The Effect of Chromosome Arm 1BS on the Fertility of Alloplasmic Recombinant Lines in Bread Wheat with the Hordeum vulgare Cytoplasm
Abstract
1. Introduction
2. Materials and Methods
2.1. Alloplasmic Recombinant Lines (H. vulgare)-T. aestivum
2.2. Control Genotypes
2.3. Assessment of the Fertility of Alloplasmic Lines and Hybrids between Alloplasmic Lines and the Line Om29-1RS.1BL
2.4. Studying of mt and cpDNA Regions
2.5. Identification of 1RS Chromosome Arm
3. Results
3.1. Study of the Level of Fertility of Alloplasmic Lines
3.2. PCR Analysis of 18S/5S mtDNA Repeat in Alloplasmic Recombinant and Introgression Lines (H. vulgare)-T. aestivum
3.3. Analysis of cpDNA Microsatellite Loci in Alloplasmic Recombinant and Introgression Lines (H. vulgare)-T. aestivum
3.4. Analysis of Hybrids Derived from Crosses between Alloplasmic Lines and the Line Om29-1RS.1BL
3.5. Identification of the 1RS Chromosome Arm in F1 and F2 Hybrids Derived from Alloplasmic Lines and the Om29-1RS.1BL Line Crosses
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tsunewaki, K. Plasmon analysis as the counterpart of genome analysis. In Methods of Genome Analysis in Plant; Jauhar, P.P., Ed.; CRC Press: Boca Raton, FL, USA, 1996; pp. 271–299. [Google Scholar]
- Soltani, A.; Kumar, A.; Mergoum, M.; Pirseyedi, S.M.; Hegstad, J.B.; Mazaheri, M.; Kianian, S.F. Novel nuclear-cytoplasmic interaction in wheat (Triticum aestivum) induces vigorous plants. Funct. Integr. Genom. 2016, 16, 171–182. [Google Scholar] [CrossRef]
- Crosatti, C.; Quansah, L.; Maré, C.; Giusti, L.; Roncaglia, E.; Atienza, S.G.; Cattivelli, L.; Fait, A. Cytoplasmic genome substitution in wheat affects the nuclear-cytoplasmic cross-talk leading to transcript and metabolite alterations. BMC Genom. 2013, 14, 868–889. [Google Scholar] [CrossRef] [PubMed]
- Talukder, S.K.; Vara Prasad, P.V.; Todd, T.; Babar, M.A.; Poland, J.; Bowden, R.; Fritz, A. Effect of cytoplasmic diversity on post anthesis heat tolerance in wheat. Euphytica 2015, 204, 383–394. [Google Scholar] [CrossRef]
- Terletskaya, N.V.; Shcherban, A.B.; Nesterov, M.A.; Perfil’ev, R.N.; Salina, E.A.; Altayeva, N.A.; Blavachinskaya, I.V. Drought Stress Tolerance and Photosynthetic Activity of Alloplasmic Lines T. dicoccum x T. aestivum. Int. J. Mol. Sci. 2020, 21, 3356. [Google Scholar] [CrossRef] [PubMed]
- Ekiz, H.; Safi Kiral, A.; Akçin, A.; Simsek, L. Cytoplasmic effects on quality traits of bread wheat (Triticum aestivum L.). Euphytica 1998, 100, 189–196. [Google Scholar] [CrossRef]
- Atienza, S.G.; Martin, A.; Peechioni, N.; Platani, C.; Cattivelli, L. The nuclear-cytoplasmic interaction controls carotenoid content in wheat. Euphytica 2008, 159, 325–331. [Google Scholar] [CrossRef]
- Gupta, P.K.; Balyan, H.S.; Gahlaut, V.; Saripalli, G.; Pal, B.; Basnet, B.R.; Joshi, A.K. Hybrid wheat: Past, present and future. Theor Appl Genet. 2019, 132, 2463–2483. [Google Scholar] [CrossRef]
- Liu, Y.; Tang, L.; Xu, Q.; Ma, D.; Zhao, M.; Sun, J.; Chen, W. Experimental and genomic evidence for the indica-type cytoplasmic effect in Oryza sativa L. ssp. japonica. J. Integr. Agric. 2016, 15, 2183–2191. [Google Scholar] [CrossRef]
- Tsunewaki, K.; Wang, G.-Z.; Matsuoka, Y. Plasmon analysis of Triticum (wheat) and Aegilops. 2. Characterization and classification of 47 plasmons based on their effects on common wheat phenotype. Genes Genet Syst. 2002, 77, 409–427. [Google Scholar] [CrossRef]
- Kihara, H. Importance of cytoplasm in plant genetics. Cytologia 1982, 47, 435–450. [Google Scholar] [CrossRef]
- Liu, C.; Yw, W.; Hou, H.; Zhang, C.; Zhang, Y. Value and utilization of alloplasmic common wheats with Aegilops crassa cytoplasm. Plant Breed. 2002, 121, 407–410. [Google Scholar] [CrossRef]
- Delibes, A.; Doussinault, G.; Mena, M.; Lopez-Brana, I.; Garcia-Olmedo, F. Eyespot resistance gene Pch-1 from Aegilops ventricosa is associated with a different chromosome in wheat line H-93-70 than the resistance factor in “Roazon’ wheat. Appl. Genet. 1988, 76, 573–576. [Google Scholar] [CrossRef]
- Friebe, B.; Jiang, J.; Raupp, W.J.; McIntosh, R.A.; Gill, B.S. Characterization on wheat-alien translocations conferring resistance to diseases and pests: Current status. Euphytica 1996, 91, 59–87. [Google Scholar] [CrossRef]
- Semenov, O.G.; Kaid, M.K.A. Morpho-biological characteristics of genotypes of spring forms of allocytoplasmic wheat in terms of their stress tolerance to drought. Bull. Rudn Univ. 2014, 2, 5–14. [Google Scholar] [CrossRef][Green Version]
- Klimushina, M.V.; Divashuk, M.G.; Karlov, G.I.; Mokhammed, T.A.K.; Semenov, O.G. Analysis of allelic state of genes responsible for baking properties in allocytoplasmic wheat hybrids. Russ. J. Genet. 2013, 49, 530–538. [Google Scholar] [CrossRef]
- Semenov, O.G.; Divashuk, M. GHaitembu Gerhard Shandzheshapvako, M.T.A.K. The production of genotypes of allocytoplasmic wheat with high quality of gluten on the basis of marker selection. Bull. Rudn Univ. 2016, 1, 7–14. [Google Scholar] [CrossRef][Green Version]
- Pershina, L.A.; Trubacheeva, N.V.; Sinyavskaya, M.G.; Devyatkina, E.P.; Kravtsova, L.A. Nuclear-cytoplasmic compatibility and the state of mitochondrial and chloroplast DNA regions in alloplasmic recombinant and introgressive lines (H. vulgare)-T. aestivum. Russ. J. Genet. 2014, 50, 1017–1024. [Google Scholar] [CrossRef]
- Pershina, L.A.; Belova, L.I.; Trubacheeva, N.V.; Osadchaya, T.S.; Shumny, V.K.; Belan, I.A.; Rosseeva, L.P.; Nemchenko, V.V.; Abakumov, S.N. Alloplasmic recombinant lines (H. vulgare)-T. aestivum with 1RS.1BL translocation: Initial genotypes for production of common wheat varieties. Vavilov J. Genet. Breed. 2018, 22, 544–552. [Google Scholar] [CrossRef]
- Belan, I.A.; Rosseeva, L.P.; Blokhina, N.P.; Lozhnikova, L.F.; Nemchenko, V.V.; Abakumov, S.N.; Cadikov, R.K.; Trubacheeva, N.V.; Pershina, L.A. Main directions of the spring bread wheat breeding in Western Siberia. In Current Challenges in Plant Genetics, Genomics, Bioinformatics, and Biotechnology, Proceedings of the Fifth International Scientific Conference PlantGen2019, Novosibirsk, Russia, 24–29 June 2019; Kochetov, A., Salina, E., Eds.; Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences: Novosibirsk, Russia, 2019; pp. 187–189. [Google Scholar] [CrossRef]
- Pershina, L.; Trubacheeva, N.; Badaeva, E.; Belan, I.; Rosseeva, L. Study of Androgenic Plant Families of Alloplasmic Introgression Lines (H. vulgare) –T. aestivum and the Use of Sister DH Lines in Breeding. Plants 2020, 9, 764. [Google Scholar] [CrossRef]
- Schlegel, R. Current list of Wheats with Rye and Alien Introgression. Version 08.19. Gatersleben, Germany. Available online: http://www.rye-gene-map.de/rye-introgression (accessed on 30 May 2020).
- Tahir, C.M.; Tsunewaki, K. Monosomic analysis of Triticum spelta var. duhamelianum, a fertility-restorer for T. timopheevi cytoplasm. Jpn. J. Genet. 1969, 44, 1–9. [Google Scholar] [CrossRef]
- Zhou, W.; Frederic, L.K.; Leslie, L.D.; Wang, S. SSR markers associated with fertility restoration genes against Triticum timopheevii cytoplasm in Triticum aestivum. Euphytica 2005, 141, 33–40. [Google Scholar] [CrossRef]
- Tsunewaki, K. Fine mapping of the first multi-fertility-restoring gene, Rf multi, of wheat for three Aegilops plasmons, using 1BS–1RS recombinant lines. Appl. Genet. 2015, 128, 723–732. [Google Scholar] [CrossRef]
- Pershina, L.A.; Numerova, O.M.; Belova, L.I.; Devyatkina, E.P. The peculiarities of fertility restoration in euploids and aneuploids of barley-wheat hybrids progenies. In Proceedings of the 11th EWAC Conference, Novosibirsk, Russia, 24–28 July 2000; Institute of Cytology and Genetics: Novosibirsk, Russia, 2000; pp. 31–32. [Google Scholar]
- Aksyonova, E.; Sinyavskaya, M.; Danilenko, N.; Pershina, L.; Nakamura, C.; Davydenko, O. Heteroplasmy and paternally oriented shift of the organellar DNA composition in barley-wheat hybrids during backcrosses with wheat parents. Genome 2005, 48, 761–769. [Google Scholar] [CrossRef]
- Trubacheeva, N.V.; Kravtsova, L.A.; Devyatkina, E.P.; Efremova, T.T.; Sinyavskaya, M.G.; Shumny, V.K.; Pershina, L.A. Heteroplasmic and homoplasmic states of mitochondrial and chloroplast DNA regions in progenies of distant common wheat hybrids of different origins. Russ. J. Genet. Appl. Res. 2012, 2, 494–500. [Google Scholar] [CrossRef]
- Trubacheeva, N.V.; Rosseeva, L.P.; Belan, I.A.; Osadchaya, T.S.; Kravtsova, L.A.; Kolmakov, Y.V.; Blokhina, N.P.; Pershina, L.A. Characteristics of Common Wheat Cultivars of West Siberia Carrying the Wheat–Rye 1RS.1BL Translocation. Russ. J. Genet. 2011, 47, 13–18. [Google Scholar] [CrossRef]
- Ausubel, M.L.; Brent, R.E.; Kingston, R.E.; Moor, D.D.; Seidman, J.G. Current Protocols in Molecular Biology; Greene Publishing Associates/Wiley Interscience: New York, NY, USA, 1988. [Google Scholar]
- Coulthart, M.B.; Spencer, D.F.; Gray, M.W. Comparative analysis of a recombining-repeat-sequence family in the mitochondrial genomes of wheat (Triticum aestivum L.) and rye (Secale cereale L.). Curr. Genet. 1993, 23, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Tomar, R.S.S.; Deshmukh, R.K.; Naik, K.B.; Tomar, S.M.S.; Vinod. Development of chloroplast-specific microsatellite markers for molecular characterization of alloplasmic lines and phylogenetic analysis in wheat. Plant Breed. 2014, 133, 12–18. [Google Scholar] [CrossRef]
- Chai, J.F.; Zhou, R.H.; Jia, J.Z.; Liu, X. Development and application of a new codominant PCR marker for detecting 1BL·1RS wheat-rye chromosome translocations. Plant Breed. 2006, 125, 302–304. [Google Scholar] [CrossRef]
- Kiang, A.S.; Connolly, V.; McConnell, D.J.; Kavanagh, T.A. Paternal inheritance of mitochondria and chloroplasts in Festuca pratensis–Lolium perenne intergeneric hybrids. Theor. Appl. Genet. 1994, 87, 681–688. [Google Scholar] [CrossRef]
- Hattori, N.; Kitagawa, K.; Takumi, S.; Nakamura, C. Mitochondrial DNA heteroplasmy in wheat, Aegilops and their nucleus-cytoplasm hybrids. Genetics 2002, 160, 1619–1630. [Google Scholar] [CrossRef]
- Kawaura, K.; Saeki, A.; Masumura, T.; Morita, S.; Ogihara, Y. Heteroplasmy and expression of mitochondrial genes in alloplasmic and euplasmic wheat. Genes Genet Syst 2011, 86, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, K.; Takumi, S.; Nakamura, C. Evidence of paternal transmission of mitochondrial DNA in a nucleus-cytoplasm hybrid of timopheevi wheat. Genes. Genet. Syst. 2002, 77, 243–250. [Google Scholar] [CrossRef]
- Kmiec, B.; Woloszynska, M.; Janska, H. Heteroplasmy as a common state of mitochondrial genetic information in plants and animals. Curr. Genet. 2006, 50, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Maan, S.S.; Lucken, K.A.; Bravo, J.M. Genetic analyses of male-fertility restoration in wheat. I. Chromosomal location of Rf genes. Crop Sci. 1984, 24, 17–21. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, M.; Yu, J. Mitochondrial retrograde regulation tuning fork in nuclear genes expressions of higher plants. J Genet Genom. 2008, 35, 65–71. [Google Scholar] [CrossRef]
- Caruso, C.M.; Case, A.L.; Bailey, M.F. The evolutionary ecology of cytonuclear interactions in angiosperms. Trends Plant Sci. 2012, 17, 638–643. [Google Scholar] [CrossRef]
- Pershina, L.A.; Devyatkina, E.P.; Trubacheeva, N.V.; Kravtsova, L.A.; Dobrovol’skaya, O.B. Characterization of fertility restoration in alloplasmic lines derived from hybridization of self-fertilized of spring of barley–wheat (Hordeum vulgare L. × Triticum aestivum L.) amphiploid with common wheat varieties Saratovskaya 29 and Pyrotrix 28. Russ. J. Genet. 2012, 48, 1184–1190. [Google Scholar] [CrossRef]
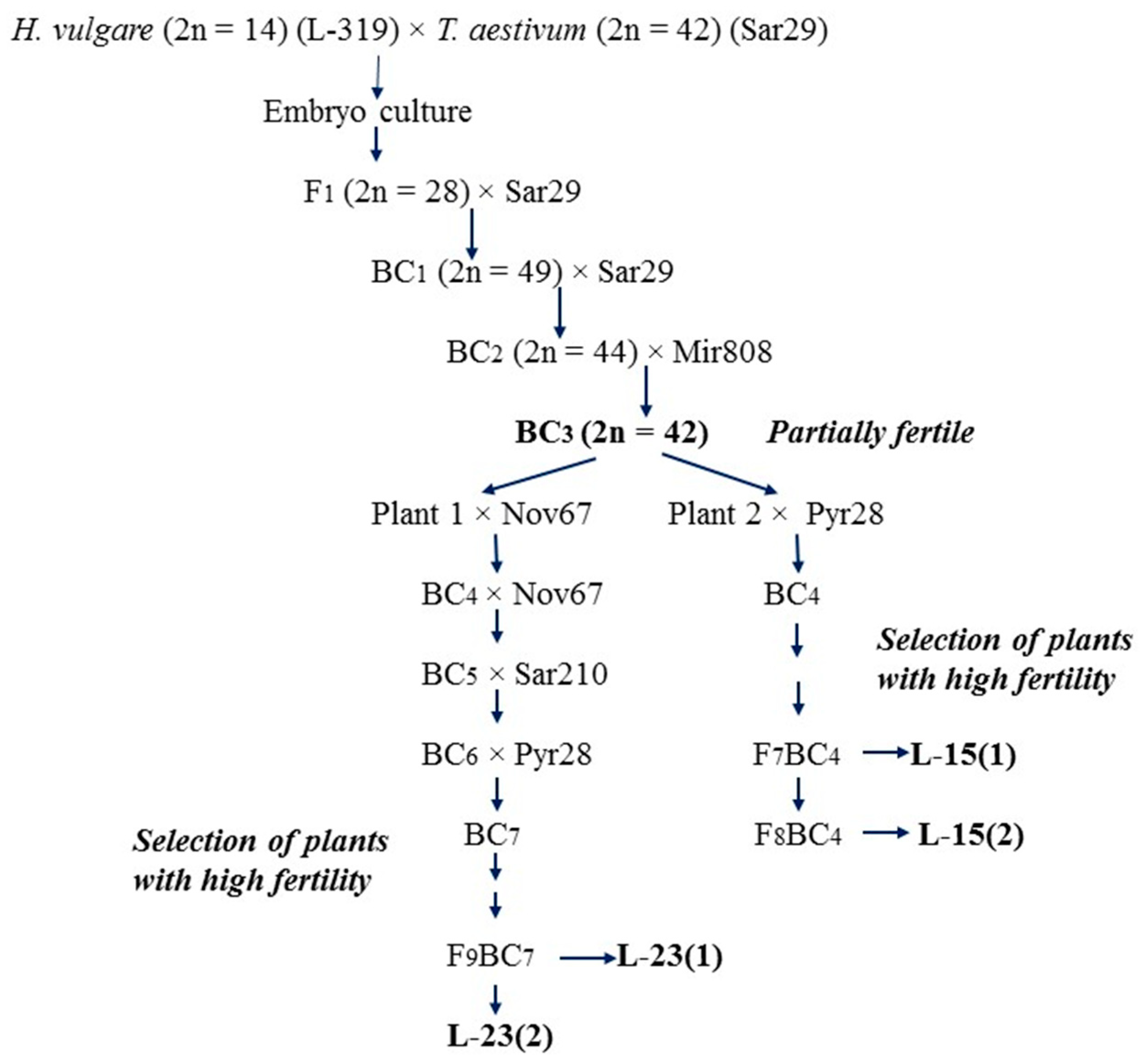


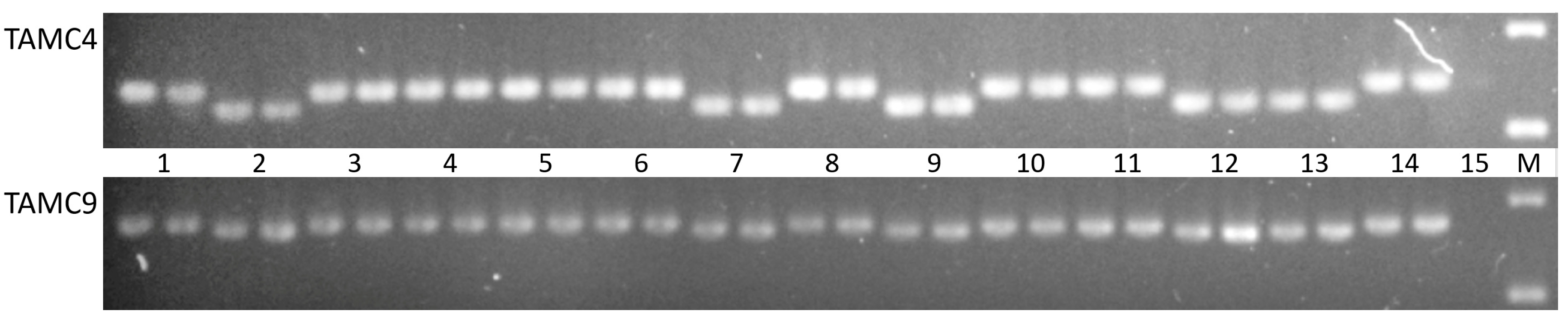
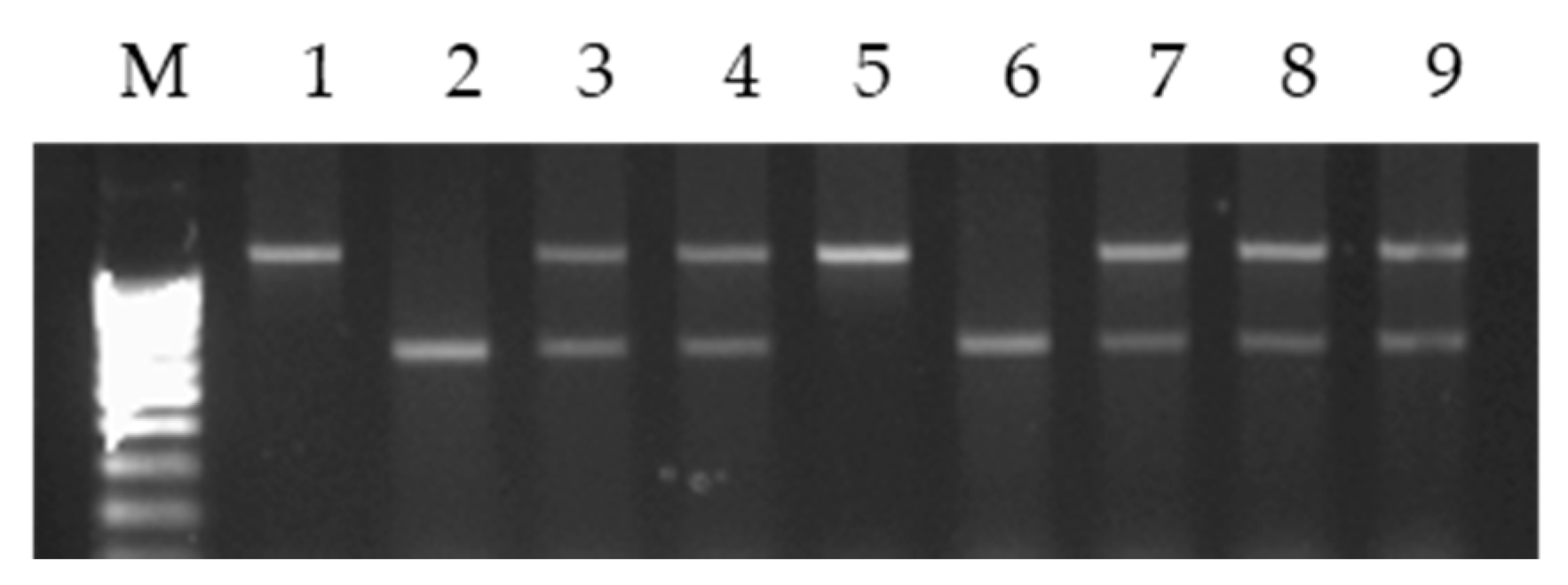
| Groups | Lines | Origin of Lines | Generation |
|---|---|---|---|
| 1 | L-15(1) | [H.v. (L-319) × T.a. (Sar29)]/Sar29/Mir808/Pyr28 | F9BC4 |
| L-15(2) | |||
| 2 | L-23(1) | [H.v. (L-319) × T.a. (Sar29)]/Sar29/Sar29/Mir808/Nov67/Nov67/Sar210/Pyr28 | F7BC7 |
| L-23(2) | F8BC7 | ||
| 3 | L-55(1) | [H.v. (Nep) × T.a. (Sar29)]/Sar29/Mir808/Pyr28/Sar29 | F9BC4 |
| L-55(2) | F10BC4 | ||
| 4 | L-55(3) | F10BC4 | |
| L-55(4) | F11BC4 | ||
| Controls | L-17(3) | [H.v. (Nep) × T.a. (Sar29)]/Mir808/Mir808/Sar29 | F14BC3 |
| L-17(3)/Om29 Om29/L-17(3) | L-17(3)/Omskaya 29 Omskaya 29/L-17(3) | F1 | |
| L-319 Om29 Sar29 | H. vulgare, line L-319 T. aestivum, var. Omskaya 29 T. aestivum, var. Saratovskaya 29 | ||
| Marker ID | Primer Sequence | Expected Size of the Product, bp |
|---|---|---|
| TaCM4 | AATCCTTGGGGTTCCAGAAT GCCACTTKGATTTCCCATTA | TA: 242 HV: 224 |
| TaCM9 | TCCAGCCAACGATGACACTA CCAAGAAAGCACATCAGATCA | TA: 269 HV: 261 |
| Groups | Lines | Self-Fertility (%) | 18S/5S mtDNA Region | SSR cpDNA Loci | |
|---|---|---|---|---|---|
| TaCM4 | TaCM9 | ||||
| 1 | L-15(1) L-15(2) | 76, 38 92, 11 *** | B + W W | B W | B W |
| 2 | L-23(1) L-23(2) | 88, 25 94, 72 * | B + W W | B W | B W |
| 3 | L-55(1) L-55(2) | 67, 05 87, 80 * | B + W W | B W | B W |
| 4 | L-55(3) L-55(4) | 90, 83 89, 81 | B + W W | B W | B W |
| Controls | L-17(3) 17(3)/Om29 Om29/17(3) | 92, 90 92, 04 89, 41 | W W W | W W W | W W W |
| L-319 Om29 Sar29 | 100 98, 11 100 | B W W | B W W | B W W | |
| Hybrid Combination | Gene-ration | Total Number of Plants | Seed Set in F1 and F2 Plants | Expected Segregation Ratio in F2 | χ2 | p Value | |
|---|---|---|---|---|---|---|---|
| No. of Fertile Plants | No. of Sterile Plants | ||||||
| L-15(1) × Om29-1RS.1BL | F1 | 12 | 12 | - | - | - | - |
| L-15(1) × Om29-1RS.1BL | F2 | 108 | 74 | 34 | 3:1 | 2.42 | 0.12 |
| L-15(2) × Om29-1RS.1BL | F1 | 16 | 16 | 0 | - | - | - |
| L-15(2) × Om29-1RS.1BL | F2 | 86 | 86 | - | - | - | - |
| L-23(1) × Om29-1RS.1BL | F1 | 10 | 10 | 0 | - | - | - |
| L-23(1) × Om29-1RS.1BL | F2 | 97 | 76 | 21 | 3:1 | 0.58 | 0.446 |
| L-23(2) × Om29-1RS.1BL | F1 | 10 | 10 | 0 | - | - | - |
| L-23(2) × Om29-1RS.1BL | F2 | 84 | 84 | 0 | - | - | - |
| L-55(1) × Om29-1RS.1BL | F1 | 60 | 0 | 60 | - | - | - |
| L-55(2) × Om29-1RS.1BL | F1 | 20 | 20 | 0 | - | - | - |
| L-55(2) × Om29-1RS.1BL | F2 | 66 | 66 | 0 | - | - | - |
| L-55(3) × Om29-1RS.1BL | F1 | 38 | 38 | 0 | - | - | - |
| L-55(3) × Om29-1RS.1BL | F2 | 75 | 58 | 17 | 3:1 | 0.22 | 0.64 |
| L-55(4) × Om29-1RS.1BL | F1 | 15 | 15 | 0 | - | - | - |
| L-55(4) × Om29-1RS.1BL | F2 | 96 | 96 | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trubacheeva, N.V.; Divashuk, M.G.; Chernook, A.G.; Belan, I.A.; Rosseeva, L.P.; Pershina, L.A. The Effect of Chromosome Arm 1BS on the Fertility of Alloplasmic Recombinant Lines in Bread Wheat with the Hordeum vulgare Cytoplasm. Plants 2021, 10, 1120. https://doi.org/10.3390/plants10061120
Trubacheeva NV, Divashuk MG, Chernook AG, Belan IA, Rosseeva LP, Pershina LA. The Effect of Chromosome Arm 1BS on the Fertility of Alloplasmic Recombinant Lines in Bread Wheat with the Hordeum vulgare Cytoplasm. Plants. 2021; 10(6):1120. https://doi.org/10.3390/plants10061120
Chicago/Turabian StyleTrubacheeva, Nataliya V., Mikhail G. Divashuk, Anastasiya G. Chernook, Igor A. Belan, Ludmila P. Rosseeva, and Lidiya A. Pershina. 2021. "The Effect of Chromosome Arm 1BS on the Fertility of Alloplasmic Recombinant Lines in Bread Wheat with the Hordeum vulgare Cytoplasm" Plants 10, no. 6: 1120. https://doi.org/10.3390/plants10061120
APA StyleTrubacheeva, N. V., Divashuk, M. G., Chernook, A. G., Belan, I. A., Rosseeva, L. P., & Pershina, L. A. (2021). The Effect of Chromosome Arm 1BS on the Fertility of Alloplasmic Recombinant Lines in Bread Wheat with the Hordeum vulgare Cytoplasm. Plants, 10(6), 1120. https://doi.org/10.3390/plants10061120






