Polymorphism in the Chloroplast ATP Synthase Beta-Subunit Is Associated with a Maternally Inherited Enhanced Cold Recovery in Cucumber
Abstract
1. Introduction
2. Results
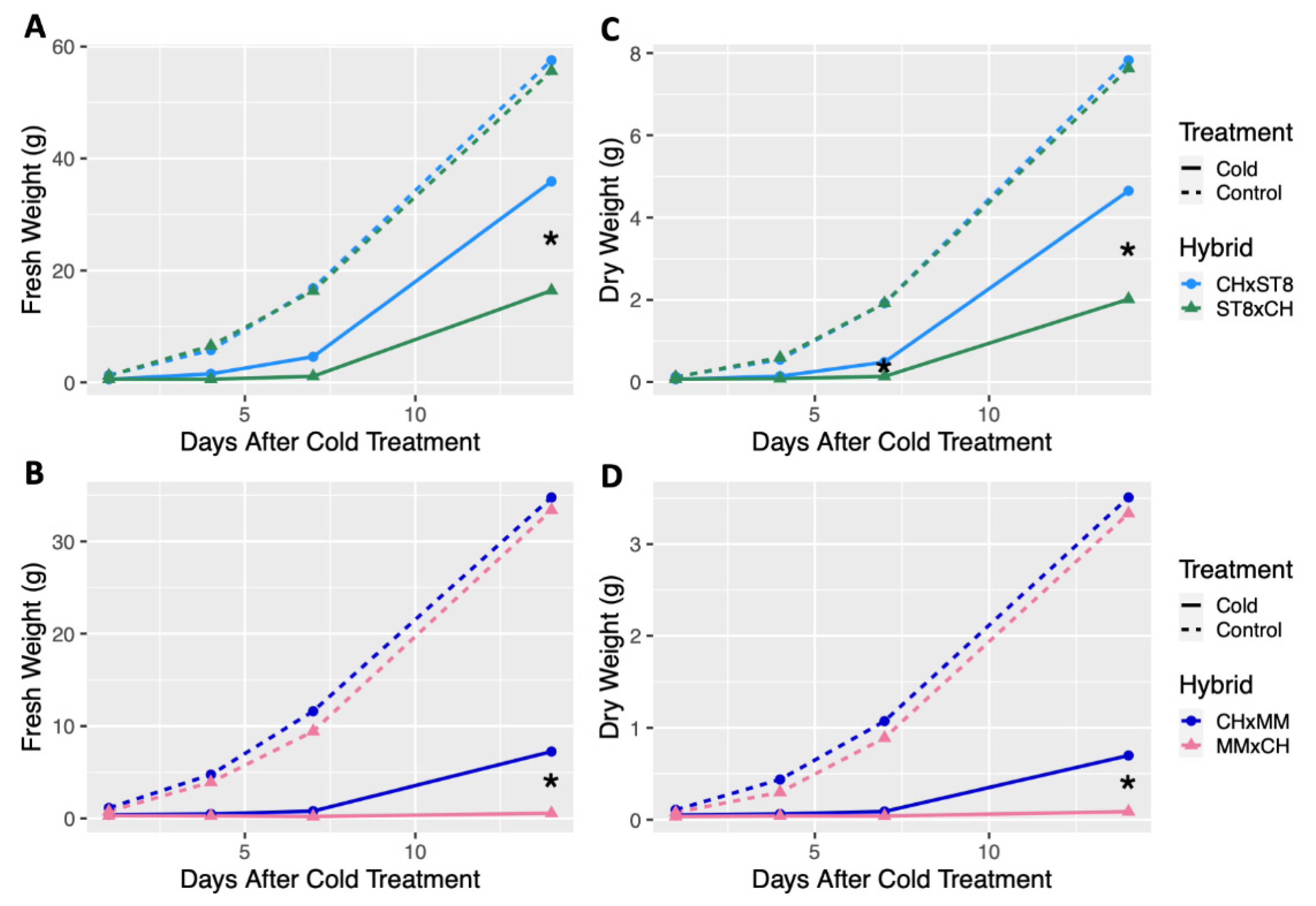
2.1. CH Shows a Maternally Transmitted, Cold Recovery Phenotype
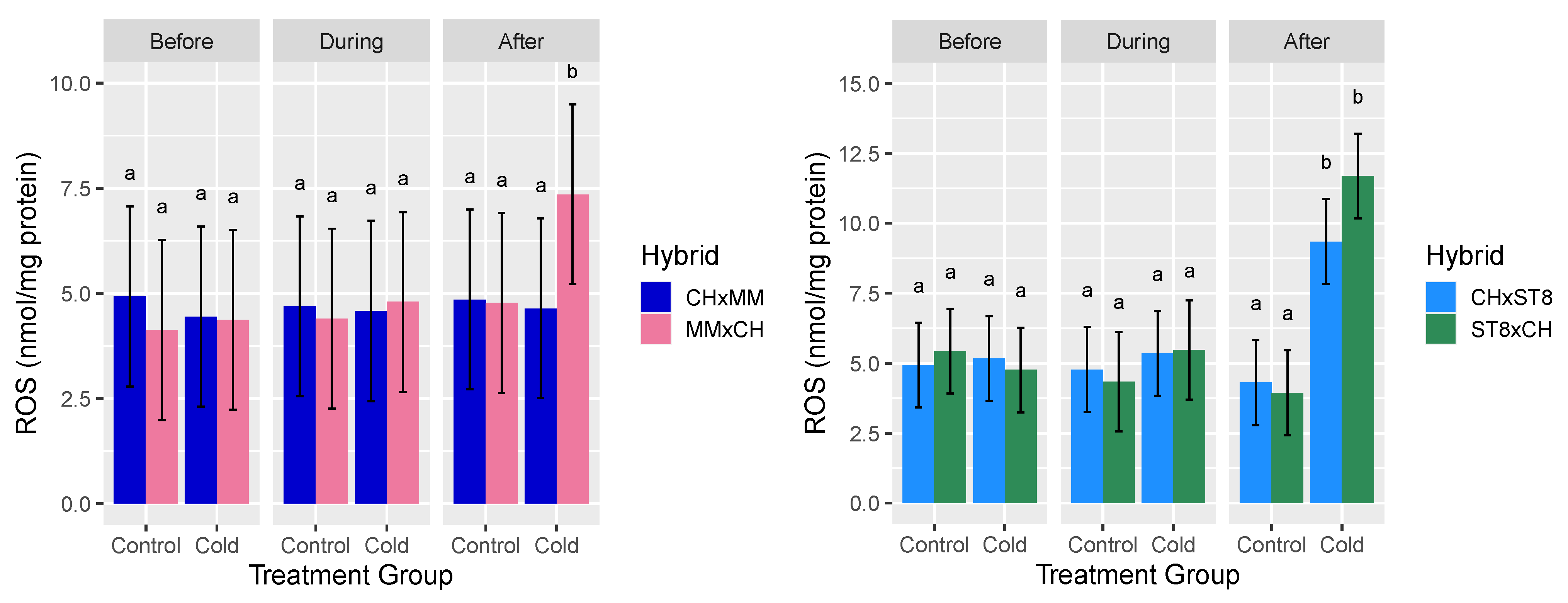
2.2. ROS Levels Across Cold Tolerant and Susceptible Reciprocal Hybrids Agree with Cold Recovery Phenotype
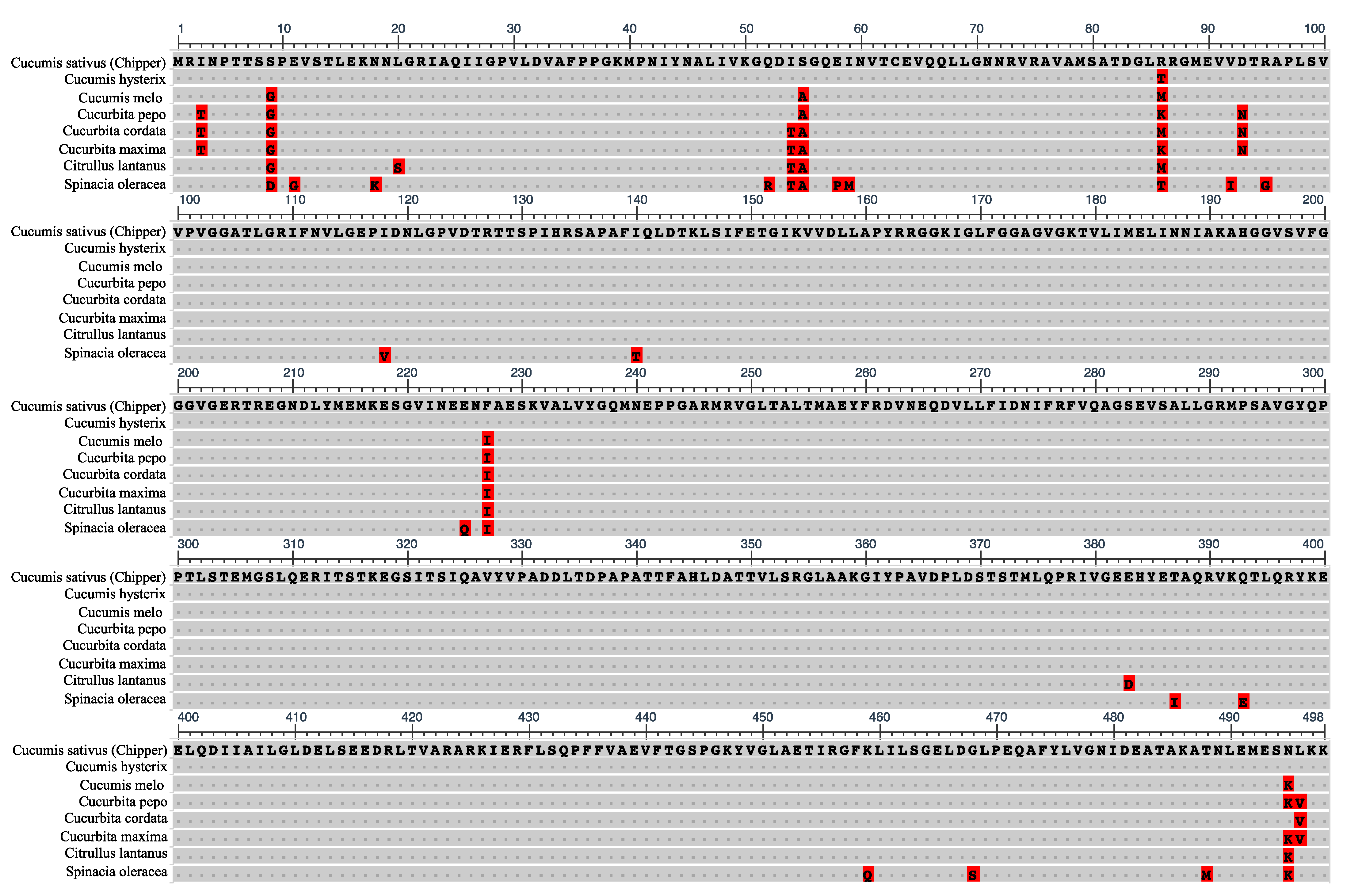
2.3. A Maternally Transmitted SNP in the CF1FO-ATPase β-Subunit Gene (atpB) Is a Marker for Cold Tolerance
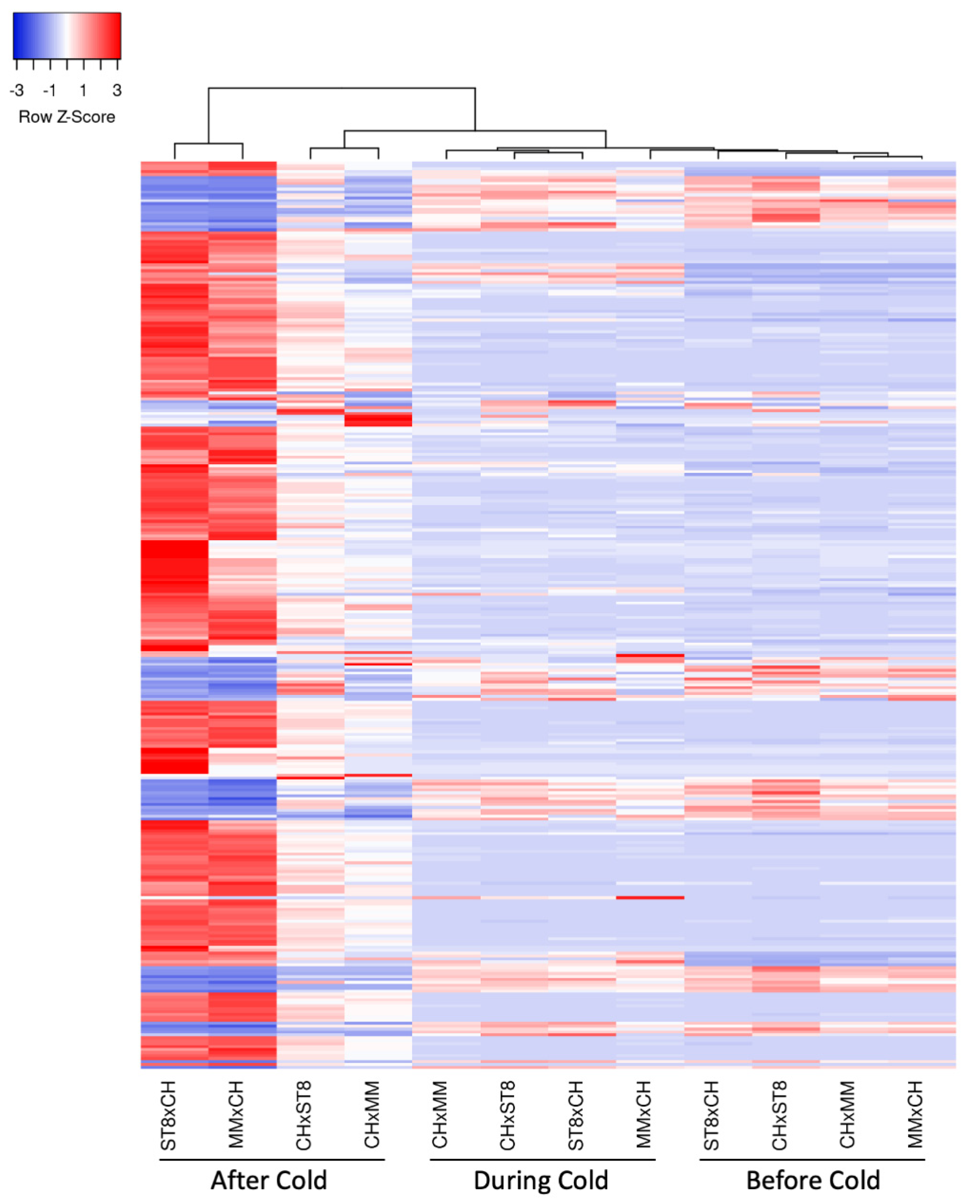
2.4. Nuclear Gene Expression Patterns Across Cold Tolerant and Susceptible Reciprocal Hybrids Agree with Cold Recovery Phenotype
3. Discussion
3.1. Chloroplast-Associated Cold Tolerance Does Not Reduce Growth under Normal Conditions
3.2. Potential Role of the Chloroplast atpB in Conferring Cold Tolerance in CH
3.3. Transcriptional Differences and ROS Levels in the Cold Recovery Phenotype
4. Materials and Methods
4.1. Plant Material
4.2. Cold Treatments
4.3. Hydrogen Peroxide (H2O2) Assays
4.4. Sequencing of Chloroplast DNA and RNA
4.5. Chloroplast Protein Modeling and Homology Comparisons
4.6. Nuclear mRNA Sequencing and Expression Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smeets, L.; Wehner, T.C. Environmental effects on genetic variation of chilling resistance in cucumber. Euphytica 1997, 97, 217–225. [Google Scholar] [CrossRef]
- Chung, S.; Staub, J.E.; Fazio, G. Inheritance of chilling injury: A maternally inherited trait in Cucumber. J. Am. Soc. Hortic. Sci. 2003, 128, 526–530. [Google Scholar] [CrossRef]
- Senaratna, T.; Mackay, C.E.; McKersie, B.D.; Fletcher, R.A. Uniconazole-induced chilling tolerance in tomato and its relationship to antioxidant content. J. Plant Physiol. 1988, 133, 56–61. [Google Scholar] [CrossRef]
- Chung, S.-M.; Gordon, V.S.; Staub, J.E. Sequencing cucumber (Cucumis sativus L.) chloroplast genomes identifies differences between chilling-tolerant and -susceptible cucumber lines. Genome 2007, 50, 215–225. [Google Scholar] [CrossRef]
- Wells, O.; Loy, J. Rowcovers and high tunnels enhance crop production in the northeastern United States. HortTechnology 1993, 3, 92–95. [Google Scholar] [CrossRef]
- Andaya, V.C.; Mackill, D.J. Mapping of QTLs associated with cold tolerance during the vegetative stage in rice. J. Exp. Bot. 2003, 54, 2579–2585. [Google Scholar] [CrossRef]
- Burow, G.; Burke, J.J.; Xin, Z.; Franks, C.D. Genetic dissection of early-season cold tolerance in sorghum (Sorghum bicolor (L.) Moench). Mol. Breed. 2011, 28, 391–402. [Google Scholar] [CrossRef]
- Allam, M.; Revilla, P.; Djemel, A.; Tracy, W.F.; Ordás, B. Identification of QTLs involved in cold tolerance in sweet × field corn. Euphytica 2016, 208, 353–365. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, T.; Ge, H.; Pang, W.; Gao, L.; Ren, L.; Chen, H. SSR mapping of QTLs conferring cold tolerance in an interspecific cross of tomato. Int. J. Genom. 2016, 2016, 3219276. [Google Scholar] [CrossRef]
- Revilla, P.; Rodríguez, V.M.; Ordás, A.; Rincent, R.; Charcosset, A.; Giauffret, C.; Melchinger, A.E.; Schön, C.C.; Bauer, E.; Altmann, T.; et al. Association mapping for cold tolerance in two large maize inbred panels. BMC Plant Biol. 2016, 16, 1–10. [Google Scholar] [CrossRef]
- Chopra, R.; Burow, G.; Hayes, C.; Emendack, Y.; Xin, Z.; Burke, J. Transcriptome profiling and validation of gene based single nucleotide polymorphisms (SNPs) in sorghum genotypes with contrasting responses to cold stress. BMC Genom. 2015, 16, 1040. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef]
- Anunanthini, P.; Manoj, V.M.; Sarath Padmanabhan, T.S.; Dhivya, S.; Narayan, J.A.; Appunu, C.; Sathishkumar, R. In silico characterization and functional validation of chilling tolerant divergence 1 (COLD1) gene in monocots during abiotic stress. Funct. Plant Biol. 2019, 46, 596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Pan, Y.; Li, J.; Zhou, L.; Shi, H.; Zeng, Y.; Guo, H.; Yang, S.; Zheng, W.; et al. Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat. Commun. 2017, 8, 14788. [Google Scholar] [CrossRef]
- Kozik, E.U.; Wehner, T.C. A single dominant gene Ch for chilling resistance in cucumber seedlings. J. Am. Soc. Hortic. Sci. 2008, 133, 225–227. [Google Scholar] [CrossRef]
- Thomashow, M.F. Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 1998, 118, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dang, P.; Liu, L.; He, C. Cold acclimation by the CBF–COR pathway in a changing climate: Lessons from Arabidopsis thaliana. Plant Cell Rep. 2019, 38, 511–519. [Google Scholar] [CrossRef]
- Wang, F.; Chen, X.; Dong, S.; Jiang, X.; Wang, L.; Yu, J.; Zhou, Y. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 2020, 18, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Sanghera, G.; Wani, S.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genom. 2011, 12, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Havey, M.J. Predominant paternal transmission of the mitochondrial genome in cucumber. J. Hered. 1997, 88, 232–235. [Google Scholar] [CrossRef]
- Gillham, N.W.; Boynton, J.E.; Hauser, C.R. Translational regulation of gene expression in chloroplasts and mitochondria. Annu. Rev. Genet. 1994, 28, 71–93. [Google Scholar] [CrossRef] [PubMed]
- Maier, R.M.; Zeltz, P.; Kössel, H.; Bonnard, G.; Gualberto, J.M.; Grienenberger, J.M. RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 1996, 32, 343–365. [Google Scholar] [CrossRef]
- del Campo, E.M. Post-transcriptional control of chloroplast gene expression. Gene Regul. Syst. Biol. 2009, 3, 31–47. [Google Scholar] [CrossRef]
- Guzowska-Nowowiejska, M.; Fiedorowicz, E.; Plader, W. Cucumber, melon, pumpkin, and squash: Are rules of editing in flowering plants chloroplast genes so well known indeed? Gene 2009, 434, 1–8. [Google Scholar] [CrossRef]
- Kuroda, H.; Maliga, P. Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol. 2001, 125, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Fernández, A.P.; Strand, Å. Retrograde signaling and plant stress: Plastid signals initiate cellular stress responses. Curr. Opin. Plant Biol. 2008, 11, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Gleason, C.; Huang, S.; Thatcher, L.F.; Foley, R.C.; Anderson, C.R.; Carroll, A.J.; Millar, A.H.; Singh, K.B. Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc. Natl. Acad. Sci. USA 2011, 108, 10768–10773. [Google Scholar] [CrossRef]
- Crosatti, C.; Rizza, F.; Badeck, F.W.; Mazzucotelli, E.; Cattivelli, L. Harden the chloroplast to protect the plant. Physiol. Plant. 2013, 147, 55–63. [Google Scholar] [CrossRef]
- Tang, H.; Zhang, D.; Yuan, S.; Zhu, F.; Xu, F.; Fu, F.-Q.; Wang, S.; Lin, H.-H. Plastid signals induce ALTERNATIVE OXIDASE expression to enhance the cold stress tolerance in Arabidopsis thaliana. Plant Growth Regul. 2014, 74, 275–283. [Google Scholar] [CrossRef]
- Bobik, K.; Burch-Smith, T.M. Chloroplast signaling within, between and beyond cells. Front. Plant Sci. 2015, 6, 781. [Google Scholar] [CrossRef]
- Yoo, Y.H.; Hong, W.J.; Jung, K.H. A systematic view exploring the role of chloroplasts in plant abiotic stress responses. BioMed Res. Int. 2019, 2019, 6534745. [Google Scholar] [CrossRef] [PubMed]
- Laloi, C.; Apel, K.; Danon, A. Reactive oxygen signalling: The latest news. Curr. Opin. Plant Biol. 2004, 7, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Galvez-Valdivieso, G.; Mullineaux, P.M. The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol. Plant. 2010, 138, 430–439. [Google Scholar] [CrossRef]
- Havey, M.J.; McCreight, J.D.; Rhodes, B.; Taurick, G. Differential transmission of the Cucumis organellar genomes. Theor. Appl. Genet. 1998, 97, 122–128. [Google Scholar] [CrossRef]
- Kim, J.-S.; Jung, J.D.; Lee, J.-A.; Park, H.-W.; Oh, K.-H.; Jeong, W.-J.; Choi, D.-W.; Liu, J.R.; Cho, K.Y. Complete sequence and organization of the cucumber (Cucumis sativus L. cv. Baekmibaekdadagi) chloroplast genome. Plant Cell Rep. 2006, 25, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Plader, W.; Yukawa, Y.; Sugiura, M.; Malepszy, S. The complete structure of the cucumber (Cucumis sativus L.) chloroplast genome: Its composition and comparative analysis. Cell. Mol. Biol. Lett. 2007, 12, 584–594. [Google Scholar] [CrossRef]
- Alverson, A.J.; Rice, D.W.; Dickinson, S.; Barry, K.; Palmer, J.D. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell 2011, 23, 2499–2513. [Google Scholar] [CrossRef]
- Yang, L.; Koo, D.-H.; Li, Y.; Zhang, X.; Luan, F.; Havey, M.J.; Jiang, J.; Weng, Y. Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 2012, 71, 895–906. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, D.; Gao, L.-Z. The complete chloroplast genome sequence of Cucumis sativus var. Hardwickii, the wild progenitor of cultivated cucumber. Mitochondrial DNA A DNA Mapp Seq Anal. 2016, 27, 4627–4628. [Google Scholar] [CrossRef]
- Li, Q.; Li, H.; Huang, W.; Xu, Y.; Zhou, Q.; Wang, S.; Ruan, J.; Huang, S.; Zhang, Z. A chromosome-scale genome assembly of cucumber (Cucumis sativus L.). GigaScience 2019, 8, giz072. [Google Scholar] [CrossRef]
- Osipowski, P.; Pawełkowicz, M.; Wojcieszek, M.; Skarzyńska, A.; Przybecki, Z.; Pląder, W. A high-quality cucumber genome assembly enhances computational comparative genomics. Mol. Genet. Genom. 2020, 295, 177–193. [Google Scholar] [CrossRef]
- Gordon, V.S.; Staub, J.E. Comparative analysis of chilling response in cucumber through plastidic and nuclear genetic effects component analysis. J. Am. Soc. Hortic. Sci. 2011, 136, 256–264. [Google Scholar] [CrossRef]
- Hsieh, T.H.; Lee, J.T.; Yang, P.T.; Chiu, L.H.; Charng, Y.Y.; Wang, Y.C.; Chan, M.T. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 2002, 129, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Woessner, J.P.; Gillham, N.W.; Boynton, J.E. The sequence of the chloroplast atpB gene and its flanking regions in Chlamydomonas reinhardtii. Gene 1986, 44, 17–28. [Google Scholar] [CrossRef]
- Palmer, J.D.; Stein, D.B. Conservation of chloroplast genome structure among vascular plants. Curr. Genet. 1986, 10, 823–833. [Google Scholar] [CrossRef]
- Androsiuk, P.; Jastrzębski, J.P.; Paukszto, Ł.; Makowczenko, K.; Okorski, A.; Pszczółkowska, A.; Chwedorzewska, K.J.; Górecki, R.; Giełwanowska, I. Evolutionary dynamics of the chloroplast genome sequences of six Colobanthus species. Sci. Rep. 2020, 10, 11522. [Google Scholar] [CrossRef]
- Daniell, H.; Lee, S.B.; Grevich, J.; Saski, C.; Quesada-Vargas, T.; Guda, C.; Tomkins, J.; Jansen, R.K. Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor. Appl. Genet. 2006, 112, 1503–1518. [Google Scholar] [CrossRef] [PubMed]
- Pawson, T.; Scott, J.D. Protein phosphorylation in signaling–50 years and counting. Trends Biochem. Sci. 2005, 30, 286–290. [Google Scholar] [CrossRef]
- Strotmann, H.; Schumann, J. Structure, function and regulation of chloroplast ATPase. Physiol. Plant. 1983, 57, 375–382. [Google Scholar] [CrossRef]
- Hahn, A.; Vonck, J.; Mills, D.J.; Meier, T.; Kühlbrandt, W. Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 2018, 360, eaat4318. [Google Scholar] [CrossRef]
- Yang, J.H.; Williams, D.; Kandiah, E.; Fromme, P.; Chiu, P.L. Structural basis of redox modulation on chloroplast ATP synthase. Commun. Biol. 2020, 3, 482. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Zhou, M.; Marriott, H.; Morgner, N.; Politis, A.; Robinson, C.V. Comparative cross-linking and mass spectrometry of an intact F-type ATPase suggest a role for phosphorylation. Nat. Commun. 2013, 4, 1985. [Google Scholar] [CrossRef]
- Schmidt, C.; Beilsten-Edmands, V.; Mohammed, S.; Robinson, C.V. Acetylation and phosphorylation control both local and global stability of the chloroplast F1 ATP synthase. Sci. Rep. 2017, 7, 44068. [Google Scholar] [CrossRef]
- Lou, W.; Tan, X.; Song, K.; Zhang, S.; Luan, G.; Li, C.; Lu, X. A specific single nucleotide polymorphism in the ATP synthase gene significantly improves environmental stress tolerance of Synechococcus elongatus. Appl. Environ. Microbiol. 2018, 84, e01222-18. [Google Scholar] [CrossRef]
- Nie, G.-Y.; Long, S.P.; Baker, N.R. The effects of development at sub-optimal growth temperatures on photosynthetic capacity and susceptibility to chilling-dependent photoinhibition in Zea mays. Physiol. Plant. 1992, 85, 554–560. [Google Scholar] [CrossRef]
- Tambussi, E.A.; Bartoli, C.G.; Guiamet, J.J.; Beltrano, J.; Araus, J.L. Oxidative stress and photodamage at low temperatures in soybean (Glycine max L. Merr.) leaves. Plant Sci. 2004, 167, 19–26. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Y.; Xiao, J.; Bao, F. Effects of chilling on the structure, function and development of chloroplasts. Front. Plant Sci. 2018, 9, 1715. [Google Scholar] [CrossRef] [PubMed]
- Murata, N.; Nishiyama, Y. ATP is a driving force in the repair of photosystem II during photoinhibition. Plant Cell Environ. 2018, 41, 285–299. [Google Scholar] [CrossRef]
- Martz, F.; Sutinen, M.L.; Kiviniemi, S.; Palta, J.P. Changes in freezing tolerance, plasma membrane H+-ATPase activity and fatty acid composition in Pinus resinosa needles during cold acclimation and de-acclimation. Tree Physiol. 2006, 26, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Muzi, C.; Camoni, L.; Visconti, S.; Aducci, P. Cold stress affects H+-ATPase and phospholipase D activity in Arabidopsis. Plant Physiol. Biochem. 2016, 108, 328–336. [Google Scholar] [CrossRef]
- Janicka-Russak, M.; Kabała, K.; Wdowikowska, A.; Kłobus, G. Response of plasma membrane H+-ATPase to low temperature in cucumber roots. J. Plant Res. 2012, 125, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, D.; Morris, E.R.; Walker, J.C. 14-3-3 and FHA domains mediate phosphoprotein interactions. Annu. Rev. Plant Biol. 2009, 60, 67–91. [Google Scholar] [CrossRef]
- Gökirmak, T.; Paul, A.-L.; Ferl, R.J. Plant phosphopeptide-binding proteins as signaling mediators. Curr. Opin. Plant Biol. 2010, 13, 527–532. [Google Scholar] [CrossRef]
- Kanekatsu, M.; Saito, H.; Motohashi, K.; Hisabori, T. The β subunit of chloroplast ATP synthase (CF0CF1-ATPase) is phosphorylated by casein kinase II. Biochem. Mol. Biol. Int. 1998, 46, 99–105. [Google Scholar] [CrossRef]
- Bunney, T.D.; van Walraven, H.S.; de Boer, A.H. 14-3-3 Protein is a regulator of the mitochondrial and chloroplast ATP synthase. Proc. Natl. Acad. Sci. USA 2001, 98, 4249–4254. [Google Scholar] [CrossRef]
- Barrero-Sicilia, C.; Silvestre, S.; Haslam, R.P.; Michaelson, L.V. Unravelling the response to cold stress in Arabidopsis and its extremophile relative Eutrema salsugineum. Plant Sci. 2017, 263, 194–200. [Google Scholar] [CrossRef]
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Egierszdorff, S.; Kacperska, A. Low temperature effects on growth and actin cytoskeleton organisation in suspension cells of winter oilseed rape. Plant Cell. Tissue Organ Cult. 2001, 65, 149–158. [Google Scholar] [CrossRef]
- Örvar, B.L.; Sangwan, V.; Omann, F.; Dhindsa, R.S. Early steps in cold sensing by plant cells: The role of actin cytoskeleton and membrane fluidity. Plant J. 2000, 23, 785–794. [Google Scholar] [CrossRef]
- Nick, P. Microtubules, signalling and abiotic stress. Plant J. 2013, 75, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Rymen, B.; Fiorani, F.; Kartal, F.; Vandepoele, K.; Inzé, D.; Beemster, G.T.S. Cold nights impair leaf growth and cell cycle progression in maize through transcriptional changes of cell cycle genes. Plant Physiol. 2007, 143, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Coulter, J.A.; Liu, L.; Zhao, Y.; Chang, Y.; Pu, Y.; Zeng, X.; Xu, Y.; Wu, J.; Fang, Y.; et al. Transcriptome analysis reveals key cold-stress-responsive genes in winter rapeseed (Brassica rapa L.). Int. J. Mol. Sci. 2019, 20, 1071. [Google Scholar] [CrossRef]
- Mahalingam, R.; Fedoroff, N. Stress response, cell death and signalling: The many faces of reactive oxygen species. Physiol. Plant. 2003, 119, 56–68. [Google Scholar] [CrossRef]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef]
- Suzuki, N.; Miller, G.; Salazar, C.; Mondal, H.A.; Shulaev, E.; Cortes, D.F.; Shuman, J.L.; Luo, X.; Shah, J.; Schlauch, K.; et al. Temporal-spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell 2013, 25, 3553–3569. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Carvalho, F.E.L.; Silveira, J.A.G. H2O2-retrograde signaling as a pivotal mechanism to understand priming and cross stress tolerance in plants. In Priming-Mediated Stress and Cross-Stress Tolerance in Crop Plants; Hossain, M.A., Liu, F., Burritt, D.J., Fujita, M., Huang, B., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 57–78. [Google Scholar]
- Dirks, R. Method for the production of double-haploid cucumbers. U.S. Patent 5,492,827; filed 11 January 1995 and issued 20 February 1996,
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 3 June 2019).
- Lenth, R.; Singmann, H.; Love, J.; Buerkner, P.; Herve, M. Estimated Marginal Means, Aka Least-Squares Means; R Package Version 1.5.1; Available online: https://CRAN.R-project.org/package=emmeans (accessed on 3 June 2019).
- Bates, D.; Mächler, M.; Bolker, B.M.; Walker, S.C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. Lmertest Package: Tests in linear mixed effects models. J. Stat. Softw. 2017, 82. [Google Scholar] [CrossRef]
- van Wijk, K.J.; Peltier, J.B.; Giacomelli, L. Isolation of chloroplast proteins from Arabidopsis thaliana for proteome analysis. In Plant Proteomics; Humana Press: Totowa, NJ, USA, 2007; Volume 355, pp. 43–48. [Google Scholar]
- Clarke, J.D. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb. Protoc. 2009, 4, pdb.prot5177. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Lei, R.; Ding, S.W.; Zhu, S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 2014, 15, 182. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Reiner, A.; Yekutieli, D.; Benjamini, Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 2003, 19, 368–375. [Google Scholar] [CrossRef]
- Oliveros, J.C. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://ci.nii.ac.jp/naid/20001505977 (accessed on 9 November 2020).
- Thomas, P.D.; Kejariwal, A.; Guo, N.; Mi, H.; Campbell, M.J.; Muruganujan, A.; Lazareva-Ulitsky, B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006, 34, W645–W650. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]




| Source | Mean Sq | DF | F Value | Prob > F | |
|---|---|---|---|---|---|
| ST8xCH and CHxST8 | |||||
| Hybrid | 0.98 | 1 | 1.00 | 0.324 | |
| Time | 27.24 | 2 | 27.87 | <0.001 | *** |
| Treatment | 62.24 | 1 | 63.67 | <0.001 | *** |
| HybridTime | 1.42 | 2 | 1.45 | 0.249 | |
| Hybrid × Treatment | 1.74 | 1 | 1.78 | 0.191 | |
| Time × Treatment | 49.59 | 2 | 50.73 | <0.001 | *** |
| Hybrid × Time × Treatment | 3.30 | 2 | 3.37 | 0.046 | * |
| MMxCH and CHxMM | |||||
| Hybrid | 0.92 | 1 | 2.29 | 0.139 | |
| Time | 4.05 | 2 | 10.08 | <0.001 | *** |
| Treatment | 1.96 | 1 | 4.89 | 0.034 | * |
| Hybrid × Time | 3.37 | 2 | 8.40 | 0.001 | ** |
| Hybrid × Treatment | 5.41 | 1 | 13.46 | <0.001 | *** |
| Time × Treatment | 1.91 | 2 | 4.75 | 0.015 | * |
| Hybrid × Time × Treatment | 1.60 | 2 | 3.98 | 0.028 | * |
| Base Pair Position a | |||||
|---|---|---|---|---|---|
| Sample | 56560 | 59151 | 122965 | 126348 | |
| cpDNA | CH | C | A | C | C |
| ST8 | G | C | Tb | C | |
| MM | G | A | C | T | |
| cpRNA | CHxST8 | C | A | C | C |
| ST8xCH | G | C | T b | C | |
| CHxMM | C | A | C | C | |
| MMxCH | G | A | C | T | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oravec, M.W.; Havey, M.J. Polymorphism in the Chloroplast ATP Synthase Beta-Subunit Is Associated with a Maternally Inherited Enhanced Cold Recovery in Cucumber. Plants 2021, 10, 1092. https://doi.org/10.3390/plants10061092
Oravec MW, Havey MJ. Polymorphism in the Chloroplast ATP Synthase Beta-Subunit Is Associated with a Maternally Inherited Enhanced Cold Recovery in Cucumber. Plants. 2021; 10(6):1092. https://doi.org/10.3390/plants10061092
Chicago/Turabian StyleOravec, Madeline W., and Michael J. Havey. 2021. "Polymorphism in the Chloroplast ATP Synthase Beta-Subunit Is Associated with a Maternally Inherited Enhanced Cold Recovery in Cucumber" Plants 10, no. 6: 1092. https://doi.org/10.3390/plants10061092
APA StyleOravec, M. W., & Havey, M. J. (2021). Polymorphism in the Chloroplast ATP Synthase Beta-Subunit Is Associated with a Maternally Inherited Enhanced Cold Recovery in Cucumber. Plants, 10(6), 1092. https://doi.org/10.3390/plants10061092






