Comparative Transcriptome Profiling of Kappaphycus alvarezii (Rhodophyta, Solieriaceae) in Response to Light of Different Wavelengths and Carbon Dioxide Enrichment
Abstract
1. Introduction
2. Results
2.1. Transcriptome Sequencing and De Novo Assembly
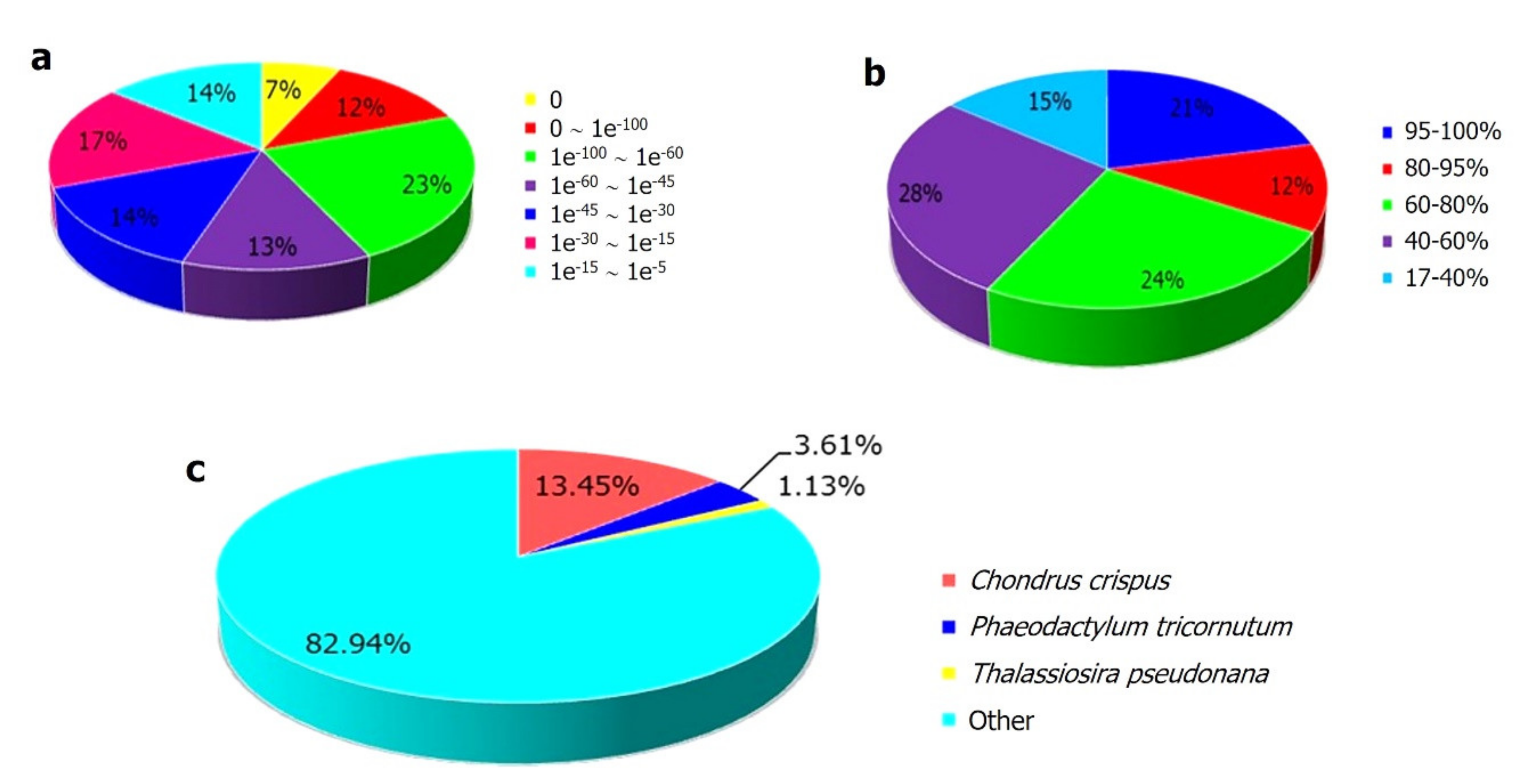
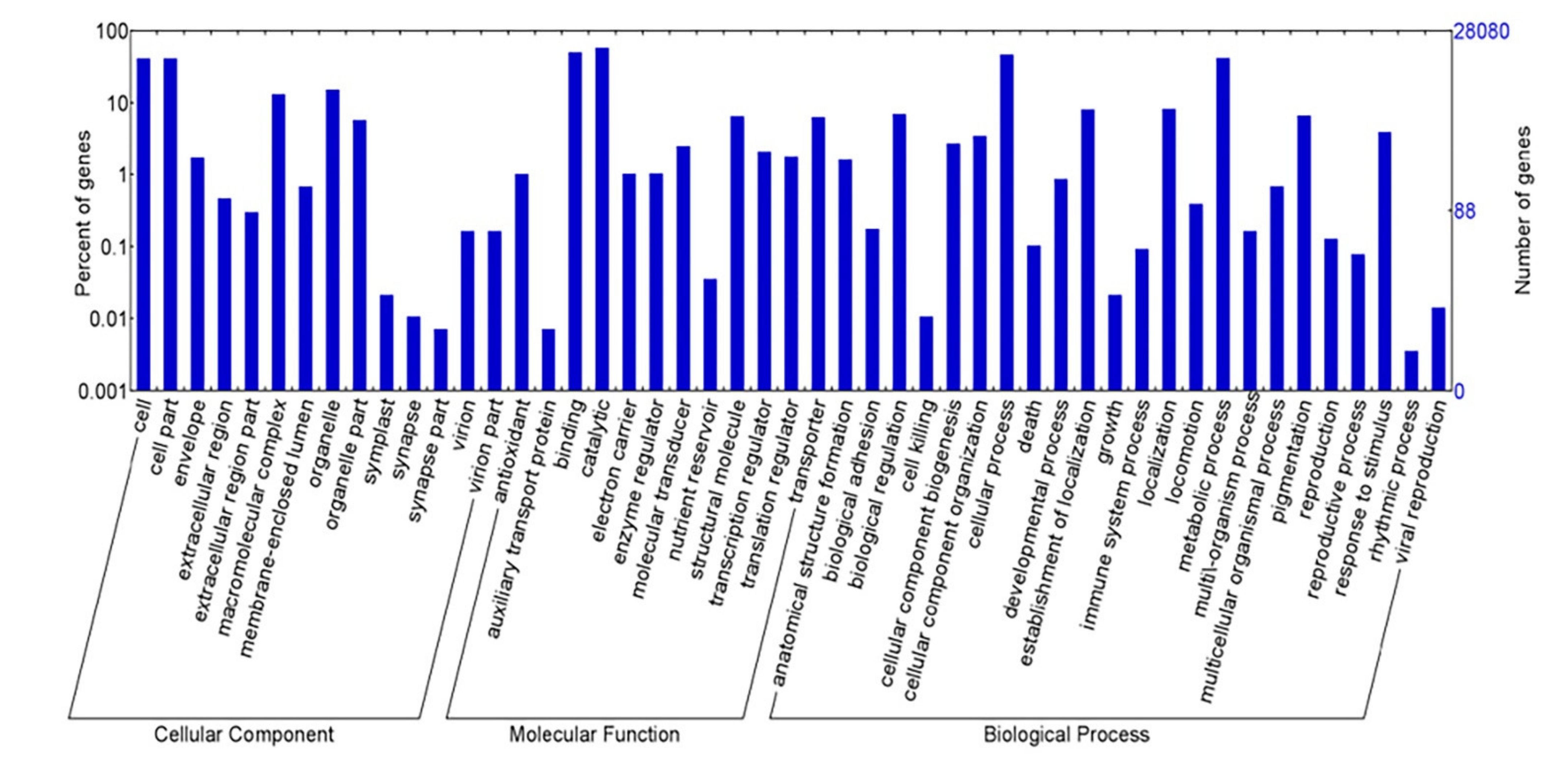
2.2. Functional Anotation of the K. alvarezii Transcriptome
2.3. Identification of Candidate Transcripts Associated with light and CO2 Responses
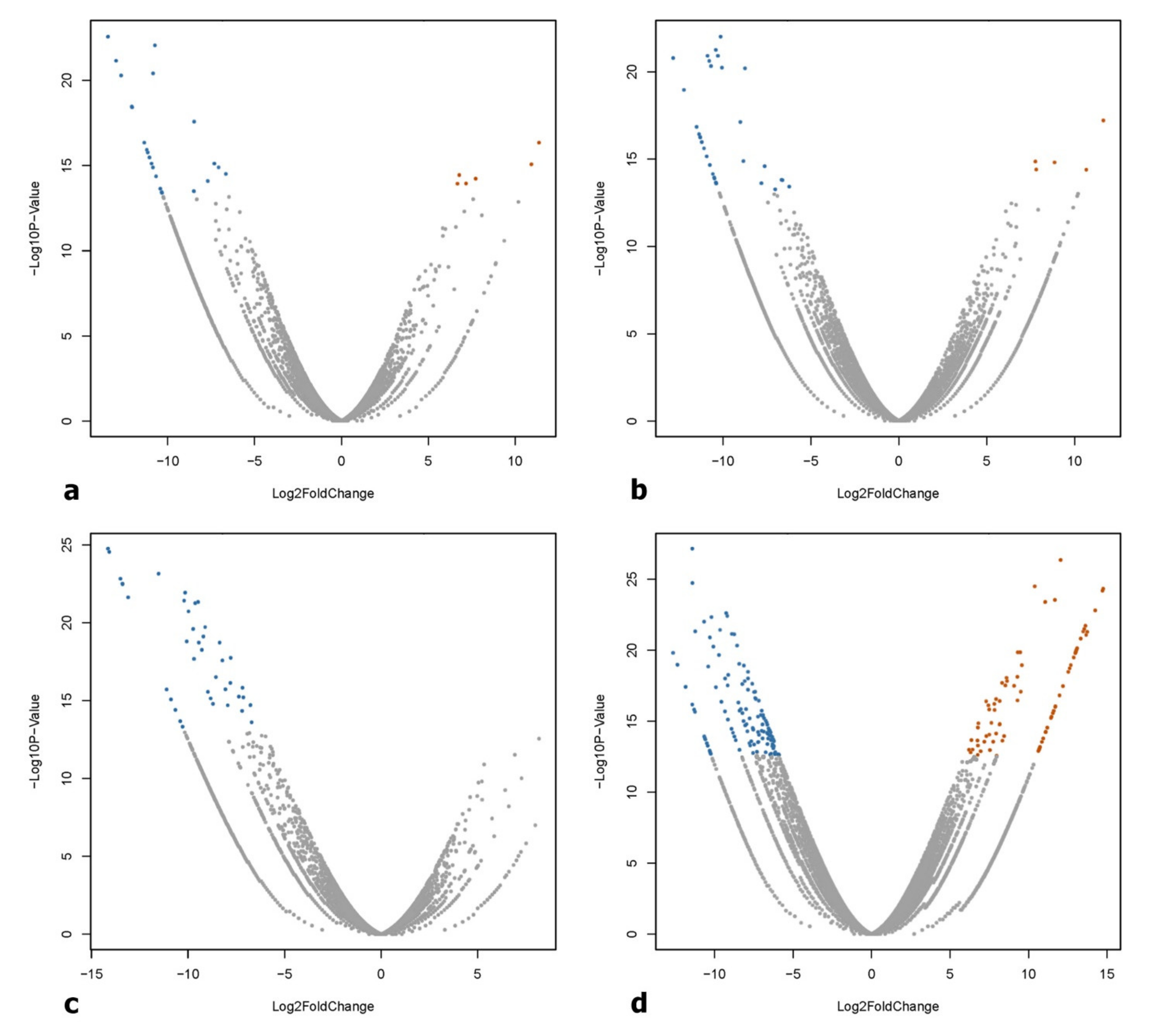
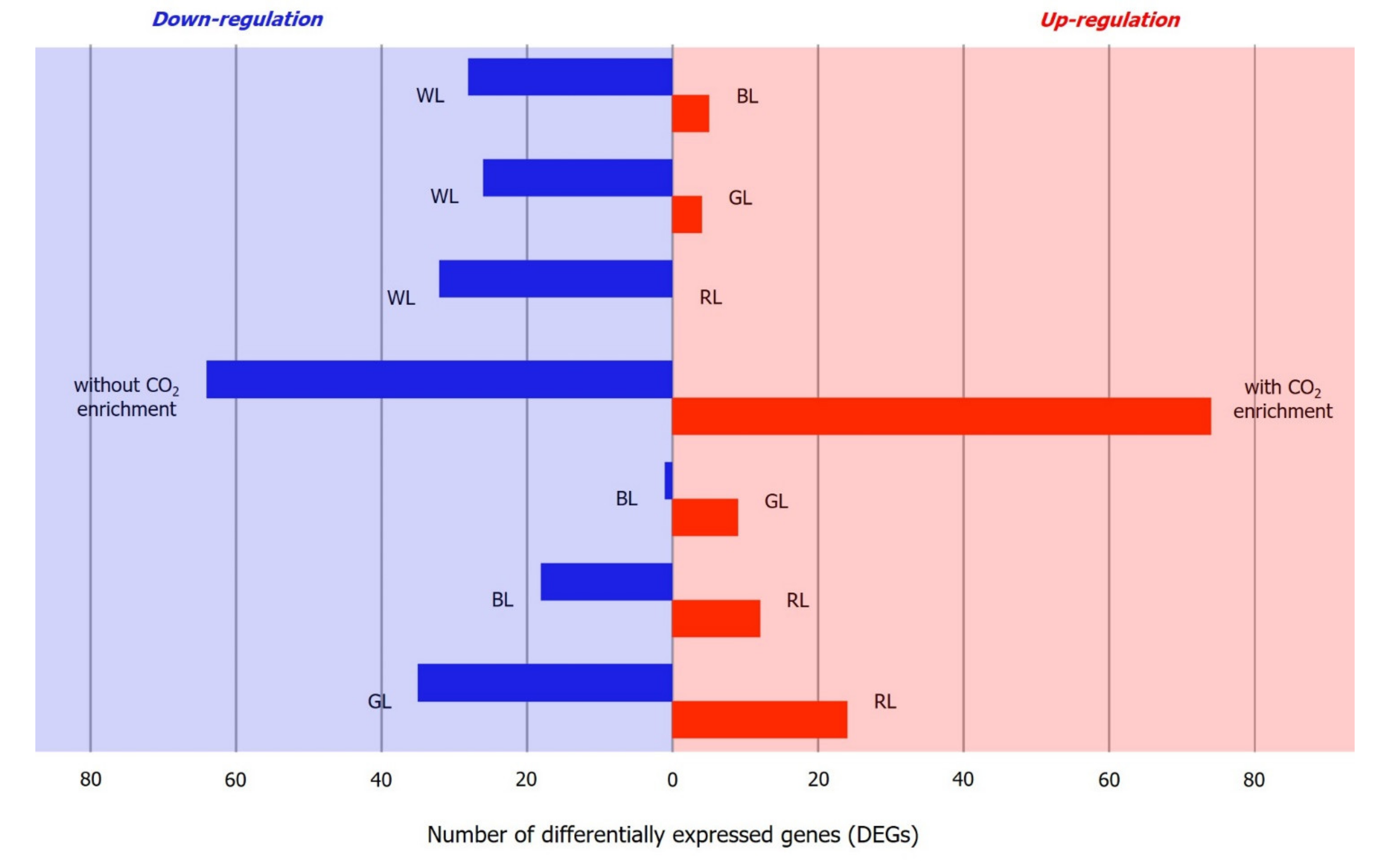
2.4. Identification of Differentially Expressed Genes (DEGs)
3. Discussion
4. Materials and Methods
4.1. Seaweed Materials
4.2. Culture Conditions
4.3. RNA Isolation
4.4. Preparation of cDNA Library
4.5. Quality Control and De Novo Transcriptome Assembly
4.6. Transcriptome Analysis
4.7. Identification of DEGs
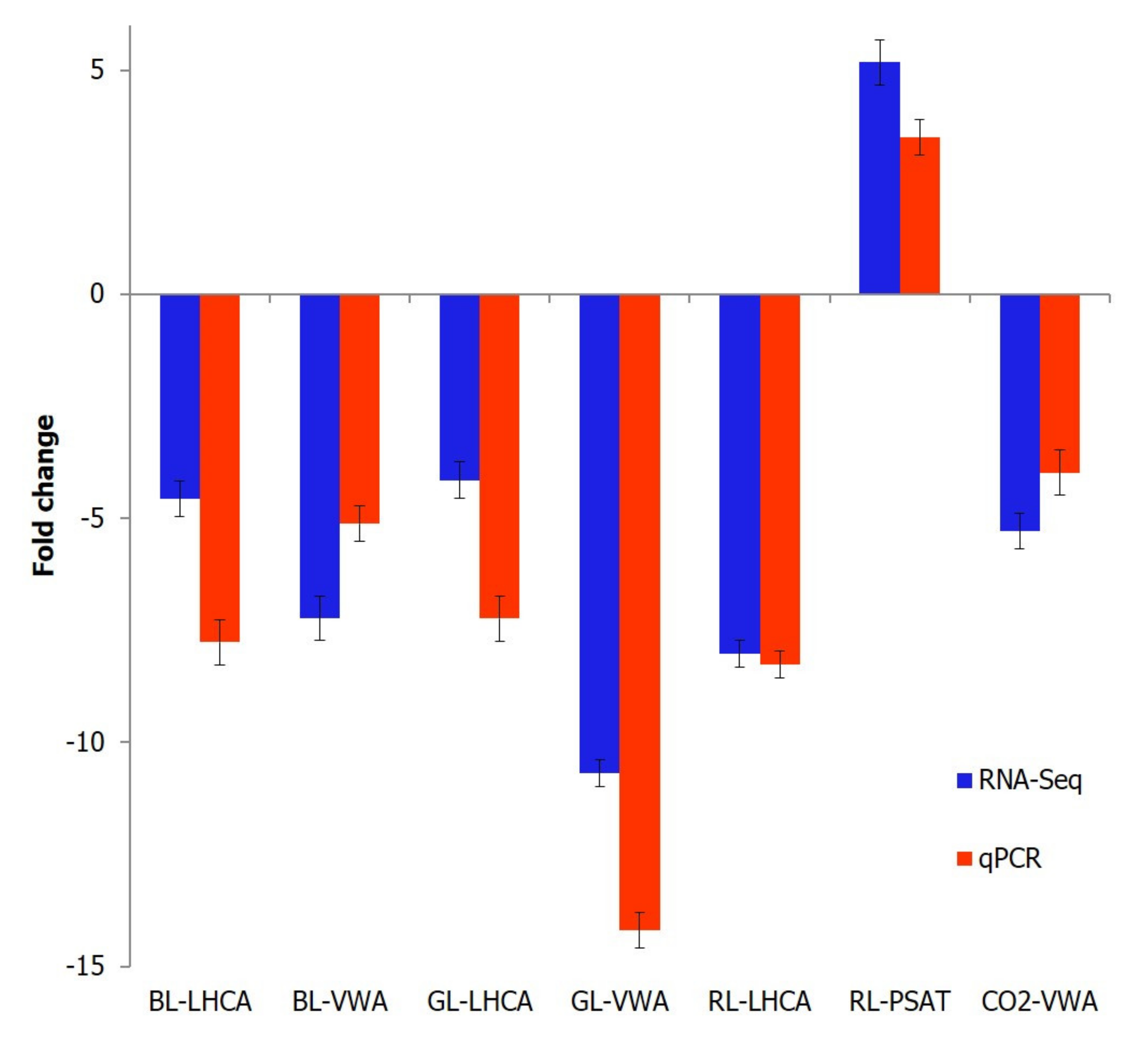
4.8. Quantitative Real-Time PCR Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bindu, M.S.; Levine, I.A. The commercial red seaweed Kappaphycus alvarezii—An overview on farming and environment. J. Appl. Phycol. 2011, 23, 789–796. [Google Scholar] [CrossRef]
- Eggertsen, M.; Halling, C. Knowledge gaps and management recommendations for future paths of sustainable seaweed farming in the Western Indian Ocean. Ambio 2020, in press. [Google Scholar] [CrossRef]
- Dring, M.J. Chromatic adaptation of photosynthesis in benthic marine algae: An examination of its ecological significance using a theoretical model. Limnol. Oceanogr. 1981, 26, 271–284. [Google Scholar] [CrossRef]
- Lobban, C.S.; Harrison, P.J. Seaweed Ecology and Physiology; Cambridge University Press: Cambridge, UK, 1994. [Google Scholar]
- Sagert, S.; Forster, R.M.; Feuerpfeil, P.; Schubert, H. Daily course of photosynthesis and photoinhibition in Chondrus crispus (Rhodophyta) from different shore levels. Eur. J. Phycol. 1997, 32, 363–371. [Google Scholar] [CrossRef][Green Version]
- Deng, Y.; Yao, J.; Wang, X.; Guo, H.; Duan, D. Transcriptome sequencing and comparative analysis of Saccharina japonica (Laminariales, Phaeophyceae) under blue light induction. PLoS ONE 2017, 7, e39704. [Google Scholar] [CrossRef]
- Wang, W.J.; Wang, F.J.; Sun, X.T.; Liu, F.L.; Liang, Z.R. Comparison of transcriptome under red and blue light culture of Saccharina japonica (Phaeophyceae). Planta 2013, 237, 1123–1133. [Google Scholar] [CrossRef]
- Wu, S.; Sun, J.; Chi, S.; Wang, L.; Wang, X.; Liu, C.; Li, X.; Yin, J.; Liu, T.; Yu, J. Transcriptome sequencing of essential marine brown and red algal species in China and its significance in algal biology and phylogeny. Acta Oceanol. Sin. 2014, 33, 1–12. [Google Scholar] [CrossRef]
- Song, L.; Wu, S.; Sun, J.; Wang, L.; Liu, T.; Chi, S.; Liu, C.; Li, X.; Yin, J.; Wang, X.; et al. De novo sequencing and comparative analysis of three red algal species of Family Solieriaceae to discover putative genes associated with carrageenan biosynthesis. Acta Oceanol. Sin. 2014, 33, 45–53. [Google Scholar] [CrossRef]
- Zhang, Z.; Pang, T.; Li, Q.; Zhang, L.; Li, L.; Liu, J. Transcriptome sequencing and characterization for Kappaphycus alvarezii. Eur. J. Phycol. 2015, 50, 400–407. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, L.; Pang, T.; Liu, J. Comparative transcriptome profiling of Kappaphycus alvarezii (Rhodophyta, Gigartinales) in response to two extreme temperature treatments: An RNA-seq-based resource for photosynthesis research. Eur. J. Phycol. 2019, 54, 162–174. [Google Scholar] [CrossRef]
- Gowik, U.; Westhoff, P. The path from C3 to C4 photosynthesis. Plant Physiol. 2011, 155, 56–63. [Google Scholar] [CrossRef]
- Robinson, S.J.; Parkin, I.A.P. Differential SAGE analysis in Arabidopsis uncovers increased transcriptome complexity in response to low temperature. BMC Genom. 2008, 9, 434. [Google Scholar] [CrossRef]
- Ehleringer, J.R.; Cerling, T.E.; Helliker, B.R. C4 photosynthesis, atmospheric CO2, and climate. Oecologia 1997, 112, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Yamori, W.; Hikosaka, K.; Way, D.A. Temperature response of photosynthesis in C3, C4, and CAM plants: Temperature acclimation and temperature adaptation. Photosynth. Res. 2014, 119, 101–117. [Google Scholar] [CrossRef]
- Beer, S.; Israel, A. Photosynthesis of Ulva sp. III. O2 effects, carboxylase activities, and the CO2 incorporation pattern . Plant Physiol. 1986, 81, 937–938. [Google Scholar] [CrossRef]
- Bidwell, R.G.S.; McLachlan, J. Carbon nutrition of seaweeds: Photosynthesis, photorespiration and respiration. J. Exp. Mar. Biol. Ecol. 1985, 86, 15–46. [Google Scholar] [CrossRef]
- Levchenko, E.V. Carbon metabolism transitions during the development of marine macroalga Gracilaria verrucose. Russ. J. Plant Physiol. 2003, 50, 68–72. [Google Scholar] [CrossRef]
- Derelle, E.; Ferraz, C.; Rombauts, S.; Rouzé, P.; Worden, A.Z.; Robbens, S.; Partensky, F.; Degroeve, S.; Echeynié, S.; Cooke, R.; et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features . Proc. Natl. Acad. Sci. USA 2006, 103, 11647–11652. [Google Scholar] [CrossRef] [PubMed]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef]
- Keeley, J.E. Photosynthetic pathway diversity in a seasonal pool community. Funct. Ecol. 1999, 13, 106–118. [Google Scholar] [CrossRef]
- Xu, J.; Fan, X.; Zhang, X.; Xu, D.; Mou, S.; Cao, S.; Zheng, Z.; Miao, J.; Ye, N. Evidence of coexistence of C3 and C4 photosynthetic pathways in green-tide-forming alga, Ulva prolifera. PLoS ONE 2012, 7, e37438. [Google Scholar] [CrossRef]
- Figueroa, F.L.; Israel, A.; Neori, A.; Martínez, B.; Malta, E.J.; Put, A.; Inken, S.; Marquardt, R.; Abdala, R.; Korbee, N. Effect of nutrient supply on photosynthesis and pigmentation to short-term stress (UV radiation) in Gracilaria conferta (Rhodophyta). Mar. Pollut. Bull. 2010, 60, 1768–1778. [Google Scholar] [CrossRef] [PubMed]
- Mercado, J.M.; Javier, F.; Gordillo, L.; Niell, F.X.; Figueroa, F.L. Effects of different levels of CO2 on photosynthesis and cell components of the red alga Porphyra leucosticta. J. Appl. Phycol. 1999, 11, 455–461. [Google Scholar] [CrossRef]
- Nejrup, L.B.; Staehr, P.A.; Thomsen, M.S. Temperature- and light-dependent growth and metabolism of the invasive red algae Gracilaria vermiculophylla–a comparison with two native macroalgae. Eur. J. Phycol. 2013, 48, 295–308. [Google Scholar] [CrossRef]
- Aguilera, J.; Gordillo, F.J.L.; Karsten, U.; Figueroa, F.L.; Niell, F.X. Light quality effect on photosynthesis and efficiency of carbon assimilation in the red alga Porphyra leucosticta. J. Plant Physiol. 2000, 157, 86–92. [Google Scholar] [CrossRef]
- Su, H.N.; Xie, B.B.; Zhang, X.Y.; Zhou, B.C.; Zhang, Y.Z. The supramolecular architecture, function, and regulation of thylakoid membranes in red algae: An overview. Photosynth. Res. 2010, 106, 73–87. [Google Scholar] [CrossRef]
- Zubia, M.; Freile-Pelegrín, Y.; Robledo, D. Photosynthesis, pigment composition and antioxidant defences in the red alga Gracilariopsis tenuifrons (Gracilariales, Rhodophyta) under environmental stress. J. Appl. Phycol. 2014, 26, 2001–2010. [Google Scholar] [CrossRef]
- Tandeau de Marsac, N. Phycobiliproteins and phycobilisomes: The early observations. Photosynth. Res. 2003, 76, 193–205. [Google Scholar] [CrossRef]
- Grossman, A.R.; Schaefer, M.R.; Chiang, G.G.; Collier, J.L. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol. Rev. 1993, 57, 725–749. [Google Scholar] [CrossRef]
- Roleda, M.Y.; Hurd, C.L. Seaweed nutrient physiology: Application of concepts to aquaculture and bioremediation. Phycologia 2019, 58, 552–562. [Google Scholar] [CrossRef]
- Van der Loos, L.M.; Schmid, M.; Leal, P.P.; McGraw, C.M.; Britton, D.; Revill, A.T.; Virtue, P.; Nichols, P.D.; Hurd, C.L. Responses of macroalgae to CO2 enrichment cannot be inferred solely from their inorganic carbon uptake strategy. Ecol. Evol. 2019, 9, 125–140. [Google Scholar] [CrossRef]
- Zou, D.; Gao, K. Effects of elevated CO2 on the red seaweed Gracilaria lemaneiformis (Gigartinales, Rhodophyta) grown at different irradiance levels. Phycologia 2009, 48, 510–517. [Google Scholar] [CrossRef]
- Kaneko, T.; Tanaka, A.; Sato, S.; Kotani, H.; Sazuka, T.; Miyajima, N.; Sugiura, M.; Tabata, S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 1995, 2, 153–166. [Google Scholar] [CrossRef]
- Stress tolerance in intertidal seaweeds. J. Phycol. 1996, 32, 197–211. [CrossRef]
- Jong, L.W.; Thien, V.Y.; Yong, Y.S.; Rodrigues, K.F.; Yong, W.T.L. Micropropagation and protein profile analysis by SDS-PAGE of Gracilaria changii (Rhodophyta, Solieriaceae). Aquacult. Rep. 2015, 1, 10–14. [Google Scholar] [CrossRef]
- Tee, M.Z.; Yong, Y.S.; Rodrigues, K.F.; Yong, W.T.L. Growth rate analysis and protein identification of Kappaphycus alvarezii (Rhodophyta, Gigartinales) under pH induced stress culture. Aquacult. Rep. 2015, 2, 112–116. [Google Scholar] [CrossRef]
- Collén, J.; Porcel, B.; Carré, W.; Ball, S.G.; Chaparro, C.; Tonon, T.; Barbeyron, T.; Michel, G.; Noel, B.; Valentin, K.; et al. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc. Natl. Acad. Sci. USA 2013, 110, 5247–5252. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Jimenez, P.; Llorens, C.; Roig, F.J.; Robaina, R.R. Analysis of the transcriptome of the red seaweed Grateloupia imbricata with emphasis on reproductive potential. Mar. Drugs 2018, 16, 490. [Google Scholar] [CrossRef]
- Ho, C.L.; Lee, W.K.; Lim, E.L. Unraveling the nuclear and chloroplast genomes of an agar producing red macroalga, Gracilaria changii (Rhodophyta, Gracilariales). Genomics 2018, 110, 124–133. [Google Scholar] [CrossRef]
- Carney, L.T.; Lane, T.W. Parasites in algae mass culture. Front. Microbiol. 2014, 5, 278. [Google Scholar] [CrossRef]
- Solis, M.J.L.; Draeger, S.; dela Cruz, T.E.E. Marine-derived fungi from Kappaphycus alvarezii and K. striatum as potential causative agents of ice-ice disease in farmed seaweeds. Bot. Mar. 2010, 53, 587–594. [Google Scholar] [CrossRef]
- Gao, D.; Kong, F.; Sun, P.; Bi, G.; Mao, Y. Transcriptome-wide identification of optimal reference genes for expression analysis of Pyropia yezoensis responses to abiotic stress. BMC Genom. 2018, 19, 251. [Google Scholar] [CrossRef]
- Martins, P.K.; Mafra, V.; de Souza, W.R.; Ribeiro, A.P.; Vinecky, F.; Basso, M.F.; da Cunha, B.A.D.B.; Kobayashi, A.K.; Molinari, H.B.C. Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis. Sci. Rep. 2016, 6, 28348. [Google Scholar] [CrossRef]
- Wan, Q.; Chen, S.; Shan, Z.; Yang, Z.; Chen, L.; Zhang, C.; Yuan, S.; Hao, Q.; Zhang, X.; Qiu, D.; et al. Stability evaluation of reference genes for gene expression analysis by RT-qPCR in soybean under different conditions. PLoS ONE 2017, 12, e0189405. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Huang, A.; Xu, M.; Wang, C.; Jia, Z.; Wang, G.; Niu, J. Variation of expression levels of seven housekeeping genes at different life-history stages in Porphyra yezoensis. PLoS ONE 2013, 8, e60740. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, C.; Xu, Y.; Ji, D.; Xie, C. Validation of housekeeping genes as internal controls for studying the gene expression in Pyropia haitanensis (Bangiales, Rhodophyta) by quantitative real-time PCR. Acta Oceanol. Sin. 2014, 33, 152–159. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.; Shim, E.; Kim, G.H.; Han, J.W.; Zuccarello, G.C. Keeping house: Evaluation of housekeeping genes for real-time PCR in the red alga, Bostrychia moritziana (Florideophyceae). Algae 2016, 31, 167–174. [Google Scholar] [CrossRef]
- Komine, Y.; Kwong, L.; Anguera, M.C.; Schuster, G.; Stern, D.B. Polyadenylation of three classes of chloroplast RNA in Chlamydomonas reinhardtii. RNA 2000, 6, 598–607. [Google Scholar] [CrossRef]
- Nishimura, Y.; Kikis, E.A.; Zimmer, S.L.; Komine, Y.; Stern, D.B. Antisense transcript and RNA processing alterations suppress instability of polyadenylated mRNA in Chlamydomonas chloroplasts. Plant Cell 2004, 16, 2849–2869. [Google Scholar] [CrossRef]
- Simpson, C.L.; Stern, D.B. The treasure trove of algal chloroplast genome. Surprises in architecture and gene content, and their functional implications. Plant Physiol. 2002, 129, 957–966. [Google Scholar] [CrossRef]
- Záhonová, K.; Hadariová, L.; Vacula, R.; Yurchenko, V.; Eliáš, M.; Krajčovič, J.; Vesteg, M. A small portion of plastid transcripts is polyadenylated in the flagellate Euglena gracilis. FEBS Lett. 2014, 588, 783–788. [Google Scholar] [CrossRef]
- Lisitsky, I.; Klaff, P.; Schuster, G. Addition of poly(A)-rich sequences to endonuclease cleavage sites during the degradation of chloroplast mRNA. Proc. Natl. Acad. Sci. USA 1996, 93, 13398–13403. [Google Scholar] [CrossRef] [PubMed]
- Schuster, G.; Lisitsky, I.; Klaff, P. Polyadenylation and degradation of mRNA in the chloroplast. Plant Physiol. 1999, 120, 937–944. [Google Scholar] [CrossRef][Green Version]
- Shi, C.; Wang, S.; Xia, E.H.; Jiang, J.J.; Zeng, F.C.; Gao, L.Z. Full transcription of the chloroplast genome in photosynthetic eukaryotes. Sci. Rep. 2016, 6, 30135. [Google Scholar] [CrossRef]
- Gantt, E.; Grabowski, B.; Cunningham, F.X. Antenna systems of red algae: Phycobilisomes with photosystem II and chlorophyll complexes with photosystem. In Light-Harvesting Antennas in Photosynthesis, Advances in Photosynthesis and Respiration; Green, B.R., Parson, W.W., Eds.; Springer: Dordrecht, The Netherlands, 2003; Volume 13, pp. 307–322. [Google Scholar]
- Green, B.R.; Kühlbrandt, W. Sequence conservation of light-harvesting and stress-response proteins in relation to the three-dimensional molecular structure of LHCII. Photosynth. Res. 1995, 44, 139–148. [Google Scholar] [CrossRef]
- Tan, S.; Ducret, A.; Aebersold, R.; Gantt, E. Red algal LHC I genes have similarities with both Chl a/b- and a/c-binding proteins: A 21 kDa polypeptide encoded by LhcaR2 is one of the six LHC I polypeptides. Photosynth. Res. 1997, 53, 129–140. [Google Scholar] [CrossRef]
- Wolfe, G.R.; Cunningham, F.X.; Cunningham, F.X.; Durnford, D.; Green, B.R.; Gantt, E. Evidence for a common origin of chloroplasts with light-harvesting complexes of different pigmentation. Nature 1994, 367, 566–568. [Google Scholar] [CrossRef]
- Maxwell, D.P.; Laudenbach, D.E.; Huner, N.P.A. Redox regulation of light-harvesting complex II and cab mRNA abundance in Dunaliella salina. Plant Physiol. 1995, 109, 787–795. [Google Scholar] [CrossRef]
- Teramoto, H.; Nakamori, A.; Minagawa, J.; Ono, T. Light-intensity-dependent expression of Lhc gene family encoding light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Physiol. 2002, 130, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Tonkyn, J.C.; Deng, X.W.; Gruissem, W. Regulation of plastid gene expression during photooxidative stress. Plant Physiol. 1992, 99, 1406–1415. [Google Scholar] [CrossRef]
- Dittami, S.M.; Scornet, D.; Petit, J.L.; Ségurens, B.; Da Silva, C.; Corre, E.; Dondrup, M.; Glatting, K.H.; König, R.; Sterck, L.; et al. Global expression analysis of the brown alga Ectocarpus siliculosus (Phaeophyceae) reveals large-scale reprogramming of the transcriptome in response to abiotic stress . Genome Biol. 2009, 10, R66. [Google Scholar] [CrossRef]
- Pearson, G.A.; Hoarau, G.; Lago-Leston, A.; Coyer, J.A.; Kube, M.; Reinhardt, R.; Henckel, K.; Serrão, E.T.A.; Corre, E.; Olsen, J.L. An expressed sequence tag analysis of the intertidal brown seaweeds Fucus serratus (L.) and F. vesiculosus (L.) (Heterokontophyta, Phaeophyceae) in response to abiotic stressors. Mar. Biotechnol. 2010, 12, 195–213. [Google Scholar] [CrossRef] [PubMed]
- Sturm, S.; Engelken, J.; Gruber, A.; Vugrinec, S.; Kroth, P.G.; Adamska, I.; Lavaud, J. A novel type of light-harvesting antenna protein of red algal origin in algae with secondary plastids. BMC Evol. Biol. 2013, 13, 159. [Google Scholar] [CrossRef]
- Blankenship, R.E. Molecular Mechanisms of Photosynthesis, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2014. [Google Scholar]
- Thien, V.Y.; Rodrigues, K.F.; Wong, C.M.V.L.; Yong, W.T.L. Investigation of growth rate and phycocolloid content from Kappaphycus alvarezii (Rhodophyta, Solieriaceae) under different light conditions using vibrational spectroscopy. J. Appl. Biotechnol. 2016, 4, 2. [Google Scholar] [CrossRef]
- Franklin, L.A.; Kräbs, G.; Kuhlenkamp, R. Blue light and UV-A radiation control the synthesis of mycosporine-like amino acids in Chondrus crispus (Florideophyceae). J. Phycol. 2002, 37, 257–270. [Google Scholar] [CrossRef]
- Godínez-Ortega, J.L.; Snoeijs, P.; Robledo, D.; Freile-Pelegrín, Y.; Pedersén, M. Growth and pigment composition in the red alga Halymenia floresii cultured under different light qualities. J. Appl. Phycol. 2008, 20, 253–260. [Google Scholar] [CrossRef]
- López-Figueroa, F.; Niell, F.X. Effects of light quality of chlorophyll and biliprotein accumulation in seaweeds. Mar. Biol. 1990, 104, 321–327. [Google Scholar] [CrossRef]
- Talarico, L.; Cortese, A. Response of Audouinella saviana (Meneghini) Woelkerling (Nemaliales, Rhodophyta) cultures to monochromatic light . Hydrobiologia 1993, 260, 477–484. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Cunningham, F.X., Jr.; Dennenberg, R.J.; Mustardy, L.; Jursinic, P.A.; Gantt, E. Stoichiometry of photosystem I, photosystem II, and phycobilisomes in the red alga Porphyridium cruentum as a function of growth irradiance. Plant Physiol. 1989, 91, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Gantt, E. Pigmentation and photoacclimation. In Biology of the Red Algae; Cole, K.M., Sheath, R.G., Eds.; Cambridge University Press: Cambridge, UK, 1990; pp. 203–219. [Google Scholar]
- Ritz, M.; Thomas, J.C.; Spilar, A.; Etienne, A.L. Kinetics of photoacclimation in response to a shift to high light of the red alga Rhodella violacea adapted to low irradiance. Plant Physiol. 2000, 123, 1415–1426. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zucchi, M.R.; Necchi, O., Jr. Effects of temperature, irradiance and photoperiod on growth and pigment content in some freshwater red algae in culture. Phycol. Res. 2001, 49, 103–114. [Google Scholar] [CrossRef]
- Frost-Christensen, H.; Sand-Jensen, K. The quantum efficiency of photosynthesis in macroalgae and submerged angiosperms. Ecologic 1992, 91, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Danon, A.; Coll, N.S.; Apel, K. Cryptochrome-1-dependent execution of programmed cell death induced by singlet oxygen in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA 2006, 103, 17036–17041. [Google Scholar] [CrossRef]
- Lopez, L.; Carbone, F.; Bianco, L.; Giuliano, G.; Facella, P.; Perrotta, G. Tomato plants overexpressing cryptochrome 2 reveals altered expression of energy and stress-related gene products in response to diurnal cues. Plant Cell Environ. 2012, 35, 994–1012. [Google Scholar] [CrossRef] [PubMed]
- Folta, K.M.; Maruhnich, S.A. Green light: A signal to slow down or stop. J. Exp. Bot. 2007, 58, 3099–3111. [Google Scholar] [CrossRef] [PubMed]
- Karekar, M.D.; Joshi, G.V. Photosynthetic carbon metabolism in marine algae. Bot. Mar. 1973, 16, 216–220. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, X.; Ye, N.; Zheng, Z.; Mou, S.; Dong, M.; Xu, D.; Miao, J. Activities of principal photosynthetic enzymes in green macroalga Ulva linza: Functional implication of C4 pathway in CO2 assimilation. Sci. China Life Sci. 2013, 56, 571–580. [Google Scholar] [CrossRef]
- Weber, A.P.M.; von Caemmerer, S. Plastid transport and metabolism of C3 and C4 plants–comparative analysis and possible biotechnological exploitation. Curr. Opin. Plant Biol. 2010, 13, 257–265. [Google Scholar] [CrossRef]
- Bowman, J.L. Stomata: Active portals for flourishing on land. Curr. Biol. 2011, 21, R540–R541. [Google Scholar] [CrossRef]
- Raven, J.A. Selection pressures on stomatal evolution. New Phytol. 2002, 153, 371–386. [Google Scholar] [CrossRef]
- Moroney, J.V.; Bartlett, S.G.; Samuelsson, G. Carbonic anhydrases in plants and algae. Plant Cell Environ. 2001, 24, 141–153. [Google Scholar] [CrossRef]
- He, L.; Zhang, X.; Wang, G. Expression analysis of phosphoenolpyruvate carboxykinase in Porphyra haitanensis (Rhodophyta) sporophytes and gametophytes. Physiol. Res. 2013, 61, 172–179. [Google Scholar] [CrossRef]
- Yong, W.T.L.; Ting, S.H.; Yong, Y.S.; Thien, V.Y.; Wong, S.H.; Chin, W.L.; Rodrigues, K.F.; Anton, A. Optimization of culture conditions for the direct regeneration of Kappaphycus alvarezii (Rhodophyta, Solieriaceae). J. Appl. Phycol. 2014, 26, 1597–1606. [Google Scholar] [CrossRef]
- Kang, E.J.; Kim, K.Y. Effects of future climate conditions on photosynthesis and biochemical component of Ulva pertusa (Chlorophyta). Algae 2016, 31, 49–59. [Google Scholar] [CrossRef]
- Kawaroe, M.; Bengen, D.G.; Barat, W.O.B. Utilization of carbon dioxide (CO2) for the optimization of the growth of seaweed kappaphycus alvarezii. Omni-Akuatika 2012, 6, 78–90. [Google Scholar]
- Kawaroe, M.; Sunuddin, A.; Augustine, D.; Lestari, D. The effect of CO2 injection on macroalgae Gelidium latifolium biomass growth rate and carbohydrate content. Ilmu Kelaut. 2016, 21, 85–92. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualisation and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalisation method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Primer3. Available online: http://frodo.wi.mit.edu/primer3/ (accessed on 15 January 2019).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Pathway | Number of Transcripts | Pathway ID |
|---|---|---|
| Metabolic pathways | 814 | Ko01100 |
| Biosynthesis of secondary metabolites | 337 | Ko01110 |
| Biosynthesis of amino acids | 131 | Ko01230 |
| Carbon metabolism | 117 | Ko01200 |
| Ribosome | 111 | Ko03010 |
| Purine metabolism | 95 | Ko00230 |
| Oxidative phosphorylation | 94 | Ko00190 |
| Pyrimidine metabolism | 73 | Ko00240 |
| RNA transport | 70 | Ko03013 |
| Spliceosome | 65 | Ko03040 |
| Pathway | Enzyme | Number of Transcripts |
|---|---|---|
| C4-CCM | Carbonic anhydrase (CA) | 23 |
| Alpha-carbonic anhydrase (α-CA) | 2 | |
| Beta-carbonic anhydrase (β-CA) | 1 | |
| Gamma-carbonic anhydrase (γ-CA) | 3 | |
| Malate dehydrogenase (MDH) | 3 | |
| Malate dehydrogenase (MDH2) | 1 | |
| Malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+) | 7 | |
| Aspartate aminotransferase, cytoplasmic (GOT1) | 3 | |
| Aspartate aminotransferase, mitochondrial (GOT2) | 3 | |
| Pyruvate orthophosphate dikinase (PPDK) | 5 | |
| Phosphoenolpyruvate carboxylase (PEPC) | 6 | |
| Phosphoenolpyruvate carboxykinase (PEPCK) | 1 | |
| C3 | Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) | 10 |
| Transketolase | 10 | |
| Phosphoribulokinase (PRK) | 1 | |
| Phosphoglycerate kinase (PGK) | 6 | |
| Sedoheptulose-bisphosphatase (SBPase) | 2 | |
| Ribulose-bisphosphate carboxylase large chain (rbcL) | 3 | |
| Fructose-bisphosphate aldolase, class I (ALDO) | 13 | |
| Fructose-bisphosphate aldolase, class II (FBA) | 6 | |
| Ribulose-phosphate 3-epimerase (rpe) | 6 | |
| Triosephosphate isomerase (TIM) | 6 | |
| Ribose 5-phosphate isomerase A (rpiA) | 2 | |
| Fructose-1,6-bisphosphatase I (FBP) | 6 |
| Gene Name 1 | Forward Primer (5′-3′) | Reverse Primer (3′-5′) | Amplicon Length (bp) | Annealing Temperature (°C) |
|---|---|---|---|---|
| LHCA | GTGCAAACACGCGCACCAGAGATGG | CAGCTCCCTTTCAACGCACAACAGCG | 138 | 60 |
| VWA | AGACTGCGTTCCTATCACCGCCAGC | CCGGCAGCAACATTGGGTCATCAGC | 141 | 60 |
| PSAT | ATTCGGTCGAGAGTGCGTGCAAGGG | ACAGAGGCGCAGAAGAGAGGGTTGC | 195 | 60 |
| 18S | CTGCCTTCCTAGACGGACTG | CGAGCGGATTTAGAGATTGG | 159 | 60 |
| Condition 1 1 | Condition 2 1 | All transcripts (No Filters) | Significant DEG (p-adj < 1 × 10−10 & log2 Fold Change of 2) | ||||
|---|---|---|---|---|---|---|---|
| Total | Upregulation 2 | Downregulation 3 | Total | Upregulation 2 | Downregulation 3 | ||
| WL | BL | 40,472 | 11,621 | 28,851 | 33 | 5 | 28 |
| WL | GL | 49,189 | 21,293 | 27,896 | 30 | 4 | 26 |
| WL | RL | 40,340 | 11,570 | 28,770 | 32 | 0 | 32 |
| WL | CO2 | 41,831 | 12,211 | 29,620 | 138 | 74 | 64 |
| BL | GL | 31,042 | 20,000 | 11,042 | 10 | 9 | 1 |
| BL | RL | 21,571 | 10,331 | 11,240 | 30 | 12 | 18 |
| GL | RL | 31,673 | 11,427 | 20,246 | 59 | 24 | 35 |
| BL | WL + GL + RL | 52,342 | 40,459 | 11,883 | 14 | 11 | 3 |
| GL | WL + BL + RL | 52,342 | 30,861 | 21,481 | 332 | 25 | 307 |
| RL | WL + BL + GL | 52,342 | 40,323 | 12,019 | 49 | 31 | 18 |
| WL | BL + GL + RL | 52,342 | 21,730 | 30,612 | 582 | 4 | 578 |
| BL + GL | RL + WL | 52,342 | 29,643 | 22,699 | 2 | 2 | 0 |
| BL + RL | GL + WL | 52,342 | 38,999 | 13,343 | 0 | 0 | 0 |
| GL + RL | BL + WL | 52,342 | 31,097 | 20,435 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thien, V.Y.; Rodrigues, K.F.; Voo, C.L.Y.; Wong, C.M.V.L.; Yong, W.T.L. Comparative Transcriptome Profiling of Kappaphycus alvarezii (Rhodophyta, Solieriaceae) in Response to Light of Different Wavelengths and Carbon Dioxide Enrichment. Plants 2021, 10, 1236. https://doi.org/10.3390/plants10061236
Thien VY, Rodrigues KF, Voo CLY, Wong CMVL, Yong WTL. Comparative Transcriptome Profiling of Kappaphycus alvarezii (Rhodophyta, Solieriaceae) in Response to Light of Different Wavelengths and Carbon Dioxide Enrichment. Plants. 2021; 10(6):1236. https://doi.org/10.3390/plants10061236
Chicago/Turabian StyleThien, Vun Yee, Kenneth Francis Rodrigues, Christopher Lok Yung Voo, Clemente Michael Vui Ling Wong, and Wilson Thau Lym Yong. 2021. "Comparative Transcriptome Profiling of Kappaphycus alvarezii (Rhodophyta, Solieriaceae) in Response to Light of Different Wavelengths and Carbon Dioxide Enrichment" Plants 10, no. 6: 1236. https://doi.org/10.3390/plants10061236
APA StyleThien, V. Y., Rodrigues, K. F., Voo, C. L. Y., Wong, C. M. V. L., & Yong, W. T. L. (2021). Comparative Transcriptome Profiling of Kappaphycus alvarezii (Rhodophyta, Solieriaceae) in Response to Light of Different Wavelengths and Carbon Dioxide Enrichment. Plants, 10(6), 1236. https://doi.org/10.3390/plants10061236







