Transcriptomic Reprograming of Xanthomonas campestris pv. campestris after Treatment with Hydrolytic Products Derived from Glucosinolates
Abstract
:1. Introduction
2. Results
2.1. Bacterial Growth
2.2. GO Analysis
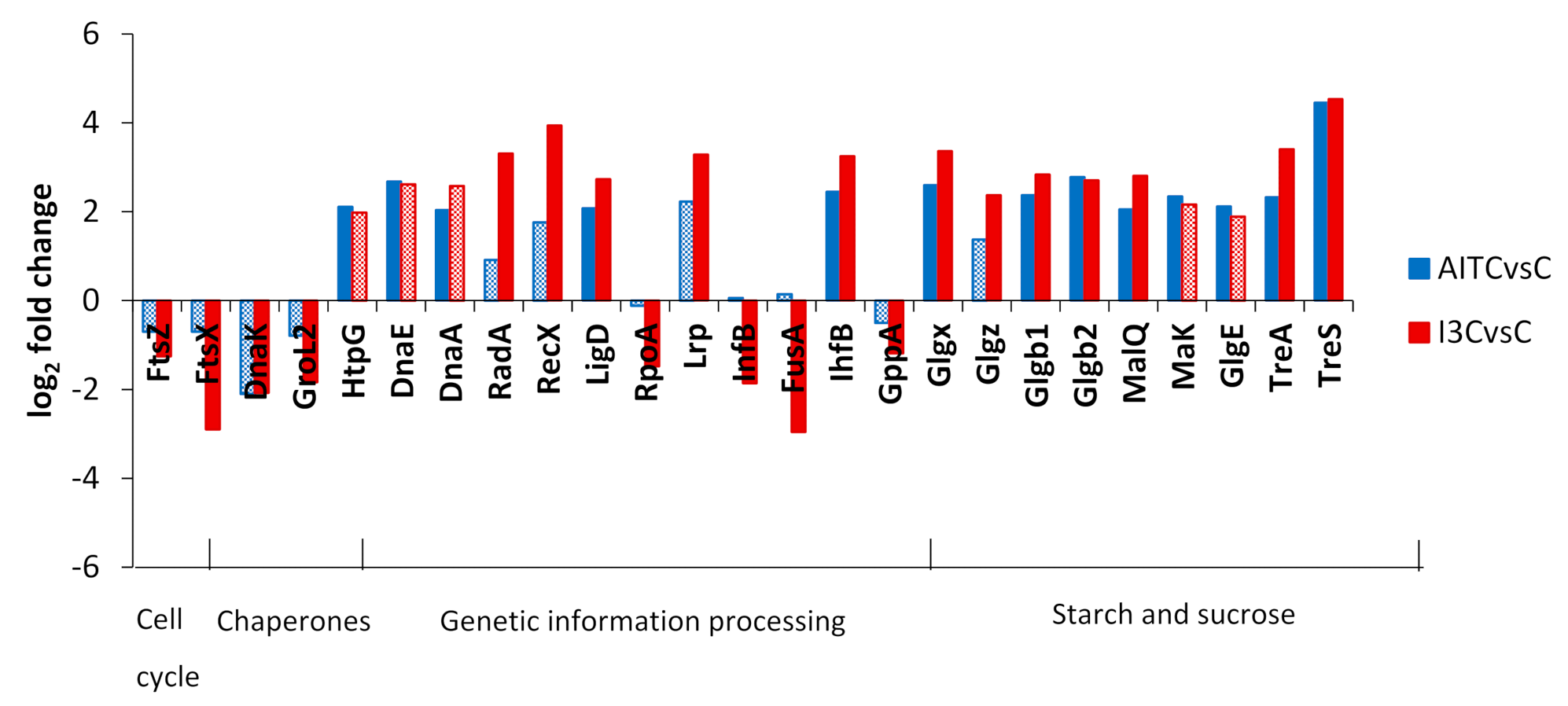
2.3. Differentially Expressed Genes
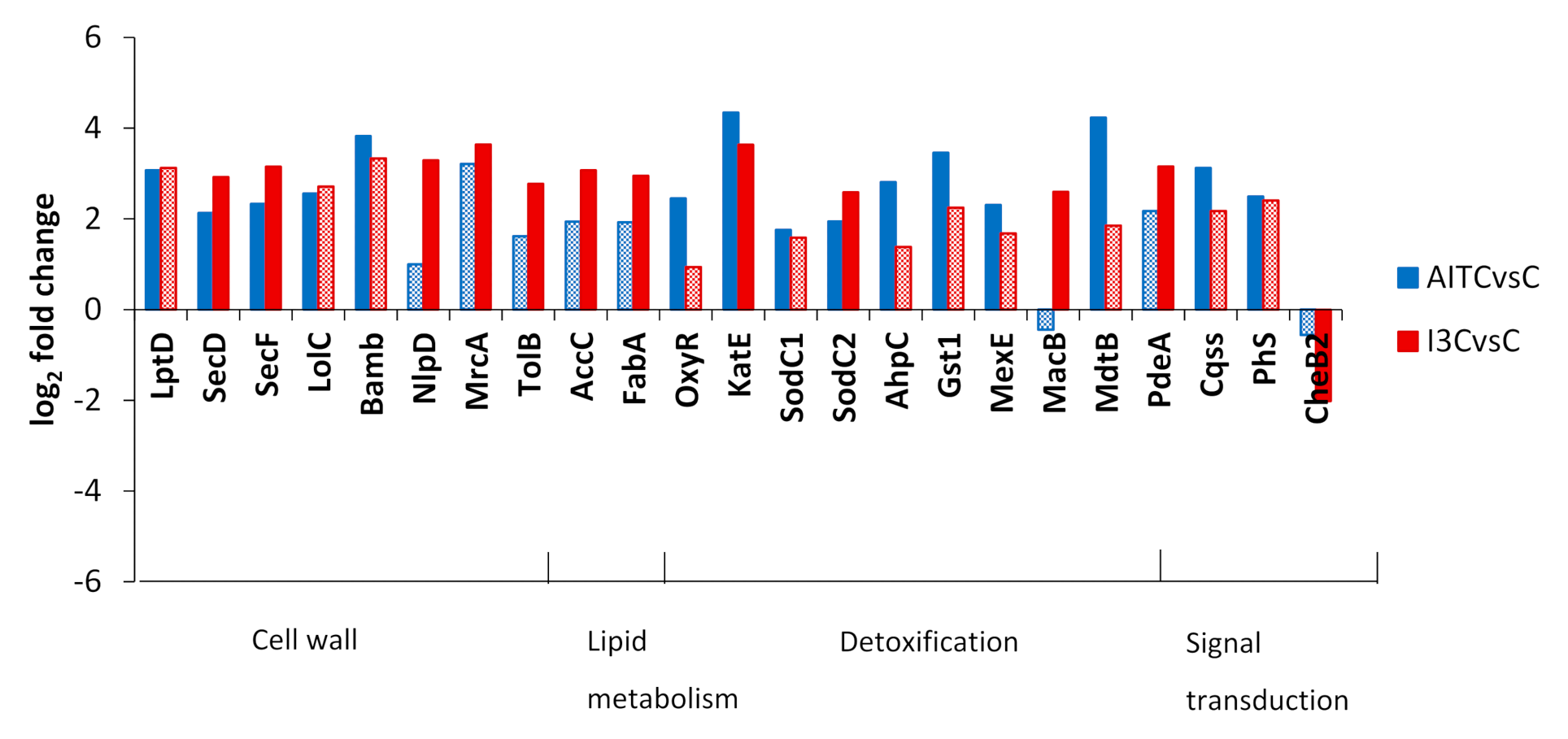
3. Discussion
3.1. Transfer of Information in the Cell
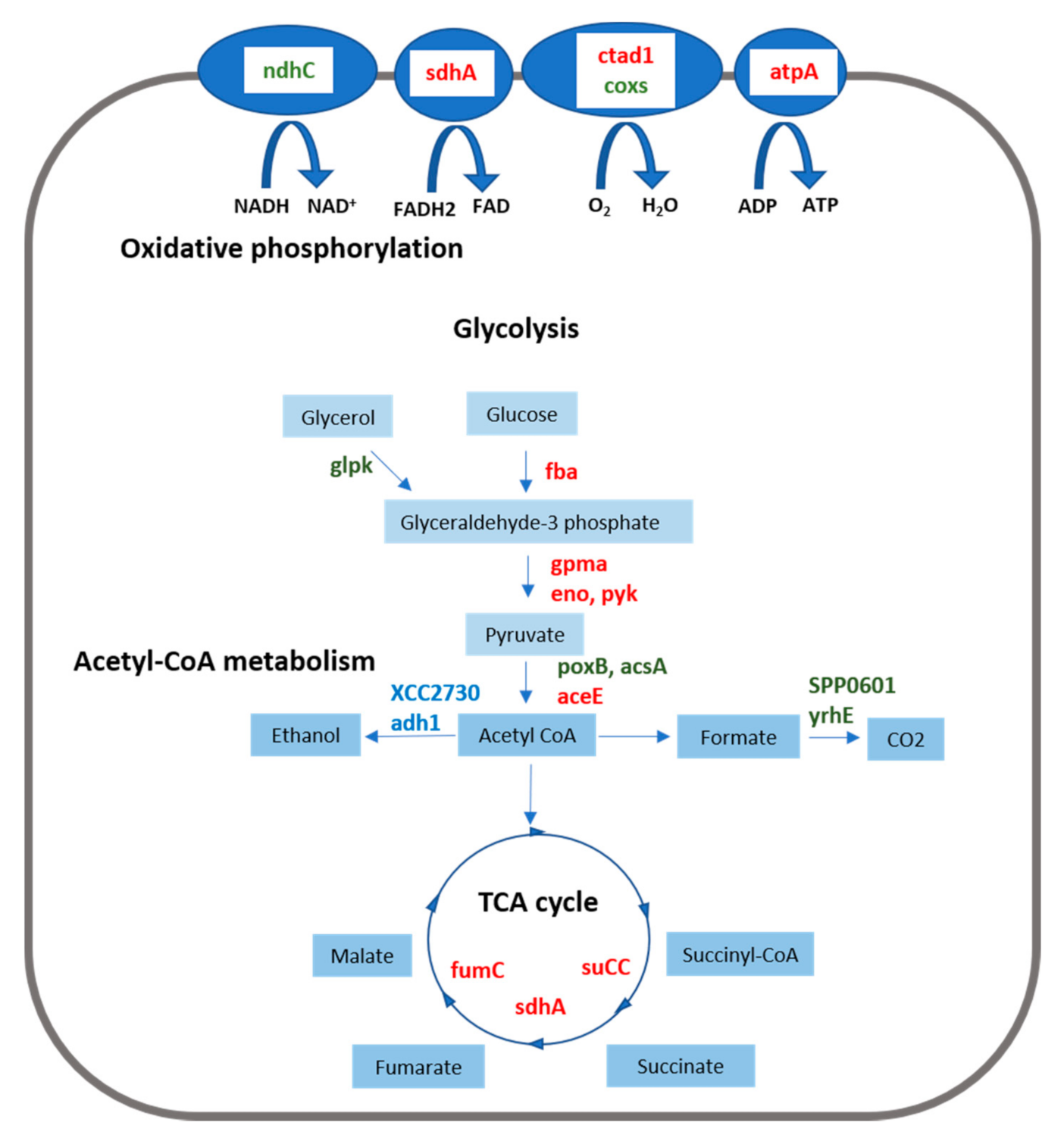
3.2. Energy Metabolism
3.3. Cell Envelope
3.4. Detoxification and Biofilm Formation
4. Materials and Methods
4.1. Bacteria Culture and GHP Treatment
4.2. RNA Isolation, Library Preparation, and Sequencing
4.3. Functional Analysis
4.4. Quantitative Reverse-Transcription-PCR (RT-qPCR) Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, S.; Dey, S.S.; Bhatia, R.; Batley, J.; Kumar, R. Molecular breeding for resistance to black rot [Xanthomonas campestris pv. campestris (Pammel) Dowson] in Brassicas: Recent advances. Euphytica 2018, 214, 196. [Google Scholar] [CrossRef]
- Rubel, M.H.; Abuyusuf, M.; Nath, U.K.; Robin, A.H.K.; Jung, H.J.; Kim, H.T.; Park, J.I.; Nou, I.S. Glucosinolate profile and glucosinolate biosynthesis and breakdown gene expression manifested by black rot disease infection in cabbage. Plants 2020, 9, 1121. [Google Scholar] [CrossRef]
- Taylor, J.D.; Conway, J.; Roberts, S.J.; Astley, D.; Vicente, J.G. Sources and origin of resistance to Xanthomonas campestris pv. campestris in Brassica genomes. Phytopathology 2002, 92, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Vicente, J.G.; Conway, J.; Roberts, S.J.; Taylor, J.D. Identification and origin of Xanthomonas campestris pv. campestris races and related pathovars. Phytopathology 2001, 91, 492–499. [Google Scholar] [CrossRef] [Green Version]
- Fargier, E.; Manceau, C. Pathogenicity assays restrict the species Xanthomonas campestris into three pathovars and reveal nine races within X. campestris pv. campestris. Plant Pathol. 2007, 56, 805–818. [Google Scholar] [CrossRef]
- Cruz, J.; Tenreiro, R.; Cruz, L. Assessment of diversity of Xanthomonas campestris pathovars affecting cruciferous plants in Portugal and disclosure of two novel X. campestris pv. campestris races. J. Plant Pathol. 2017, 99, 403–414. [Google Scholar] [CrossRef]
- Tortosa, M.; Cartea, M.E.; Velasco, P.; Soengas, P.; Rodriguez, V.M. Calcium-signaling proteins mediate the plant transcriptomic response during a well-established Xanthomonas campestris pv. campestris Infection. Hortic. Res. 2019, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Zhang, E.; Liu, Y.; Xu, Z.; Hui, M.; Zhang, X.; Cai, M. Transcriptome analysis of two lines of Brassica oleracea in response to early infection with Xanthomonas campestris pv. campestris. Can. J. Plant Pathol. 2021, 43, 127–139. [Google Scholar] [CrossRef]
- Aires, A.; Dias, C.S.P.; Carvalho, R.; Oliveira, M.H.; Monteiro, A.A.; Simões, M.V.; Rosa, E.A.S.; Bennett, R.N.; Saavedra, M.J. Correlations between disease severity, glucosinolate profiles and total phenolics and Xanthomonas campestris pv. campestris inoculation of different Brassicaceae. Sci. Hortic. 2011, 129, 503–510. [Google Scholar] [CrossRef]
- Madloo, P.; Lema, M.; Francisco, M.; Soengas, P. Role of Major Glucosinolates in the defense of kale against Sclerotinia Sclerotiorum and Xanthomonas campestris pv. campestris. Phytopathology 2019, 109, 1246–1256. [Google Scholar] [CrossRef] [PubMed]
- Calmes, B.; N’Guyen, G.; Dumur, J.; Brisach, C.A.; Campion, C.; Lacomi, B.; PignÉ, S.; Dias, E.; Macherel, D.; Guillemette, T.; et al. Glucosinolate-derived isothiocyanates impact mitochondrial function in fungal cells and elicit an oxidative stress response necessary for growth recovery. Front. Plant Sci. 2015, 6, 414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burow, M.; Losansky, A.; Müller, R.; Plock, A.; Kliebenstein, D.J.; Wittstock, U. The genetic basis of constitutive and herbivore-induced ESP-independent nitrile formation in Arabidopsis. Plant Physiol. 2009, 149, 561–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sotelo, T.; Lema, M.; Soengas, P.; Cartea, M.E.; Velasco, P. In vitro activity of glucosinolates and their degradation products against Brassica-pathogenic bacteria and fungi. Appl. Environ. Microbiol. 2015, 81, 432–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastorczyk, M.; Bednarek, P. The function of glucosinolates and related metabolites in plant innate immunity. Adv. Bot. Res. 2016, 80, 171–198. [Google Scholar] [CrossRef]
- Aires, A.; Mota, V.R.; Saavedra, M.J.; Monteiro, A.A.; Simões, M.; Rosa, E.A.S.; Bennett, R.N. Initial in vitro evaluations of the antibacterial activities of glucosinolate enzymatic hydrolysis products against plant pathogenic bacteria. J. Appl. Microbiol. 2009, 106, 2096–2105. [Google Scholar] [CrossRef]
- Velasco, P.; Lema, M.; Francisco, M.; Soengas, P.; Elena Cartea, M. In vivo and in vitro effects of secondary metabolites against Xanthomonas campestris pv. campestris. Molecules 2013, 18, 11131–11143. [Google Scholar] [CrossRef] [Green Version]
- Borges, A.; Simoes, L.C.; Saavedra, M.J.; Simoes, M. The action of selected isothiocyanates on bacterial biofilm prevention and control. Int. Biodeterior. Biodegrad. 2014, 86, 25–33. [Google Scholar] [CrossRef] [Green Version]
- Borges, A.; Serra, S.; Cristina Abreu, A.; Saavedra, M.J.; Salgado, A.; Simões, M. Evaluation of the effects of selected phytochemicals on quorum sensing inhibition and in vitro cytotoxicity. Biofouling 2014, 30, 183–195. [Google Scholar] [CrossRef]
- Nowicki, D.; Maciąg-Dorszyńska, M.; Bogucka, K.; Szalewska-Pałasz, A.; Herman-Antosiewicz, A. Various modes of action of dietary phytochemicals, sulforaphane and phenethyl isothiocyanate, on pathogenic bacteria. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Dufour, V.; Stahl, M.; Rosenfeld, E.; Stintzi, A.; Baysse, C. Insights into the mode of action of benzyl isothiocyanate on Campylobacter Jejuni. Appl. Environ. Microbiol. 2013, 79, 6958–6968. [Google Scholar] [CrossRef] [Green Version]
- Dufour, V.; Stahl, M.; Baysse, C. The antibacterial properties of isothiocyanates. Microbiology 2015, 161, 229–243. [Google Scholar] [CrossRef] [Green Version]
- Borges, A.; Abreu, A.C.; Ferreira, C.; Saavedra, M.J.; Simões, L.C.; Simões, M. Antibacterial activity and mode of action of selected glucosinolate hydrolysis products against bacterial pathogens. J. Food Sci. Technol. 2015, 52, 4737–4748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burby, P.E.; Simmons, L.A. Regulation of cell division in bacteria by monitoring genome integrity and DNA replication status. J. Bacteriol. 2020, 202, e00408-19. [Google Scholar] [CrossRef] [PubMed]
- Margolin, W. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 2005, 6, 862–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calloni, G.; Chen, T.; Schermann, S.M.; Chang, H.C.; Genevaux, P.; Agostini, F.; Tartaglia, G.G.; Hayer-Hartl, M.; Hartl, F.U. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 2012, 1, 251–264. [Google Scholar] [CrossRef] [Green Version]
- Calvo, J.M.; Matthews, R.G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 1994, 58, 466–490. [Google Scholar] [CrossRef]
- Caldas, T.; Laalami, S.; Richarme, G. Chaperone properties of bacterial elongation factor EF-G and initiation factor IF2. J. Biol. Chem. 2000, 275, 855–860. [Google Scholar] [CrossRef] [Green Version]
- Roghanian, M.; Semsey, S.; Løbner-Olesen, A.; Jalalvand, F. (P)PpGpp-mediated stress response induced by defects in outer membrane biogenesis and ATP production promotes survival in Escherichia coli. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Denapoli, J.; Tehranchi, A.K.; Wang, J.D. Dose-dependent reduction of replication elongation rate by (p)PpGpp in Escherichia coli and Bacillus subtilis. Mol. Microbiol. 2013, 88, 93–104. [Google Scholar] [CrossRef] [Green Version]
- Gourse, R.L.; Chen, A.Y.; Gopalkrishnan, S.; Sanchez-Vazquez, P.; Myers, A.; Ross, W. Transcriptional responses to ppGpp and DksA. Annu. Rev. Microbiol. 2018, 72, 163–184. [Google Scholar] [CrossRef]
- Kassie, F.; Knasmüller, S. Genotoxic effects of allyl isothiocyanate (AITC) and phenethyl isothiocyanate (PEITC). Chem. Biol. Interact. 2000, 127, 163–180. [Google Scholar] [CrossRef]
- Ruhal, R.; Kataria, R.; Choudhury, B. Trends in bacterial trehalose metabolism and significant nodes of metabolic pathway in the direction of trehalose accumulation. Microb. Biotechnol. 2013, 6, 493–502. [Google Scholar] [CrossRef]
- Wilson, W.A.; Roach, P.J.; Montero, M.; Baroja-Fernández, E.; Muñoz, F.J.; Eydallin, G.; Viale, A.M.; Pozueta-Romero, J. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol. Rev. 2010, 34, 952–985. [Google Scholar] [CrossRef] [Green Version]
- Iwadate, Y.; Funabasama, N.; Kato, J.I. Involvement of formate dehydrogenases in stationary phase oxidative stress tolerance in Escherichia coli. FEMS Microbiol. Lett. 2017, 364, fnx193. [Google Scholar] [CrossRef]
- Lee, W.; Lee, D.G. Reactive oxygen species modulate itraconazole-induced apoptosis via mitochondrial disruption in Candida Albicans. Free Radic. Res. 2018, 52, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Cascales, E.; Lloubès, R. Deletion Analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol. Microbiol. 2004, 51, 873–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okuda, S.; Sherman, D.J.; Silhavy, T.J.; Ruiz, N.; Kahne, D. Lipopolysaccharide transport and assembly at the outer membrane: The PEZ model. Nat. Rev. Microbiol. 2016, 14, 798. [Google Scholar] [CrossRef] [Green Version]
- Polissi, A.; Sperandeo, P. The lipopolysaccharide export pathway in Escherichia coli: Structure, organization and regulated assembly of the Lpt machinery. Mar. Drugs 2014, 12, 1023–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pletnev, P.; Osterman, I.; Sergiev, P.; Bogdanov, A.; Dontsova, O. Survival guide: Escherichia coli in the stationary phase. Acta Nat. 2015, 7, 22–33. [Google Scholar] [CrossRef]
- Bernadac, A.; Gavioli, M.; Lazzaroni, J.C.; Raina, S.; Lloubès, R. Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 1998, 180, 4872–4878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Cronan, J.E. Escherichia coli unsaturated fatty acid synthesis. Complex trancription of the fabA gene and in vivo identification of the essential reaction catalyzed by fabB. J. Biol. Chem. 2009, 284, 29526–29535. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.H.; Zeng, X.A.; Wang, M.S.; Brennan, C.S.; Gong, D. Modification of membrane properties and fatty acids biosynthesis-related genes in Escherichia coli and Staphylococcus aureus: Implications for the antibacterial mechanism of naringenin. Biochim. Biophys. Acta-Biomembr. 2018, 1860, 481–490. [Google Scholar] [CrossRef]
- Miko, M.; Chance, B. Isothiocyanates. A new class of uncouplers. BBA-Bioenergetics 1975, 396, 165–174. [Google Scholar] [CrossRef]
- Kubo, E.; Chhunchha, B.; Singh, P.; Sasaki, H.; Singh, D.P. Sulforaphane reactivates cellular antioxidant defense by inducing Nrf2/ARE/Prdx6 activity during aging and oxidative stress. Sci. Rep. 2017, 7, 14130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakobsen, T.H.; Bragason, S.K.; Phipps, R.K.; Christensen, L.D.; van Gennip, M.; Alhede, M.; Skindersoe, M.; Larsen, T.O.; Høiby, N.; Bjarnsholt, T.; et al. Food as a source for quorum sensing inhibitors: Iberin from horseradish revealed as a quorum sensing inhibitor of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2012, 78, 2410–2421. [Google Scholar] [CrossRef] [Green Version]
- Orr, M.W.; Donaldson, G.P.; Severin, G.B.; Wang, J.; Sintim, H.O.; Waters, C.M.; Lee, V.T. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that Is required for cyclic-di-GMP turnover. Proc. Natl. Acad. Sci. USA 2015, 112, E5048–E5057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yonekawa, H.; Hayashi, H.; Parkinson, J.S. Requirement of the cheB function for sensory adaptation in Escherichia coli. J. Bacteriol. 1983, 156, 1228–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, X.; Zhang, Q.; Zou, J.; He, C.; Tao, J. Selection of optimized reference genes for QRT-PCR normalization in Xanthomonas campestris pv. campestris cultured in different media. Curr. Microbiol. 2019, 76, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Ramakers, C.; Ruijter, J.M.; Lekanne Deprez, R.H.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madloo, P.; Lema, M.; Rodríguez, V.M.; Soengas, P. Transcriptomic Reprograming of Xanthomonas campestris pv. campestris after Treatment with Hydrolytic Products Derived from Glucosinolates. Plants 2021, 10, 1656. https://doi.org/10.3390/plants10081656
Madloo P, Lema M, Rodríguez VM, Soengas P. Transcriptomic Reprograming of Xanthomonas campestris pv. campestris after Treatment with Hydrolytic Products Derived from Glucosinolates. Plants. 2021; 10(8):1656. https://doi.org/10.3390/plants10081656
Chicago/Turabian StyleMadloo, Pari, Margarita Lema, Victor Manuel Rodríguez, and Pilar Soengas. 2021. "Transcriptomic Reprograming of Xanthomonas campestris pv. campestris after Treatment with Hydrolytic Products Derived from Glucosinolates" Plants 10, no. 8: 1656. https://doi.org/10.3390/plants10081656
APA StyleMadloo, P., Lema, M., Rodríguez, V. M., & Soengas, P. (2021). Transcriptomic Reprograming of Xanthomonas campestris pv. campestris after Treatment with Hydrolytic Products Derived from Glucosinolates. Plants, 10(8), 1656. https://doi.org/10.3390/plants10081656






