Fine Mapping of a Major Pleiotropic QTL Associated with Sesamin and Sesamolin Variation in Sesame (Sesamum indicum L.)
Abstract
:1. Introduction
2. Results
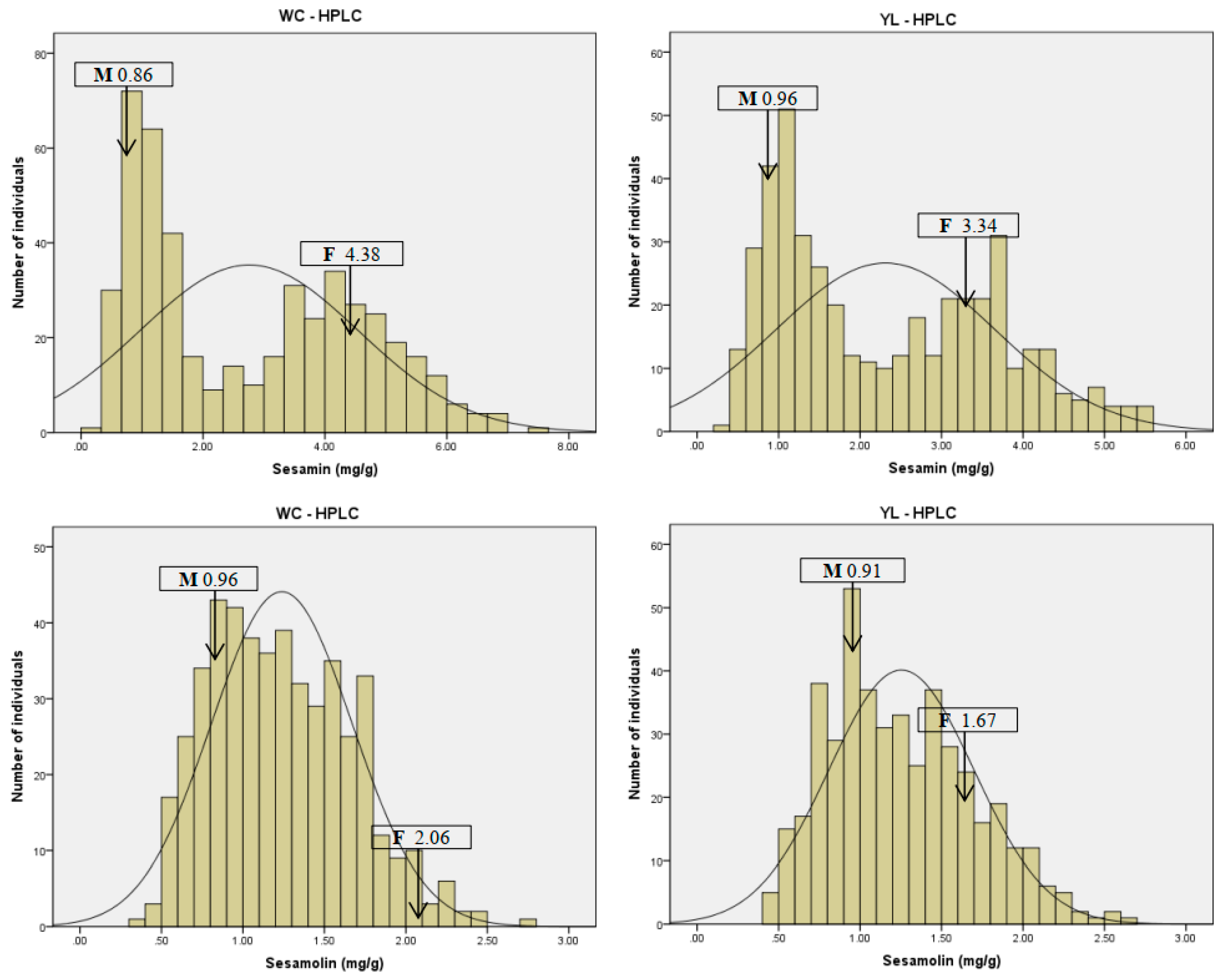
2.1. Seed Sesamin and Sesamolin Content Variation in the RILs Population
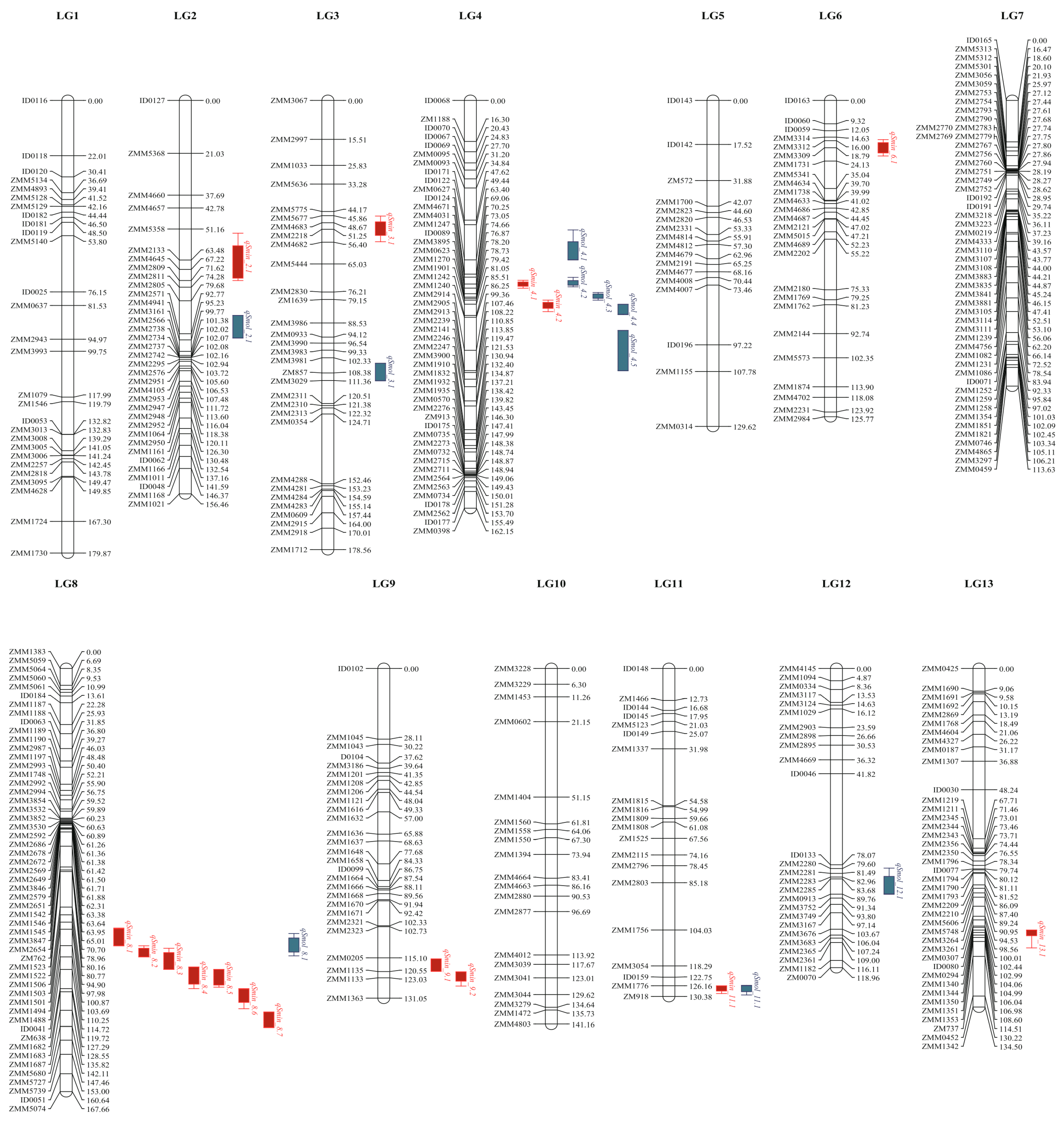
2.2. The QTL for Sesamin and Sesamolin Content Variations
2.3. Digenic Epistatic Interactions Analysis of the Predicted Loci
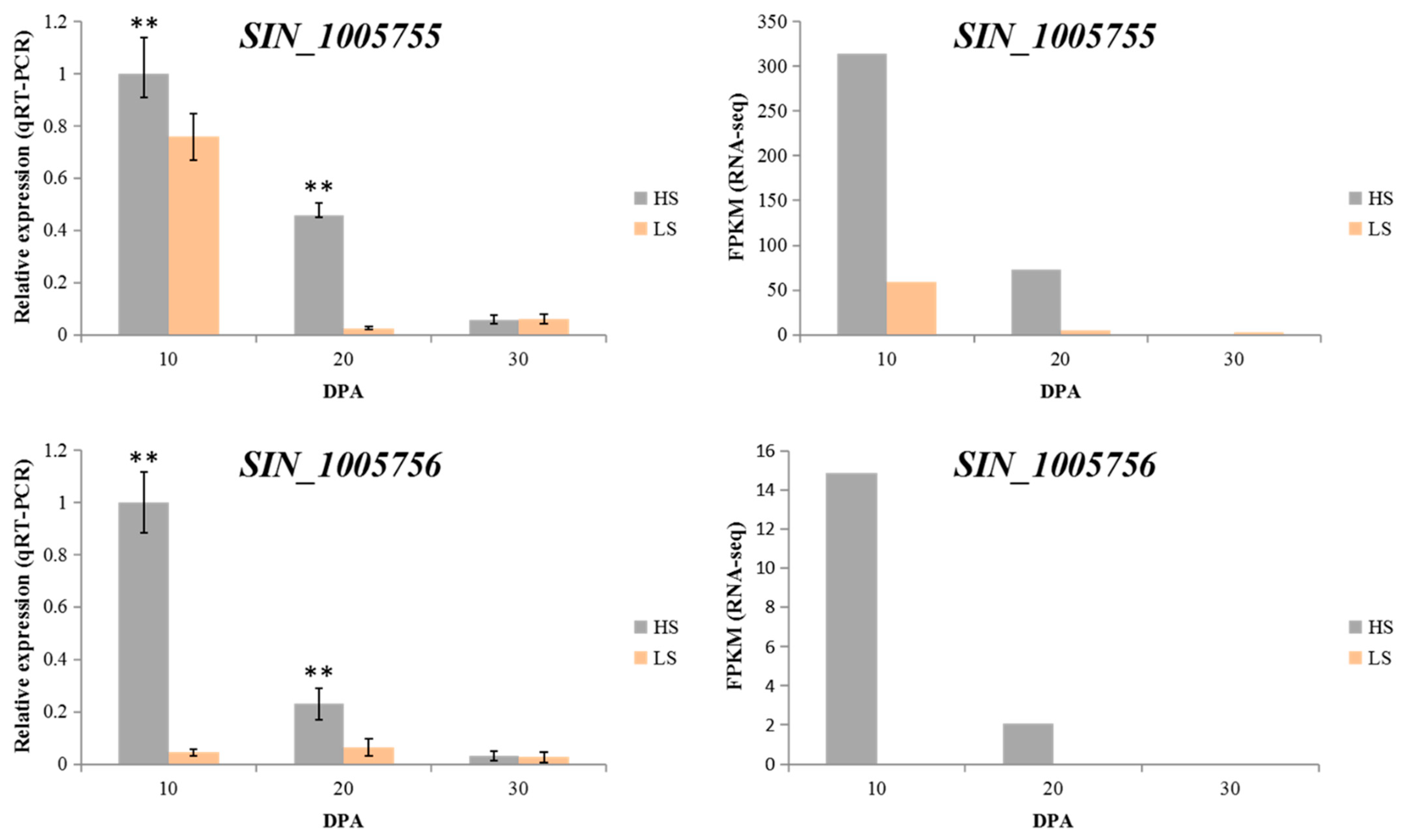
2.4. Candidate Genes under the Major QTL Region
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Experimental Design and Sampling
4.3. Sesamin and Sesamolin Extraction from Seeds
4.4. HPLC Analysis of Sesamin and Sesamolin
4.5. Phenotypic Data Analysis
4.6. QTL Analysis
4.7. RNA Isolation and RNA-seq Analysis
4.8. qRT-PCR Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Wan, Y.; Li, H.; Fu, G.; Chen, X.; Chen, F.; Xie, M. The relationship of antioxidant components and antioxidant activity of sesame seed oil. J. Sci. Food Agric. 2015, 95, 2571–2578. [Google Scholar] [CrossRef]
- Satake, H.; Koyama, T.; Bahabadi, S.E.; Matsumoto, E.; Ono, E.; Murata, J. Essences in metabolic engineering of lignan biosynthesis. Metabolites 2015, 5, 270–290. [Google Scholar] [CrossRef] [Green Version]
- Pathak, N.; Rai, A.K.; Kumari, R.; Bha, K.V. Value addition in sesame: A perspective on bioactive components for enhancing utility and profitability. Pharmacogn. Rev. 2014, 8, 147–155. [Google Scholar] [CrossRef] [Green Version]
- Kim, A.Y.; Yun, C.I.; Lee, J.G.; Kim, Y.J. Determination and Daily Intake Estimation of Lignans in Sesame Seeds and Sesame Oil Products in Korea. Foods 2020, 9, 394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davin, L.B.; Lewis, N.G. Dirigent phenoxy radical coupling: Advances and challenges. Curr. Opin. Biotechnol. 2005, 16, 398–406. [Google Scholar] [CrossRef]
- Budowski, P. Recent Research on Sesamin, Sesamolin, and Related Compounds. J. Am. Oil Chem. Soc. 1964, 41, 280–285. [Google Scholar] [CrossRef]
- Majdalawieh, A.F.; Dalibalta, S.; Yousef, S.M. Effects of sesamin on fatty acid and cholesterol metabolism, macrophage cholesterol homeostasis and serum lipid profile: A comprehensive review. Eur. J. Pharmacol. 2020, 885, 173417. [Google Scholar] [CrossRef]
- Majdalawieh, A.F.; Massri, M.; Nasrallah, G.K. A comprehensive review on the anti-cancer properties and mechanisms of action of sesamin, a lignan in sesame seeds (Sesamum indicum). Eur. J. Pharmacol. 2017, 815, 512–521. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kim, E.Y.; Rodriguez, I.; Nam, Y.H.; Jeong, S.Y.; Hong, B.N.; Choung, S.Y.; Kang, T.H. Sesamum indicum L. Oil and Sesamin Induce Auditory-Protective Effects through Changes in Hearing Loss-Related Gene Expression. J. Med. Food 2020, 23, 491–498. [Google Scholar] [CrossRef]
- Abe-Kanoh, N.; Kunimoto, Y.; Takemoto, D.; Ono, Y.; Shibata, H.; Ohnishi, K.; Kawai, Y. Sesamin Catechol Glucuronides Exert Anti-inflammatory Effects by Suppressing Interferon β and Inducible Nitric Oxide Synthase Expression through Deconjugation in Macrophage-like J774.1 Cells. J. Agric. Food Chem. 2019, 67, 7640–7649. [Google Scholar] [CrossRef]
- Kim, K.-S.; Lee, J.-R.; Lee, J.-S. Determination of sesamin and sesamolin in sesame (Sesamum indicum L.) seeds using UV spectrophotometer and HPLC. Korean J. Crop Sci. 2006, 51, 95–100. [Google Scholar]
- Dossa, K.; Diouf, D.; Wang, L.; Wei, X.; Zhang, Y.; Niang, M.; Fonceka, D.; Yu, J.; Mmadi, M.A.; Yehouessi, L.W.; et al. The Emerging Oilseed Crop Sesamum indicum Enters the “Omics” Era. Front. Plant Sci. 2017, 8, 1154. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, T. Diversity in lignan biosynthesis. Phytochem. Rev. 2003, 2, 371–390. [Google Scholar] [CrossRef]
- Ke, T.; Dong, C.; Mao, H.; Zhao, Y.; Chen, H.; Liu, H.; Dong, X.; Tong, C.; Liu, S. Analysis of expression sequence tags from a full-length-enriched cDNA library of developing sesame seeds (Sesamum indicum). BMC Plant Biol. 2011, 11, 180. [Google Scholar] [CrossRef] [Green Version]
- Usman, S.M.; Viswanathan, P.L.; Manonmani, S.; Uma, D. Genetic studies on sesamin and sesamolin content and other yield attributing characters in sesame (Sesamum indicum L.). Electron. J. Plant Breed. 2020, 11, 132–138. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Li, P.; Wang, X.; Zhang, W.; Wei, W.; Zhang, X. HPLC Analysis of Seed Sesamin and Sesamolin Variation in a Sesame Germplasm Collection in China. J. Am. Oil Chem. Soc. 2012, 89, 1011–1020. [Google Scholar] [CrossRef]
- Ajit, G.; Uma, D.; Manonmani, S.; Vinothkumar, B.; Rajesh, S. Diversity Analysis of Sesame Lignans in 40 Sesame Collections in Tamil Nadu, India. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 2329–2336. [Google Scholar] [CrossRef]
- Kim, S.-U.; Oh, K.-W.; Lee, M.-H.; Lee, B.-K.; Pae, S.-B.; Hwang, C.-D.; Kim, M.-S.; Baek, I.-Y.; Lee, J.-D. Variation of Lignan Content for Sesame Seed Across Origin and Growing Environments. Korean J. Crop Sci. 2014, 59, 151–161. [Google Scholar] [CrossRef]
- Dar, A.A.; Kancharla, P.K.; Chandra, K.; Sodhi, Y.S.; Arumugam, N. Assessment of variability in lignan and fatty acid content in the germplasm of Sesamum indicum L. J. Food Sci. Technol. 2019, 56, 976–986. [Google Scholar] [CrossRef]
- Li, C.; Miao, H.; Wei, L.; Zhang, T.; Han, X.; Zhang, H. Association mapping of seed oil and protein content in Sesamum indicum L. using SSR markers. PLoS ONE 2014, 9, e105757. [Google Scholar] [CrossRef] [Green Version]
- Kancharla, P.K.; Arumugam, N. Variation of Oil, Sesamin, and Sesamolin Content in the Germplasm of the Ancient Oilseed Crop Sesamum indicum L. J. Am. Oil Chem. Soc. 2020, 97, 475–483. [Google Scholar] [CrossRef]
- Wei, X.; Liu, K.; Zhang, Y.; Feng, Q.; Wang, L.; Zhao, Y.; Li, D.; Zhao, Q.; Zhu, X.; Zhu, X.; et al. Genetic discovery for oil production and quality in sesame. Nat. Commun. 2015, 6, 8609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, K.; Wu, W.-X.; Yang, M.-M.; Liu, H.-Y.; Hao, G.-C.; Zhao, Y.-Z. QTL Mapping for Oil, Protein and Sesamin Contents in Seeds of White Sesame. Acta Agron. Sin. 2017, 43, 1003–1011. [Google Scholar] [CrossRef]
- Ghotbzadeh Kermani, S.; Saeidi, G.; Sabzalian, M.R.; Gianinetti, A. Drought stress influenced sesamin and sesamolin content and polyphenolic components in sesame (Sesamum indicum L.) populations with contrasting seed coat colors. Food Chem. 2019, 289, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Matsui, K. Engineering the biosynthesis of low molecular weight metabolites for quality traits (essential nutrients, health-promoting phytochemicals, volatiles, and aroma compounds). In Plant Biotechnology and Agriculture; Academic Press: Cambridge, MA, USA, 2012; pp. 443–461. [Google Scholar]
- Ono, E.; Nakai, M.; Fukui, Y.; Tomimori, N.; Fukuchi-Mizutani, M.; Saito, M.; Satake, H.; Tanaka, T.; Katsuta, M.; Umezawa, T.; et al. Formation of two methylenedioxy bridges by a Sesamum CYP81Q protein yielding a furofuran lignan, (+)-sesamin. Proc. Natl. Acad. Sci. USA 2006, 103, 10116–10121. [Google Scholar] [CrossRef] [Green Version]
- Murata, J.; Ono, E.; Yoroizuka, S.; Toyonaga, H.; Shiraishi, A.; Mori, S.; Tera, M.; Azuma, T.; Nagano, A.J.; Nakayasu, M.; et al. Oxidative rearrangement of (+)-sesamin by CYP92B14 co-generates twin dietary lignans in sesame. Nat. Commun. 2017, 8, 2155. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, S.; Tong, C.; Zhao, Y.; Liu, Y.; Song, C.; Zhang, Y.; Zhang, X.; Wang, Y.; Hua, W.; et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014, 15, R39. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, Y.; Zhu, X.; Zhu, X.; Li, D.; Zhang, X.; Gao, Y.; Xiao, G.; Wei, X.; Zhang, X. Development of an SSR-based genetic map in sesame and identification of quantitative trait loci associated with charcoal rot resistance. Sci. Rep. 2017, 7, 8349. [Google Scholar] [CrossRef]
- Wu, K.; Liu, H.; Yang, M.; Tao, Y.; Ma, H.; Wu, W.; Zuo, Y.; Zhao, Y. High-density genetic map construction and QTLs analysis of grain yield-related traits in Sesame (Sesamum indicum L.) based on RAD-Seq techonology. BMC Plant Biol. 2014, 14, 274. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, Y.; Li, D.; Dossa, K.; Wang, M.L.; Zhou, R.; Yu, J.; Zhang, X. Gene expression profiles that shape high and low oil content sesames. BMC Genet. 2019, 20, 45. [Google Scholar] [CrossRef]
- Kulwal, P.L. Trait Mapping Approaches through Linkage Mapping in Plants. Adv. Biochem. Eng. Biotechnol. 2018, 164, 53–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Miao, H.; Wei, L.; Li, C.; Zhao, R.; Wang, C. Genetic analysis and QTL mapping of seed coat color in sesame (Sesamum indicum L.). PLoS ONE 2013, 8, e63898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, L.; Li, D.; Gao, Y.; Lu, H.; Zhang, X. Mapping of Sesame Waterlogging Tolerance QTL and Identification of Excellent Waterlogging Tolerant Germplasm. Sci. Agric. Sin. 2014, 47, 422–430. [Google Scholar]
- Rao, P.V.R.; Prasuna, K.; Anuradha, G.; Srividya, A.; Vemireddy, L.R.; Shankar, V.G.; Sridhar, S.; Jayaprada, M.; Reddy, K.R.; Reddy, N.P.E.; et al. Molecular mapping of important agro-botanic traits in sesame. Electron. J. Plant Breed. 2014, 5, 475–488. [Google Scholar]
- Wang, L.; Li, D.; Xi, X.; Zhang, Y.; Ding, X.; Wang, L.; Wei, W.; Gao, Y.; Zhang, X. Association analysis of sesamin and sesamolin in the core sesame (Sesamum indicum L.) germplasm. Chin. J. Oil. Crop. Sci. 2014, 36, 32–37. [Google Scholar]
- Tiwari, S.; Kumar, S.; Gontia, I. Biotechnological approaches for sesame (Sesamum indicum L.) and niger (Guizotia abyssinica L.f. Cass.). Asia-Pac. J. Mol. Biotechnol. 2011, 19, 2–9. [Google Scholar]
- Dar, A.A.; Arumugam, N. Lignans of sesame: Purification methods, biological activities and biosynthesis—A review. Bioorg. Chem. 2013, 50, 1–10. [Google Scholar] [CrossRef]
- Pathak, N.; Rai, A.K.; Saha, S.; Walia, S.; Sen, S.K.; Bhat, K.V. Quantitative dissection of antioxidative bioactive components in cultivated and wild sesame germplasm reveals potentially exploitable wide genetic variability. J. Crop Sci. Biotechnol. 2014, 17, 127–139. [Google Scholar] [CrossRef]
- Muthulakshmi, C.; Pavithra, S.; Selvi, S. Evaluation of sesame (Sesamum indicum L.) germplasm collection of Tamil Nadu for α-linolenic acid, sesamin and sesamol content. Afr. J. Biotechnol. 2017, 16, 1308–1313. [Google Scholar] [CrossRef] [Green Version]
- Rangkadilok, N.; Pholphana, N.; Mahidol, C.; Wongyai, W.; Saengsooksree, K.; Nookabkaew, S.; Satayavivad, J. Variation of sesamin, sesamolin and tocopherols in sesame (Sesamum indicum L.) seeds and oil products in Thailand. Food Chem. 2010, 122, 724–730. [Google Scholar] [CrossRef]
- Murata, J.; Matsumoto, E.; Morimoto, K.; Koyama, T.; Satake, H. Generation of Triple-Transgenic Forsythia Cell Cultures as a Platform for the Efficient, Stable, and Sustainable Production of Lignans. PLoS ONE 2015, 10, e0144519. [Google Scholar] [CrossRef] [PubMed]
- Pathak, N.; Bhaduri, A.; Bhat, K.V.; Rai, A.K. Tracking sesamin synthase gene expression through seed maturity in wild and cultivated sesame species—A domestication footprint. Plant Biol. 2015, 17, 1039–1046. [Google Scholar] [CrossRef]
- Tera, M.; Koyama, T.; Murata, J.; Furukawa, A.; Mori, S.; Azuma, T.; Watanabe, T.; Hori, K.; Okazawa, A.; Kabe, Y.; et al. Identification of a binding protein for sesamin and characterization of its roles in plant growth. Sci. Rep. 2019, 9, 8631. [Google Scholar] [CrossRef] [Green Version]
- Podzimska-Sroka, D.; O’Shea, C.; Gregersen, P.L.; Skriver, K. NAC Transcription Factors in Senescence: From Molecular Structure to Function in Crops. Plants 2015, 4, 412–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.L.; Wang, S.B.; Chen, R.G.; Chen, B.H.; Du, X.H.; Yin, Y.X.; Gong, Z.H.; Zhang, Y.Y. Characterization and expression profile of CaNAC2 pepper gene. Front. Plant Sci. 2015, 6, 755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.; Huang, J.; Guo, Y.; Yang, M.; Guo, Y.; Li, J.; Zhang, J.; Xu, W. A cotton NAC domain transcription factor, GhFSN5, negatively regulates secondary cell wall biosynthesis and anther development in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 146, 303–314. [Google Scholar] [CrossRef]
- Hu, P.; Zhang, K.; Yang, C. BpNAC012 Positively Regulates Abiotic Stress Responses and Secondary Wall Biosynthesis. Plant Physiol. 2019, 179, 700–717. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Yoo, C.G.; Rottmann, W.; Winkeler, K.A.; Collins, C.M.; Gunter, L.E.; Jawdy, S.S.; Yang, X.; Pu, Y.; Ragauskas, A.J.; et al. PdWND3A, a wood-associated NAC domain-containing protein, affects lignin biosynthesis and composition in Populus. BMC Plant Biol. 2019, 19, 486. [Google Scholar] [CrossRef] [Green Version]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mccouch, S.R.; Cho, Y.G.; Yano, M.; Paul, E.; Blinstrub, M.; Morishima, H.; Kinosita, T.; McCouch, S.R.; Cho, Y.G.; Yano, M.; et al. Report on QTL nomenclature. Rice Genet. Newsl. 1997, 14, 11–13. [Google Scholar]
- Yang, J.; Hu, C.; Hu, H.; Yu, R.; Xia, Z.; Ye, X.; Zhu, J. QTLNetwork: Mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 2008, 24, 721–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Traits | Location | Parent | RIL Population | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Zhongzhi No. 13 | ZZM2748 | Mean + sd | Maximum | Minimum | Kurtosis | Skew | CV (%) | ||
| Sesamin (mg/g) | Wuchang | 4.38 | 0.86 | 2.76 ± 1.8 | 7.52 | 0.33 | −1.15 | 0.39 | 65.12 |
| Yangluo | 3.34 | 0.96 | 2.31 ± 1.34 | 5.58 | 0.36 | −0.99 | 0.46 | 58.03 | |
| Mean | 3.86 | 0.91 | 2.54 ± 1.57 | 6.55 | 0.35 | −1.07 | 0.42 | 61.81 | |
| Sesamolin (mg/g) | Wuchang | 2.06 | 0.96 | 1.24 ± 0.43 | 2.70 | 0.36 | −0.31 | 0.43 | 34.85 |
| Yangluo | 1.67 | 0.91 | 1.25 ± 0.45 | 2.70 | 0.40 | −0.33 | 0.49 | 35.53 | |
| Mean | 1.86 | 0.94 | 1.25 ± 0.44 | 2.70 | 0.38 | −0.31 | 0.47 | 35.19 | |
| Trait | Location | Sesamin | Sesamolin | ||
|---|---|---|---|---|---|
| Wuchang | Yangluo | Wuchang | Yangluo | ||
| Sesamin | Wuchang | 1 | |||
| Yangluo | 0.834 ** | 1 | |||
| Sesamolin | Wuchang | 0.765 ** | 0.640 ** | 1 | |
| Yangluo | 0.668 ** | 0.789 ** | 0.728 ** | 1 | |
| Trait | QTL | Chr | Position (cM) | Left Marker | Right Marker | LOD | Add | R2 (%) | LOD_L (cM) | LOD_R (cM) | WinQTLcart | ICIMapping | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WC | YL | WC | YL | |||||||||||
| sesamin | qSmin_2.1 | 2 | 66.51–66 | ZMM2133 | ZMM4645 | 3.54 | −0.20 | 1.20 | 52.8 | 71.6 | √ | √ | ||
| qSmin_3.1 | 3 | 50.71 | ZMM4683 | ZMM2218 | 4.22 | −0.19 | 1.42 | 46.0 | 56.2 | √ | √ | |||
| qSmin_4.1 | 4 | 73–73.11 | ZMM4671 | ZMM4031 | 4.34 | 0.17 | 1.45 | 71.5 | 74.7 | √ | √ | |||
| qSmin_4.2 | 4 | 81.11 | ZMM1901 | ZMM1242 | 3.77 | 0.15 | 1.29 | 79.4 | 84.0 | √ | ||||
| qSmin_6.1 | 6 | 18.01–20 | ZMM3312 | ZMM1731 | 4.56 | 0.23 | 1.79 | 15.6 | 22.1 | √ | √ | |||
| qSmin_8.1 | 8 | 106.71 | ZMM1494 | ZMM1488 | 3.37 | 0.20 | 1.25 | 103.2 | 110.3 | √ | ||||
| qSmin_8.2 | 8 | 114.31 | ZMM1488 | ID0041 | 4.74 | 0.23 | 1.64 | 110.3 | 114.7 | √ | ||||
| qSmin_8.3 | 8 | 116.71–118 | ID0041 | ZM638 | 6.45 | 0.25 | 2.53 | 111.3 | 119.7 | √ | √ | |||
| qSmin_8.4 | 8 | 120.71 | ZM638 | ZMM1682 | 5.20 | 0.25 | 1.82 | 118.7 | 127.3 | √ | ||||
| qSmin_8.5 | 8 | 122.71 | ZM638 | ZMM1682 | 5.98 | 0.22 | 2.51 | 119.7 | 126.7 | √ | ||||
| qSmin_8.6 | 8 | 128.61 | ZMM1683 | ZMM1687 | 4.65 | 0.22 | 1.53 | 127.4 | 135.2 | √ | ||||
| qSmin_8.7 | 8 | 139 | ZMM1687 | ZMM5680 | 5.52 | 0.21 | 1.23 | 136.5 | 142.5 | √ | ||||
| qSmin_9.1 | 9 | 119 | ZMM0205 | ZMM1135 | 6.20 | −0.22 | 1.29 | 115.5 | 120.5 | √ | ||||
| qSmin_9.2 | 9 | 122.61–123.01 | ZMM1135 | ZMM1133 | 8.78 | −0.29 | 2.67 | 120.6 | 126.2 | √ | √ | √ | ||
| qSmin_11.1 | 11 | 127–127.21 | ZMM1776 | ZM918 | 108.38 | 1.27 | 67.69 | 126.2 | 129.2 | √ | √ | √ | √ | |
| qSmin_13.1 | 13 | 105.01 | ZMM1344 | ZMM1350 | 3.88 | 0.20 | 1.15 | 104.0 | 111.1 | √ | √ | |||
| Sesamolin | qSmol_2.1 | 2 | 91 | ZMM2805 | ZMM2571 | 3.19 | 0.06 | 2.02 | 85.5 | 94.5 | √ | |||
| qSmol_3.1 | 3 | 108 | ZMM3981 | ZM857 | 4.57 | 0.07 | 2.31 | 104.5 | 111.5 | √ | ||||
| qSmol_4.1 | 4 | 57.51 | ID0122 | ZMM0627 | 3.28 | 0.07 | 2.16 | 51.5 | 63.4 | √ | ||||
| qSmol_4.2 | 4 | 72.31–73 | ZMM4671 | ZMM4031 | 6.18 | 0.08 | 4.04 | 70.4 | 74.1 | √ | √ | |||
| qSmol_4.3 | 4 | 77.91 | ID0089 | ZMM3895 | 6.12 | 0.08 | 3.09 | 76.4 | 79.4 | √ | ||||
| qSmol_4.4 | 4 | 83.11–85 | ZMM1901 | ZMM1242 | 5.26 | 0.07 | 3.82 | 81.1 | 85.2 | √ | √ | |||
| qSmol_4.5 | 4 | 102 | ZMM2914 | ZMM2905 | 3.57 | 0.07 | 2.02 | 91.5 | 107.5 | √ | ||||
| qSmol_8.1 | 8 | 109.71–111 | ZMM1494 | ID0041 | 4.26 | 0.07 | 2.53 | 105.4 | 114.3 | √ | √ | |||
| qSmol_11.1 | 11 | 127–127.21 | ZMM1776 | ZM918 | 63.45 | 0.30 | 46.05 | 126.0 | 129.8 | √ | √ | √ | √ | |
| qSmol_12.1 | 12 | 87.71 | ZMM2285 | ZMM0913 | 3.52 | −0.06 | 1.87 | 79.4 | 89.8 | √ | ||||
| Trait | QTL_i | Interval_i | Position_i | Range_i | QTL_j | Interval_j | Position_j | Range_j | AA | AAE1 | AAE2 | H2 (AA), % | H2 (AAE), % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sesamin | 2–6 | ZMM2133–ZMM4645 | 66.5 | 63.5–67.2 | 6–11 | ZMM4633–ZMM4686 | 42 | 41.0–42.9 | −0.0773 | −0.0504 | 0.0506 | 0.22 | 0.28 |
| 2–6 | ZMM2133–ZMM4645 | 66.5 | 63.5–67.2 | 11–19 | ZMM1776–ZM918 | 127.2 | 126.2–128.2 | −0.0848 | −0.0001 | 0.0001 | 0.27 | 0.04 | |
| 6–6 | ZMM3312–ZMM1731 | 19.8 | 18.8–21.8 | 11–19 | ZMM1776–ZM918 | 127.2 | 126.2–128.2 | 0.0819 | 0.0446 | −0.0445 | 0.18 | 0.22 | |
| sesamolin | 3–17 | ZMM3981–ZM857 | 108.3 | 105.3–108.4 | 13–7 | ZMM4604–ZMM4327 | 26.1 | 22.1–26.2 | 0.0244 | −0.0107 | 0.0107 | 0.25 | 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, F.; Zhou, R.; Dossou, S.S.K.; Song, S.; Wang, L. Fine Mapping of a Major Pleiotropic QTL Associated with Sesamin and Sesamolin Variation in Sesame (Sesamum indicum L.). Plants 2021, 10, 1343. https://doi.org/10.3390/plants10071343
Xu F, Zhou R, Dossou SSK, Song S, Wang L. Fine Mapping of a Major Pleiotropic QTL Associated with Sesamin and Sesamolin Variation in Sesame (Sesamum indicum L.). Plants. 2021; 10(7):1343. https://doi.org/10.3390/plants10071343
Chicago/Turabian StyleXu, Fangtao, Rong Zhou, Senouwa Segla Koffi Dossou, Shengnan Song, and Linhai Wang. 2021. "Fine Mapping of a Major Pleiotropic QTL Associated with Sesamin and Sesamolin Variation in Sesame (Sesamum indicum L.)" Plants 10, no. 7: 1343. https://doi.org/10.3390/plants10071343
APA StyleXu, F., Zhou, R., Dossou, S. S. K., Song, S., & Wang, L. (2021). Fine Mapping of a Major Pleiotropic QTL Associated with Sesamin and Sesamolin Variation in Sesame (Sesamum indicum L.). Plants, 10(7), 1343. https://doi.org/10.3390/plants10071343








