How to Unravel the Key Functions of Cryptic Oomycete Elicitin Proteins and Their Role in Plant Disease
Abstract
1. Introduction
2. Distribution of Elicitins within the Oomycetes
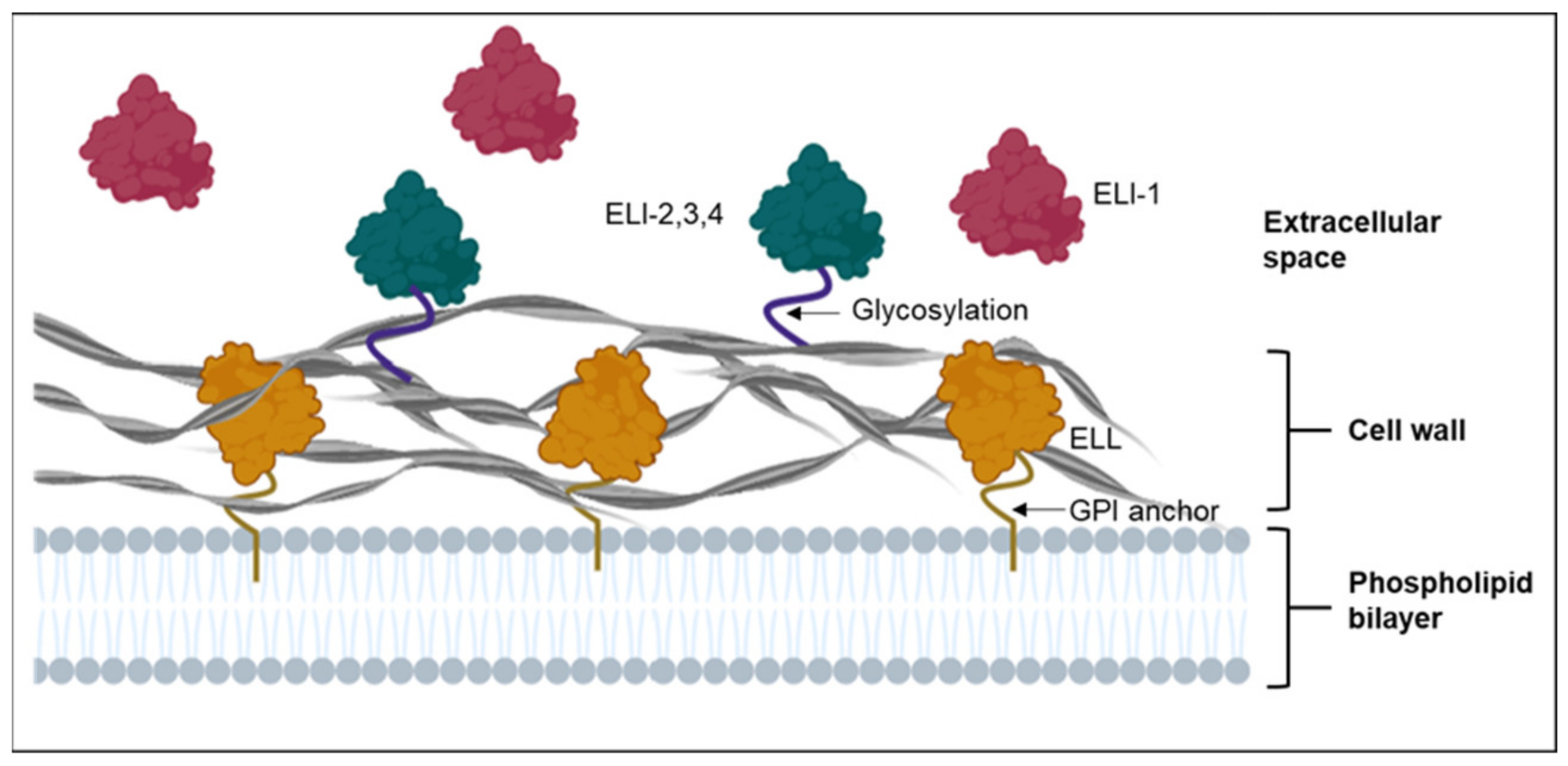
3. Cell Wall-Bound Elicitins and Elicitin-Like Proteins
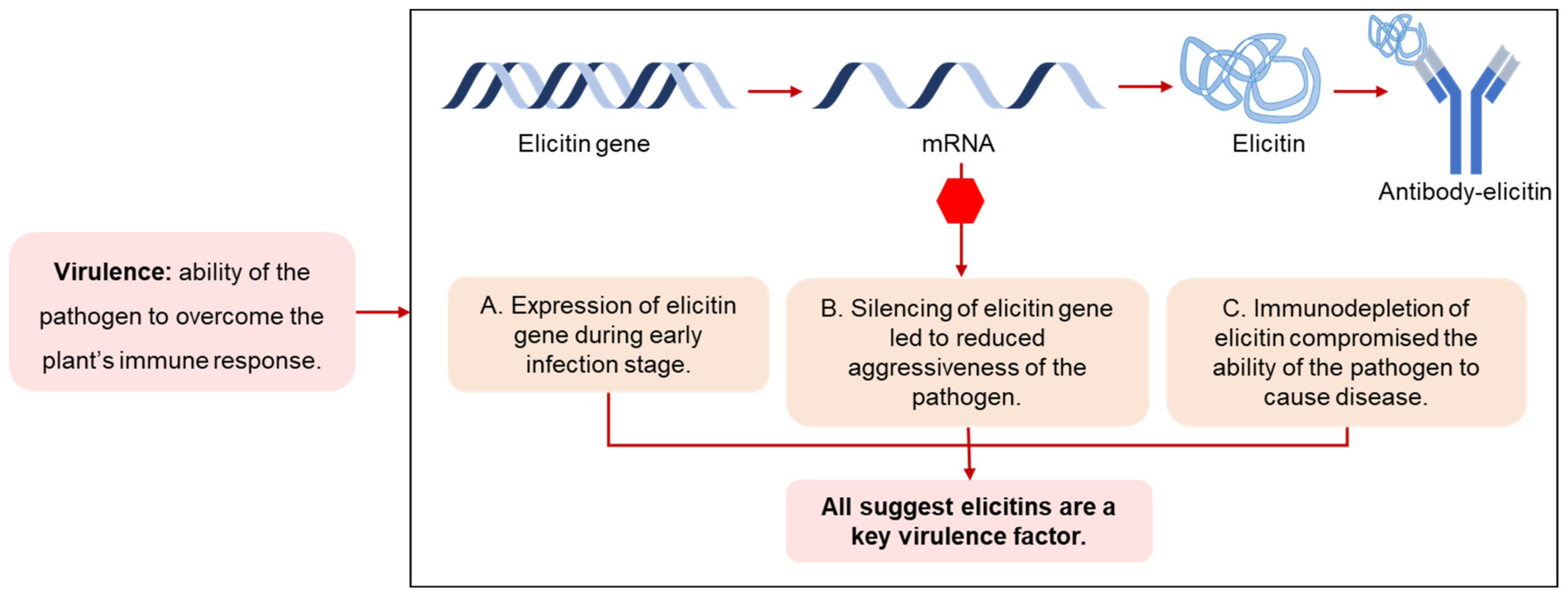
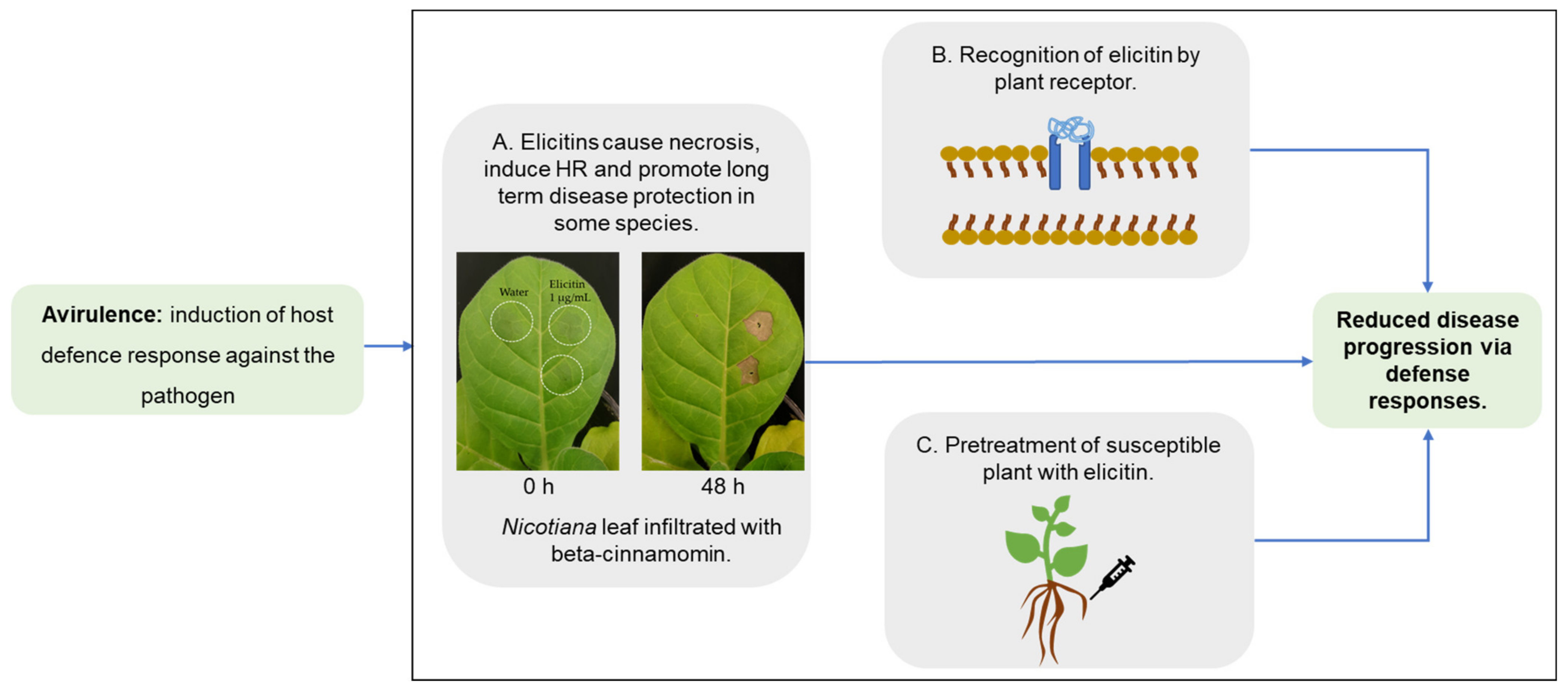
4. Elicitins as Virulence or Avirulence Factors?
5. Application of Modern Tools and Techniques for Functional Analysis of Elicitins
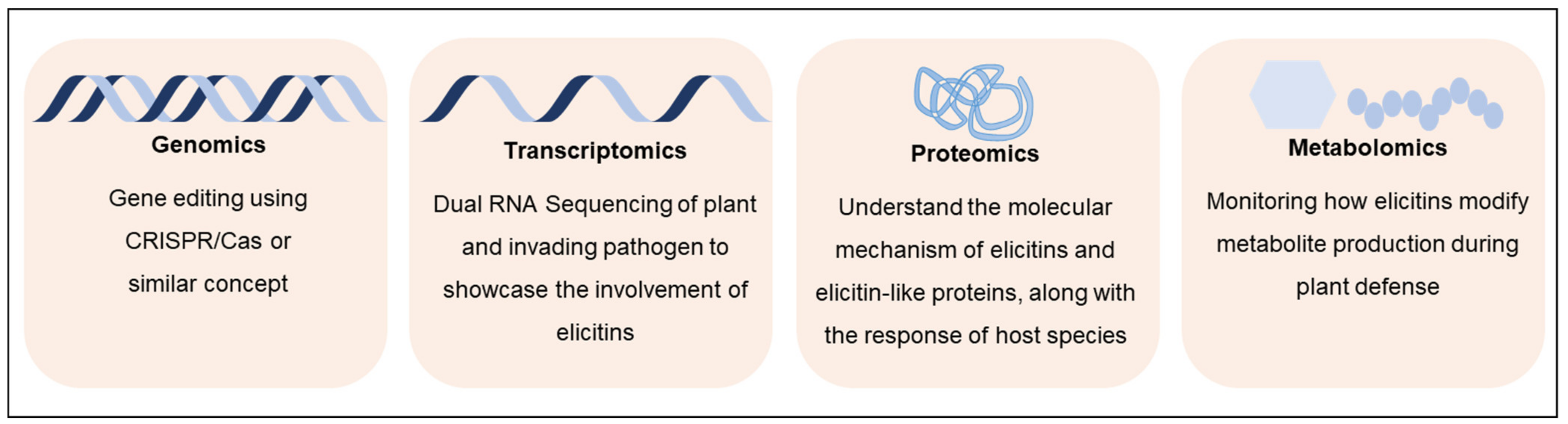
5.1. Omics Approaches for Defining Functional Roles of Elicitins
5.1.1. Genomics and Next-Generation Sequencing
5.1.2. Transcriptomics
5.1.3. Proteomic Approaches
5.1.4. Metabolomics and Cell Signaling
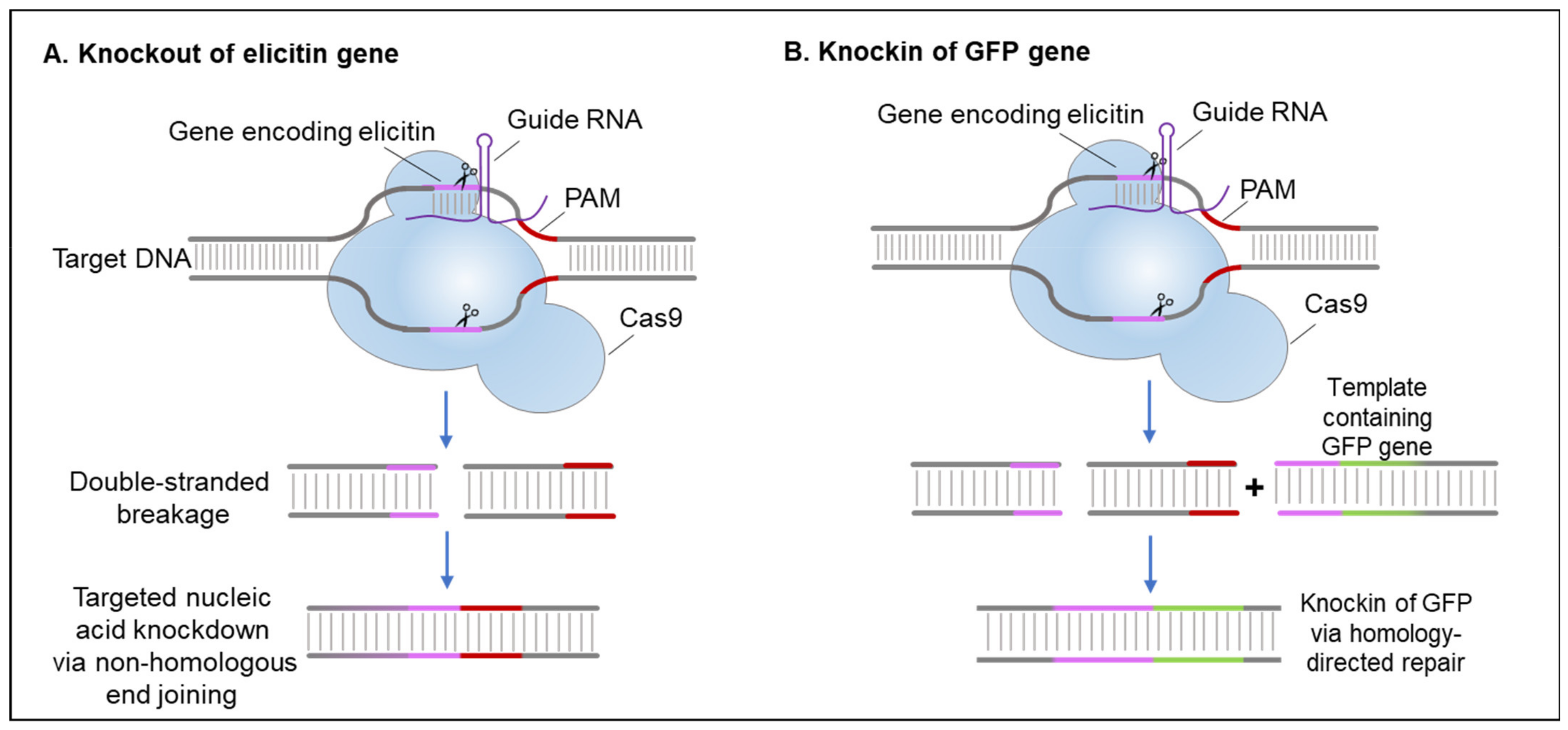
5.2. Gene Editing Using CRISPR/Cas
6. Association of Elicitins with Other Organisms
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Derevnina, L.; Dagdas, Y.F.; De la Concepcion, J.C.; Bialas, A.; Kellner, R.; Petre, B.; Domazakis, E.; Du, J.; Wu, C.H.; Lin, X. Nine things to know about elicitins. New Phytol. 2016, 212, 888–895. [Google Scholar] [CrossRef]
- Ricci, P.; Bonnet, P.; Huet, J.C.; Sallantin, M.; Beauvais-Cante, F.; Bruneteau, M.; Billard, V.; Michel, G.; Pernollet, J.C. Structure and activity of proteins from pathogenic fungi Phytophthora eliciting necrosis and acquired resistance in tobacco. Eur. J. Biochem. 1989, 183, 555–563. [Google Scholar] [CrossRef]
- Uhlikova, H.; Solansky, M.; Hrdinova, V.; Sedo, O.; Kasparovsky, T.; Hejatko, J.; Lochman, J. MAMP (microbe-associated molecular pattern)-induced changes in plasma membrane-associated proteins. J. Plant Physiol. 2017, 210, 51–57. [Google Scholar] [CrossRef]
- Mikes, V.; Milat, M.-L.; Ponchet, M.; Ricci, P.; Blein, J.-P. The fungal elicitor cryptogein is a sterol carrier protein. FEBS Lett. 1998, 416, 190–192. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; Irshad, M.K.; Qari, S.H.; Hashem, M.; Alamri, S.; AbdulMajeed, A.M.; Al-Sadi, A.M. Elicitins as molecular weapons against pathogens: Consolidated biotechnological strategy for enhancing plant growth. Crit. Rev. Biotechnol. 2020, 40, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.M. Elicitins from Phytophthora and basic resistance in tobacco. Proc. Natl. Acad. Sci. USA 1995, 92, 4088–4094. [Google Scholar] [CrossRef] [PubMed]
- Janků, M.; Činčalová, L.; Luhová, L.; Lochman, J.; Petřivalský, M. Biological effects of oomycetes elicitins. Plant Prot. Sci. 2019, 56, 1–8. [Google Scholar] [CrossRef]
- Boevink, P.C.; Birch, P.R.; Turnbull, D.; Whisson, S.C. Devastating intimacy: The cell biology of plant—Phytophthora interactions. New Phytol. 2020, 228, 445–458. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Buscaill, P.; van der Hoorn, R.A. Defeated by the nines: Nine extracellular strategies to avoid MAMP recognition in plants. Plant Cell 2021, 109. [Google Scholar] [CrossRef] [PubMed]
- Crandall, S.G.; Gold, K.M.; Jiménez-Gasco, M.d.M.; Filgueiras, C.C.; Willett, D.S. A multi-omics approach to solving problems in plant disease ecology. PLoS ONE 2020, 15, e0237975. [Google Scholar] [CrossRef]
- Khan, M.M.; Ernst, O.; Manes, N.P.; Oyler, B.L.; Fraser, I.D.; Goodlett, D.R.; Nita-Lazar, A. Multi-omics strategies uncover host–pathogen interactions. ACS Infect. Dis. 2019, 5, 493–505. [Google Scholar] [CrossRef]
- Pramanik, D.; Shelake, R.M.; Kim, M.J.; Kim, J.-Y. CRISPR-mediated engineering across the central dogma in plant biology for basic research and crop improvement. Mol. Plant 2020, 14, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Gosavi, G.; Yan, F.; Ren, B.; Kuang, Y.; Yan, D.; Zhou, X.; Zhou, H. Applications of CRISPR technology in studying plant-pathogen interactions: Overview and perspective. Phytopathol. Res. 2020, 2, 1–9. [Google Scholar] [CrossRef]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef]
- Arya, S.S.; Rookes, J.; Cahill, D.; Lenka, S.K. Next-generation metabolic engineering approaches towards development of plant cell suspension cultures as specialized metabolite producing biofactories. Biotechnol. Adv. 2020, 45, 107635. [Google Scholar] [CrossRef] [PubMed]
- Rönspies, M.; Dorn, A.; Schindele, P.; Puchta, H. CRISPR–Cas-mediated chromosome engineering for crop improvement and synthetic biology. Nat. Plants 2021, 7, 566–573. [Google Scholar] [CrossRef]
- Beakes, G.W.; Glockling, S.L.; Sekimoto, S. The evolutionary phylogeny of the oomycete “fungi”. Protoplasma 2012, 249, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Lerksuthirat, T.; Lohnoo, T.; Inkomlue, R.; Rujirawat, T.; Yingyong, W.; Khositnithikul, R.; Phaonakrop, N.; Roytrakul, S.; Sullivan, T.D.; Krajaejun, T. The elicitin-like glycoprotein, ELI025, is secreted by the pathogenic oomycete Pythium insidiosum and evades host antibody responses. PLoS ONE 2015, 10, e0118547. [Google Scholar] [CrossRef]
- Jiang, R.H.; Tyler, B.M.; Whisson, S.C.; Hardham, A.R.; Govers, F. Ancient origin of elicitin gene clusters in Phytophthora genomes. Mol. Biol. Evol. 2006, 23, 338–351. [Google Scholar] [CrossRef]
- Grenville-Briggs, L.J.; Avrova, A.O.; Hay, R.J.; Bruce, C.R.; Whisson, S.C.; Van West, P. Identification of appressorial and mycelial cell wall proteins and a survey of the membrane proteome of Phytophthora infestans. Fungal Biol. 2010, 114, 702–723. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, S.; Lindqvist, H.; Govers, F. A novel class of elicitin-like genes from Phytophthora infestans. Mol. Plant-Microbe Interact. 1997, 10, 1028–1030. [Google Scholar] [CrossRef] [PubMed]
- Huitema, E.; Vleeshouwers, V.G.; Cakir, C.; Kamoun, S.; Govers, F. Differences in intensity and specificity of hypersensitive response induction in Nicotiana spp. by INF1, INF2A, and INF2B of Phytophthora infestans. Mol. Plant-Microbe Interact. 2005, 18, 183–193. [Google Scholar] [CrossRef]
- Islam, M.; Hussain, H.; Russo, R.; Chambery, A.; Amoresano, A.; Schallmey, A.; Oßwald, W.; Nadiminti, P.; Cahill, D. Functional analysis of elicitins and identification of cell wall proteins in Phytophthora cinnamomi. Physiol. Mol. Plant Pathol. 2019, 107, 21–32. [Google Scholar] [CrossRef]
- Kunjeti, S.G.; Evans, T.A.; Marsh, A.G.; Gregory, N.F.; Kunjeti, S.; Meyers, B.C.; Kalavacharla, V.S.; Donofrio, N.M. RNA-Seq reveals infection-related global gene changes in Phytophthora phaseoli, the causal agent of lima bean downy mildew. Mol. Plant Pathol. 2012, 13, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, S.; Huitema, E.; Vleeshouwers, V.G. Resistance to oomycetes: A general role for the hypersensitive response? Trends Plant Sci. 1999, 4, 196–200. [Google Scholar] [CrossRef]
- Horta, M.; Sousa, N.; Coelho, A.C.; Neves, D.; Cravador, A. In vitro and in vivo quantification of elicitin expression in Phytophthora cinnamomi. Physiol. Mol. Plant Pathol. 2008, 73, 48–57. [Google Scholar] [CrossRef]
- Dalio, R.; Fleischmann, F.; Chambery, A.; Eichmann, R.; Massola, N., Jr.; Pascholati, S.; Osswald, W. Immunodepletion of α-plurivorin effector leads to loss of virulence of Phytophthora plurivora towards Fagus sylvatica. Forest Pathol. 2017, 47, e12362. [Google Scholar] [CrossRef]
- Du, J.; Verzaux, E.; Chaparro-Garcia, A.; Bijsterbosch, G.; Keizer, L.C.; Zhou, J.; Liebrand, T.W.; Xie, C.; Govers, F.; Robatzek, S.; et al. Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants 2015, 1, 15034. [Google Scholar] [CrossRef] [PubMed]
- Ebadzad, G.; Medeira, C.; Maia, I.; Martins, J.; Cravador, A. Induction of defence responses by cinnamomins against Phytophthora cinnamomi in Quercus suber and Quercus ilex subs. rotundifolia. Eur. J. Plant Pathol. 2015, 143, 705–723. [Google Scholar] [CrossRef]
- Churngchow, N.; Rattarasarn, M. The elicitin secreted by Phytophthora palmivora, a rubber tree pathogen. Phytochemistry 2000, 54, 33–38. [Google Scholar] [CrossRef]
- Evangelisti, E.; Gogleva, A.; Hainaux, T.; Doumane, M.; Tulin, F.; Quan, C.; Yunusov, T.; Floch, K.; Schornack, S. Time-resolved dual transcriptomics reveal early induced Nicotiana benthamiana root genes and conserved infection-promoting Phytophthora palmivora effectors. BMC Biol. 2017, 15, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Thomma, B.P.; Nürnberger, T.; Joosten, M.H. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Naveed, Z.A.; Wei, X.; Chen, J.; Mubeen, H.; Ali, G.S. The PTI to ETI Continuum in Phytophthora-Plant Interactions. Front. Plant Sci. 2020, 11, 2030. [Google Scholar] [CrossRef]
- Lherminier, J.; Benhamou, N.; Larrue, J.; Milat, M.-L.; Boudon-Padieu, E.; Nicole, M.; Blein, J.-P. Cytological characterization of elicitin-induced protection in tobacco plants infected by Phytophthora parasitica or phytoplasma. Phytopathology 2003, 93, 1308–1319. [Google Scholar] [CrossRef]
- Félix, C.; Meneses, R.; Gonçalves, M.F.; Tilleman, L.; Duarte, A.S.; Jorrín-Novo, J.V.; Van de Peer, Y.; Deforce, D.; Van Nieuwerburgh, F.; Esteves, A.C. A multi-omics analysis of the grapevine pathogen Lasiodiplodia theobromae reveals that temperature affects the expression of virulence-and pathogenicity-related genes. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Neik, T.X.; Amas, J.; Barbetti, M.; Edwards, D.; Batley, J. Understanding host–pathogen interactions in Brassica napus in the omics era. Plants 2020, 9, 1336. [Google Scholar] [CrossRef] [PubMed]
- Arafa, R.A.; Shirasawa, K. Technical review of molecular markers and next-generation sequencing technology to manage plant pathogenic oomycetes. Afr. J. Biotechnol. 2018, 17, 369–379. [Google Scholar]
- Badet, T.; Croll, D. The rise and fall of genes: Origins and functions of plant pathogen pangenomes. Curr. Opin. Plant Biol. 2020, 56, 65–73. [Google Scholar] [CrossRef]
- Bayer, P.E.; Golicz, A.A.; Scheben, A.; Batley, J.; Edwards, D. Plant pan-genomes are the new reference. Nat. Plants 2020, 6, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Reitmann, A.; Berger, D.K.; Van den Berg, N. Putative pathogenicity genes of Phytophthora cinnamomi identified via RNA-Seq analysis of pre-infection structures. Eur. J. Plant Pathol. 2017, 147, 211–228. [Google Scholar] [CrossRef]
- Judelson, H.S.; Ah-Fong, A.M.; Aux, G.; Avrova, A.O.; Bruce, C.; Cakir, C.; da Cunha, L.; Grenville-Briggs, L.; Latijnhouwers, M.; Ligterink, W. Gene expression profiling during asexual development of the late blight pathogen Phytophthora infestans reveals a highly dynamic transcriptome. Mol. Plant-Microbe Interact. 2008, 21, 433–447. [Google Scholar] [CrossRef]
- Sun, J.; Gao, Z.; Zhang, X.; Zou, X.; Cao, L.; Wang, J. Transcriptome analysis of Phytophthora litchii reveals pathogenicity arsenals and confirms taxonomic status. PLoS ONE 2017, 12, e0178245. [Google Scholar] [CrossRef] [PubMed]
- Toljamo, A.; Blande, D.; Munawar, M.; Kärenlampi, S.O.; Kokko, H. Expression of the GAF Sensor, Carbohydrate-Active Enzymes, Elicitins, and RXLRs Differs Markedly Between Two Phytophthora cactorum Isolates. Phytopathology 2019, 109, 726–735. [Google Scholar] [CrossRef]
- Rustagi, A.; Singh, G.; Agrawal, S.; Gupta, P.K. Proteomic studies revealing enigma of plant–pathogen interaction. In Molecular Aspects of Plant-Pathogen Interaction; Springer: Singapore, 2018; pp. 239–264. [Google Scholar]
- Ashwin, N.; Barnabas, L.; Sundar, A.R.; Malathi, P.; Viswanathan, R.; Masi, A.; Agrawal, G.K.; Rakwal, R. Advances in proteomic technologies and their scope of application in understanding plant–pathogen interactions. J. Plant Biochem. Biotechnol. 2017, 26, 371–386. [Google Scholar] [CrossRef]
- Rauwane, M.E.; Ogugua, U.V.; Kalu, C.M.; Ledwaba, L.K.; Woldesemayat, A.A.; Ntushelo, K. Pathogenicity and virulence factors of Fusarium graminearum including factors discovered using next generation sequencing technologies and proteomics. Microorganisms 2020, 8, 305. [Google Scholar] [CrossRef] [PubMed]
- Kan, Y.; Lyu, Q.; Jiang, N.; Han, S.; Li, J.; Burdman, S.; Luo, L. iTRAQ-based proteomic analyses of the plant-pathogenic bacterium Acidovorax citrulli during entrance into and resuscitation from the viable but nonculturable state. J. Proteom. 2020, 211, 103547. [Google Scholar] [CrossRef]
- Guo, S.; Wang, D.; Ma, Y.; Zhang, Y.; Zhao, X. Combination of RNA-Seq transcriptomics and iTRAQ proteomics reveal the mechanism involved in fresh-cut yam yellowing. Sci. Rep. 2021, 11, 1–16. [Google Scholar] [CrossRef]
- Du, J.; Guo, X.; Chen, L.; Xie, C.; Liu, J. Proteomic analysis of differentially expressed proteins of Nicotiana benthamiana triggered by INF1 elicitin from Phytophthora infestans. J. Gen. Plant Pathol. 2017, 83, 66–77. [Google Scholar] [CrossRef]
- Coelho, A.C.; Pires, R.; Schütz, G.; Santa, C.; Manadas, B.; Pinto, P. Disclosing proteins in the leaves of cork oak plants associated with the immune response to Phytophthora cinnamomi inoculation in the roots: A long-term proteomics approach. PLoS ONE 2021, 16, e0245148. [Google Scholar] [CrossRef]
- Hoehenwarter, W.; Chen, Y.; Recuenco-Munoz, L.; Wienkoop, S.; Weckwerth, W. Functional analysis of proteins and protein species using shotgun proteomics and linear mathematics. Amino Acids 2011, 41, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.L.; Youssef, N.H.; Hartson, S.; Elshahed, M.S. The extraradical proteins of Rhizophagus irregularis: A shotgun proteomics approach. Fungal Biol. 2020, 124, 91–101. [Google Scholar] [CrossRef]
- Marshall, D.D.; Powers, R. Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 100, 1–16. [Google Scholar] [CrossRef]
- Castro-Moretti, F.R.; Gentzel, I.N.; Mackey, D.; Alonso, A.P. Metabolomics as an emerging tool for the study of plant–pathogen interactions. Metabolites 2020, 10, 52. [Google Scholar] [CrossRef] [PubMed]
- Camejo, D.; Guzmán-Cedeño, A.; Vera-Macias, L.; Jiménez, A. Oxidative post-translational modifications controlling plant-pathogen interaction. Plant Physiol. Biochem. 2019, 144, 110–117. [Google Scholar] [CrossRef]
- Nejat, N.; Rookes, J.; Mantri, N.L.; Cahill, D.M. Plant–pathogen interactions: Toward development of next-generation disease-resistant plants. Crit. Rev. Biotechnol. 2017, 37, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Cui, L.; Gu, B.; Arredondo, F.; Tyler, B.M. Efficient genome editing in the oomycete Phytophthora sojae using CRISPR/Cas9. Curr. Protoc. Microbiol. 2017, 44, 21A.21.21–21A.21.26. [Google Scholar] [CrossRef] [PubMed]
- Pettongkhao, S.; Navet, N.; Schornack, S.; Tian, M.; Churngchow, N. A secreted protein of 15 kDa plays an important role in Phytophthora palmivora development and pathogenicity. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Fang, Y.; Tyler, B.M. Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 2016, 17, 127–139. [Google Scholar] [CrossRef]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.W.; Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef]
- Davidson, M.W.; Campbell, R.E. Engineered fluorescent proteins: Innovations and applications. Nat. Methods 2009, 6, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Ah-Fong, A.M.; Boyd, A.M.; Matson, M.E.; Judelson, H.S. A Cas12a-based gene editing system for Phytophthora infestans reveals monoallelic expression of an elicitor. Mol. Plant Pathol. 2021, 22, 737–752. [Google Scholar] [CrossRef]
- Miao, J.; Chi, Y.; Lin, D.; Tyler, B.M.; Liu, X. Mutations in ORP1 conferring oxathiapiprolin resistance confirmed by genome editing using CRISPR/Cas9 in Phytophthora capsici and P. sojae. Phytopathology 2018, 108, 1412–1419. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, I.V.; Sarrocco, S.; Malfatti, L.; Baroncelli, R.; Vannacci, G. CRISPR-Cas for fungal genome editing: A new tool for the management of plant diseases. Front. Plant Sci. 2019, 10, 135. [Google Scholar] [CrossRef] [PubMed]
- Brooks, S.M.; Alper, H.S. Applications, challenges, and needs for employing synthetic biology beyond the lab. Nature Comm. 2021, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.K.; Akter, T.; Islam, T. Gene editing in filamentous fungi and oomycetes using CRISPR-Cas technology. In CRISPR and RNAi Systems; Elsevier: Amsterdam, The Netherlands, 2021; pp. 723–753. [Google Scholar]
- Anutarapongpan, O.; Thanathanee, O.; Worrawitchawong, J.; Suwan-apichon, O. Role of confocal microscopy in the diagnosis of Pythium insidiosum keratitis. Cornea 2018, 37, 156–161. [Google Scholar] [CrossRef]
- Badenoch, P.R.; Mills, R.A.; Chang, J.H.; Sadlon, T.A.; Klebe, S.; Coster, D.J. Pythium insidiosum keratitis in an Australian child. Clin. Exp. Opthalmol. 2009, 37, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Krajaejun, T.; Sathapatayavongs, B.; Pracharktam, R.; Nitiyanant, P.; Leelachaikul, P.; Wanachiwanawin, W.; Chaiprasert, A.; Assanasen, P.; Saipetch, M.; Mootsikapun, P. Clinical and epidemiological analyses of human pythiosis in Thailand. Clin. Infect. Dis. 2006, 43, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Inkomlue, R.; Larbcharoensub, N.; Karnsombut, P.; Lerksuthirat, T.; Aroonroch, R.; Lohnoo, T.; Yingyong, W.; Santanirand, P.; Sansopha, L.; Krajaejun, T. Development of an anti-elicitin antibody-based immunohistochemical assay for diagnosis of pythiosis. J. Clin. Microbiol. 2016, 54, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Iberahim, N.A.; Trusch, F.; Van West, P. Aphanomyces invadans, the causal agent of epizootic ulcerative syndrome, is a global threat to wild and farmed fish. Fungal Biol. Rev. 2018, 32, 118–130. [Google Scholar] [CrossRef]
- Shen, D.; Tang, Z.; Wang, C.; Wang, J.; Dong, Y.; Chen, Y.; Wei, Y.; Cheng, B.; Zhang, M.; Grenville-Briggs, L.J. Infection mechanisms and putative effector repertoire of the mosquito pathogenic oomycete Pythium guiyangense uncovered by genomic analysis. PLoS Genet. 2019, 15, e1008116. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kharel, A.; Islam, M.T.; Rookes, J.; Cahill, D. How to Unravel the Key Functions of Cryptic Oomycete Elicitin Proteins and Their Role in Plant Disease. Plants 2021, 10, 1201. https://doi.org/10.3390/plants10061201
Kharel A, Islam MT, Rookes J, Cahill D. How to Unravel the Key Functions of Cryptic Oomycete Elicitin Proteins and Their Role in Plant Disease. Plants. 2021; 10(6):1201. https://doi.org/10.3390/plants10061201
Chicago/Turabian StyleKharel, Aayushree, Md Tohidul Islam, James Rookes, and David Cahill. 2021. "How to Unravel the Key Functions of Cryptic Oomycete Elicitin Proteins and Their Role in Plant Disease" Plants 10, no. 6: 1201. https://doi.org/10.3390/plants10061201
APA StyleKharel, A., Islam, M. T., Rookes, J., & Cahill, D. (2021). How to Unravel the Key Functions of Cryptic Oomycete Elicitin Proteins and Their Role in Plant Disease. Plants, 10(6), 1201. https://doi.org/10.3390/plants10061201






