen G, Wang W, Zhou J, Pan C, Borisjuk N.
The Dynamics of
Zhou, Yuzhen, Olena Kishchenko, Anton Stepanenko, Guimin Chen, Wei Wang, Jie Zhou, Chaozhi Pan, and Nikolai Borisjuk.
2022. "The Dynamics of
Zhou, Y., Kishchenko, O., Stepanenko, A., Chen, G., Wang, W., Zhou, J., Pan, C., & Borisjuk, N.
(2022). The Dynamics of





