Sources of Resistance to Common Bacterial Blight and Charcoal Rot Disease for the Production of Mesoamerican Common Beans in the Southern United States
Abstract
1. Introduction
2. Results
2.1. Phenotypic Differences in the Mesoamerican Diversity Panel
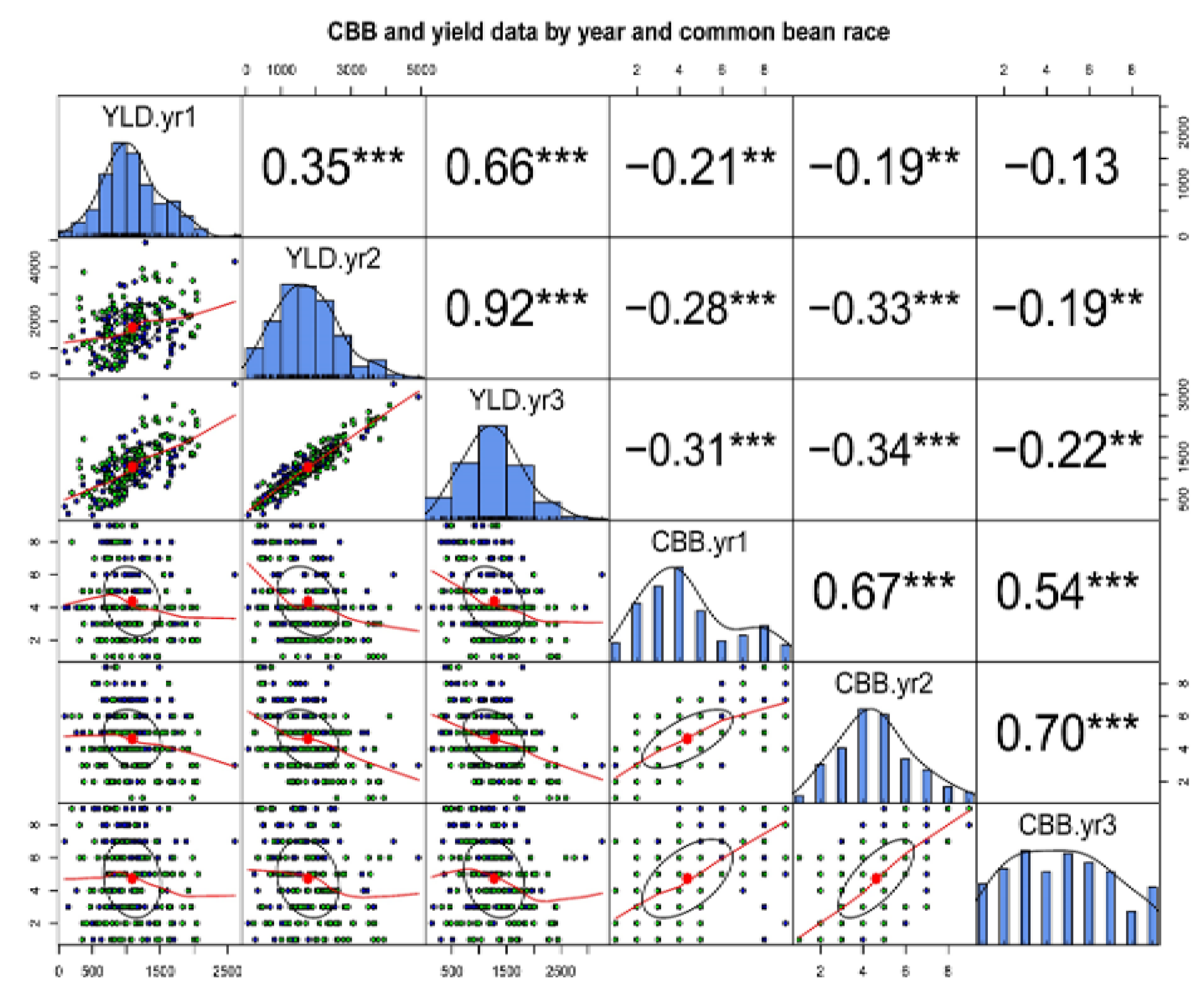
2.2. Correlation between Seasons and Climate Condition
2.3. Differential Response among Bean Races and Commercial Classes for Disease Reaction
2.4. Mean Yield Performance of Commercial Classes
2.5. Greenhouse Validation of CBB Results
2.6. Orthogonal Contrasts and Determination of Best Genotypes
2.7. Principal Component and Phylogenetic Tree Analysis
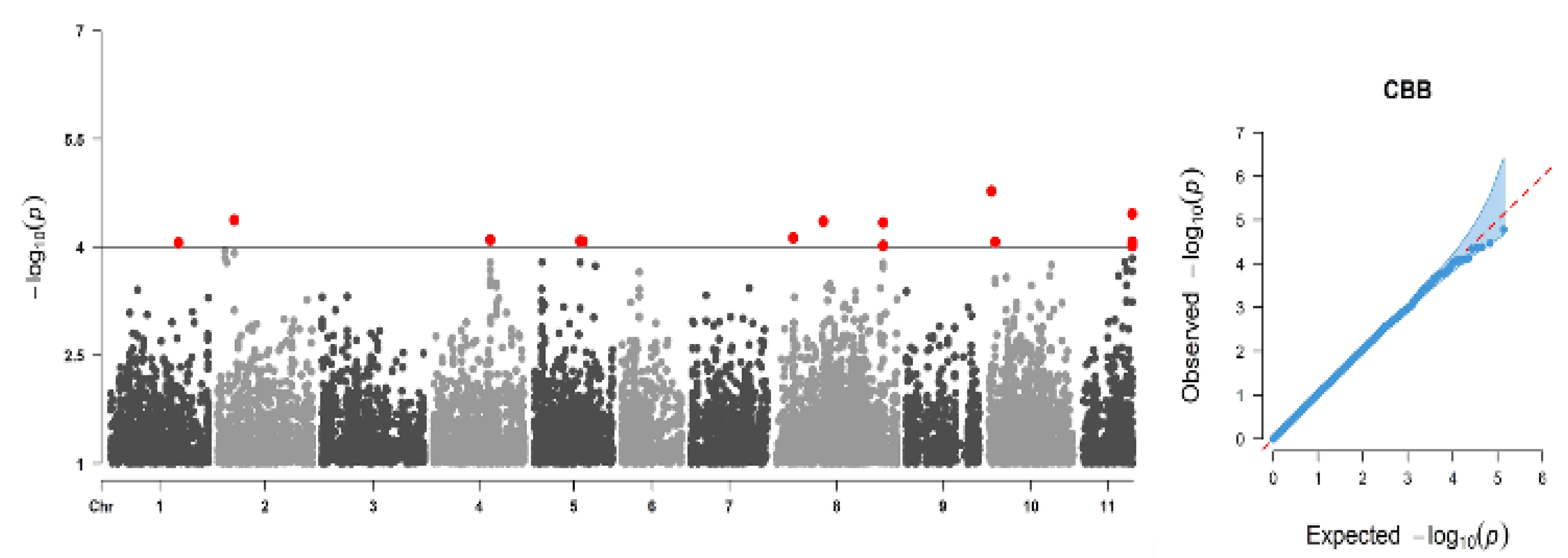
2.8. Genome-Wide Association Analysis Result and Associated Genes
3. Discussion
4. Materials and Methods
4.1. Experimental Site Description
4.2. Plant Materials
4.3. Field Trial Management and Design
4.4. Plant Trait Measurements
4.5. Greenhouse Evaluation with Field Strain
4.6. Phenotypic Data Statistical Analysis
4.7. Genotypic Data Analysis, Genotype Relationships, and Principal Component Analysis
4.8. Genome-Wide Association Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brouwer, B.; Winkler, L.; Atterberry, K.; Jones, S.; Miles, C. Exploring the role of local heirloom germplasm in expanding western Washington dry bean production. Agroecol. Sustain. Food Syst. 2016, 40, 319–332. [Google Scholar] [CrossRef]
- Hagerty, C.H.; Cuesta-Marcos, A.; Cregan, P.; Song, Q.; McClean, P.; Myers, J.R. Mapping snap bean pod and color traits, in a dry bean × snap bean recombinant inbred population. J. Am. Soc. Hortic. Sci. 2016, 141, 131–138. [Google Scholar] [CrossRef]
- Singh, B.K.; Singh, B. Breeding perspectives of snap bean (Phaseolus vulgaris L.). Veg. Sci 2015, 42, 1–17. [Google Scholar]
- Wallace, L.; Arkwazee, H.; Vining, K.; Myers, J.R. Genetic diversity within snap beans and their relation to dry beans. Genes 2018, 9, 587. [Google Scholar] [CrossRef]
- NASS. United States Department of Agriculture National Agricultural Statistics Service. 2019. [Google Scholar]
- NASS. United States Department of Agriculture-National Agricultural Statistics Serviceitle. 2020. [Google Scholar]
- Kelly, J.D. The Story of Bean Breeding White Paper Prepared for BeanCAP & PBG Works on the Topic of Dry Bean Production and Breeding Research in the U.S. Michigan; Michigan State University: East Lansing, MI, USA, 2010; Available online: https://www.canr.msu.edu/beanbreeding/_pdf/Story_of_Bean_Breeding_in_the_US.pdf (accessed on 31 March 2021).
- Blair, M.; Wu, X.; Bhandari, D.; Zhang, X.; Hao, J. Role of Legumes for and as Horticultural Crops in Sustainable Agriculture. In Organic Farming for Sustainable Agriculture Sustainable Development and Biodiversity; Nandwani, D., Ed.; Springer: Cham, Switzerland, 2016; pp. 185–211. [Google Scholar]
- Bost, S.; Canaday, C.; Hale, F.; Hensley, D.; Wszelaki, A. Crop Profile for Beans (Snap) in Tennessee; University of Tennessee: Knoxville, TN, USA, 2013. [Google Scholar]
- Harveson, R.M.; Schwartz, H.F.; Urrea, C.A.; Yonts, C.D. Bacterial wilt of dry-edible beans in the central high plains of the U.S.: Past, present, and future. Plant Dis. 2015, 99, 1665–1677. [Google Scholar] [CrossRef]
- Akhavan, A.; Bahar, M.; Askarian, H.; Lak, M.R.; Nazemi, A.; Zamani, Z. Bean common bacterial blight: Pathogen epiphytic life and effect of irrigation practices. SpringerPlus 2013, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Miklas, P.N.; Coyne, D.P.; Grafton, K.F.; Mutlu, N.; Reiser, J.; Lindgren, D.T.; Singh, S.P. A major QTL for common bacterial blight resistance derives from the common bean great northern landrace cultivar Montana No. 5. Euphytica 2003, 131, 137–146. [Google Scholar] [CrossRef]
- Miklas, P.; Kelly, J.; Beebe, S.; Blair, M. Common bean breeding for resistance against biotic and abiotic stresses: From classical to MAS breeding. Euphytica 2006, 147, 105–131. [Google Scholar] [CrossRef]
- Singh, S.P.; Schwartz, H.F. Breeding common bean for resistance to diseases: A review. Crop. Sci. 2010, 50, 2199–2223. [Google Scholar] [CrossRef]
- Bull, C.T.; Coutinho, T.A.; Denny, T.P.; Firrao, G.; Fischer-Le Saux, M.; Li, X.; Saddler, G.S.; Scortichini, M.; Stead, D.E.; Takikawa, Y. List of new names of plant pathogenic bacteria (2011–2012). J. Plant Pathol. 2014, 96, 223–226. [Google Scholar]
- Sun, S.; Wang, X.; Zhu, Z.; Wang, B.; Wang, M. Occurrence of Charcoal Rot Caused by Macrophomina phaseolina, an Emerging Disease of Adzuki Bean in China. J. Phytopathol. 2016, 164, 212–216. [Google Scholar] [CrossRef]
- Chi-Chim, W.A.; Hernández-Delgado, S.; González-Prieto, J.M.; Segura-Cabrera, A.; Mayek-Pérez, N. Phylogenetic analysis of Macrophomina phaseolina (Tassi) Goid. from common beans and other hosts. In Annual Report of the Bean Improvement Cooperative; USDA: Lincoln, NE, USA, 2017; pp. 19–20. [Google Scholar]
- Shi, C.; Navabi, A.; Yu, K. Association mapping of common bacterial blight resistance QTL in Ontario bean breeding populations. BMC Plant Biol. 2011, 11, 52. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wu, J.; Wang, L.; Blair, M.W.; Zhu, Z.; Wang, S. QTL and candidate genes associated with common bacterial blight resistance in the common bean cultivar Longyundou 5 from China. Crop. J. 2016, 4, 344–352. [Google Scholar] [CrossRef]
- Mutlu, N.; Vidaver, A.K.; Coyne, D.P.; Steadman, J.R.; Lambrecht, P.A.; Reiser, J. Differential pathogenicity of Xanthomonas campestris pv. phaseoli and X. fuscans subsp. fuscans strains on bean genotypes with common blight resistance. Plant Dis. 2008, 92, 546–554. [Google Scholar] [CrossRef]
- Mayék-Pérez, N.; López-Castañeda, C.; González-Chavira, M.; Garcia-Espinosa, R.; Acosta-Gallegos, J.; de la Vega, O.M.; Simpson, J. Variability of Mexican isolates of Macrophomina phaseolina based on pathogenesis and AFLP genotype. Physiol. Mol. Plant. Pathol. 2001, 59, 257–264. [Google Scholar] [CrossRef]
- Reyes-Franco, M.C.; Hernández-Delgado, S.; Beas-Fernández, R.; Medina-Fernández, M.; Simpson, J.; Mayek-Pérez, N. Pathogenic and genetic variability within Macrophomina phaseolina from Mexico and other countries. J. Phytopathol. 2006, 154, 447–453. [Google Scholar] [CrossRef]
- Durham, K.M.; Xie, W.; Yu, K.; Pauls, K.P.; Lee, E.; Navabi, A. Interaction of common bacterial blight quantitative trait loci in a resistant inter-cross population of common bean. Plant Breed. 2013, 132, 658–666. [Google Scholar] [CrossRef]
- Singh, S.; Miklas, P. Breeding common bean for resistance to common blight: A review. Crop. Sci. 2015, 55, 971–984. [Google Scholar] [CrossRef]
- Miklas, P.N.; Singh, S.P.; Terán, H.; Kelly, J.D.; Smith, J.R. Registration of Common Bacterial Blight Resistant Cranberry Dry Bean Germplasm Line USCR-CBB-20. J. Plant Regist. 2011, 5, 98–102. [Google Scholar] [CrossRef]
- Urrea, C.A.; Steadman, J.R.; Pastor-Corrales, M.A.; Lindgren, D.T.; Venegas, J.P. Registration of great northern common bean cultivar “Coyne” with enhanced disease resistance to common bacterial blight and bean rust. J. Plant Regist. 2009, 3, 219–222. [Google Scholar] [CrossRef]
- Beaver, J.S.; Porch, T.G.; Zapata, M. Registration of ‘Badillo’ Light Red Kidney Bean. J. Plant Regist. 2010, 4, 1–4. [Google Scholar] [CrossRef]
- Khanal, R.; Smith, T.H.; Michaels, T.E.; Pauls, K.P. Lighthouse common bean. Can. J. Plant Sci. 2017, 97, 165–168. [Google Scholar] [CrossRef][Green Version]
- Kelly, J.D.; Varner, G.V.; Miklas, P.N.; Cichy, K.A.; Wright, E.M. Registration of ‘Cayenne’ Small Red Bean. J. Plant Regist. 2018, 12, 194–198. [Google Scholar] [CrossRef]
- Urrea, C.A.; Hurtado-Gonzales, O.P.; Pastor-Corrales, M.A.; Steadman, J.R. Registration of Great Northern Common Bean Cultivar ‘Panhandle Pride’ with Enhanced Disease Resistance to Bean Rust and Common Bacterial Blight. J. Plant Regist. 2019, 13, 311–315. [Google Scholar] [CrossRef]
- Tar’an, B.; Michaels, T.E.; Pauls, K.P. Mapping genetic factors affecting the reaction to Xanthomonas axonopodis pv. phaseoli in Phaseolus vulgaris L. under field conditions. Genome 2001, 44, 1046–1056. [Google Scholar] [CrossRef]
- Miklas, P.N.; Fourie, D.; Chaves, B.; Chirembe, C. Common bacterial blight resistance QTL BC420 and SU91 effect on seed yield, seed weight, and canning quality in dry bean. Crop. Sci. 2017, 57, 802–811. [Google Scholar] [CrossRef]
- Liu, S.; Yu, K.; Park, S.J. Development of STS markers and QTL validation for common bacterial blight resistance in common bean. Plant Breed. 2008, 127, 62–68. [Google Scholar] [CrossRef]
- Viteri, D.M.; Cregan, P.B.; Trapp, J.J.; Miklas, P.N.; Singh, S.P. A New Common Bacterial Blight Resistance QTL in VAX 1 Common Bean and Interaction of the New QTL, SAP6, and SU91 with Bacterial Strains. Crop. Sci. 2014, 54, 1598–1608. [Google Scholar] [CrossRef]
- Viteri, D.M.; Terán, H.; Asensio-S.-Manzanera, M.C.; Asensio, C.; Porch, T.G.; Miklas, P.N.; Singh, S.P. Progress in Breeding Andean Common Bean for Resistance to Common Bacterial Blight. Crop. Sci. 2014, 54, 2084–2092. [Google Scholar] [CrossRef]
- Zapata, M.; Beaver, J.S.; Porch, T.G. Dominant gene for common bean resistance to common bacterial blight caused by Xanthomonasaxonopodis pv. phaseoli. Euphytica 2011, 179, 373–382. [Google Scholar] [CrossRef]
- Pastor-Corrales, M.A. Reactions of Selected Bean Accessions to Infection by Macrophomina phaseolina. Plant Dis. 1988, 72, 39. [Google Scholar] [CrossRef]
- Olaya, G.; Abawi, G.S.; Weeden, N.F. Inheritance of the resistance to Macrophomina phaseolina and identification of RAPD markers linked to the resistance genes in beans. Phytopathology 1996, 86, 674–679. [Google Scholar] [CrossRef]
- Miklas, P.; Stone, V.; Urrea, C.A.; Johnson, E.; Beaver, J.S. Inheritance and QTL Analysis of Field Resistance to Ashy Stem Blight in Common Bean. Crop. Sci. 1998, 38, 916–921. [Google Scholar] [CrossRef]
- Hernandez-Delgado, S.; Reyes-Valdés, M.H.; Rosales-Serna, R.; Mayek-Perez, N. Molecular markers associated with resistance to Macrophomina phaseolina (Tassi) Goid. In common bean. J. Plant Pathol. 2009, 91, 163–170. [Google Scholar]
- Álvarez, M.F.; Mosquera, T.; Blair, M.W. The Use of Association Genetics Approaches in Plant Breeding New Crop Species. Plant. Breed. Rev. 2015, 38, 17–65. [Google Scholar]
- Kamfwa, K.; Cichy, K.A.; Kelly, J.D. Genome-Wide Association Study of Agronomic Traits in Common Bean. Plant Genome 2015, 8, 1–12. [Google Scholar] [CrossRef]
- Moghaddam, S.M.; Mamidi, S.; Osorno, J.M.; Lee, R.; Brick, M.; Kelly, J.; Miklas, P.; Urrea, C.; Song, Q.; Cregan, P.; et al. Genome-Wide Association Study Identifies Candidate Loci Underlying Agronomic Traits in a Middle American Diversity Panel of Common Bean. Plant Genome 2016, 9, 1–21. [Google Scholar] [CrossRef]
- Zuiderveen, G.H.; Padder, B.A.; Kamfwa, K.; Song, Q.; Kelly, J.D. Genome-Wide association study of anthracnose resistance in andean beans (Phaseolus vulgaris). PLoS ONE 2016, 11, e0156391. [Google Scholar] [CrossRef]
- Perseguini, J.M.K.C.; Oblessuc, P.R.; Rosa, J.R.B.F.; Gomes, K.A.; Chiorato, A.F.; Carbonell, S.A.M.; Garcia, A.A.F.; Vianello, R.P.; Benchimol-Reis, L.L. Genome-Wide Association Studies of Anthracnose and Angular Leaf Spot Resistance in Common Bean (Phaseolus vulgaris L.). PLoS ONE 2016, 11, e0150506. [Google Scholar] [CrossRef]
- Oladzad, A.; Zitnick-Anderson, K.; Jain, S.; Simons, K.; Osorno, J.M.; McClean, P.E.; Pasche, J.S. Genotypes and Genomic Regions Associated With Rhizoctonia solani Resistance in Common Bean. Front. Plant Sci. 2019, 10, 956. [Google Scholar] [CrossRef]
- Wen, L.; Chang, H.-X.X.; Brown, P.J.; Domier, L.L.; Hartman, G.L. Genome-wide association and genomic prediction identifies soybean cyst nematode resistance in common bean including a syntenic region to soybean Rhg1 locus. Hortic. Res. 2019, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Li, M.; Guo, X.; Tang, M.; Cao, J.; Wang, Z.; Liu, R.; Zhu, K.; Guo, L.; Liu, S.; et al. Arabidopsis GDSL1 overexpression enhances rapeseed Sclerotinia sclerotiorum resistance and the functional identification of its homolog in Brassica napus. Plant Biotechnol. J. 2019, 18, 1255–1270. [Google Scholar] [CrossRef]
- Kim, H.G.; Kwon, S.J.; Jang, Y.J.; Nam, M.H.; Chung, J.H.; Na, Y.-C.; Guo, H.; Park, O.K. GDSL LIPASE1 Modulates Plant Immunity through Feedback Regulation of Ethylene Signaling. Plant Physiol. 2013, 163, 1776–1791. [Google Scholar] [CrossRef]
- Cunha, C.; Carvalho, A.; Esposito, A.; Bistoni, F.; Romani, L. DAMP signaling in fungal infections and diseases. Front. Immunol. 2012, 3, 286. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, H.; Ma, M.; Li, Q.; Kong, D.; Sun, J.; Ma, X.; Wang, B.; Chen, C.; Xie, Y.; et al. Arabidopsis FHY3 and FAR1 Regulate the Balance between Growth and Defense Responses under Shade Conditions. Plant Cell 2019, 31, 2089–2106. [Google Scholar] [CrossRef]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Raffaele, S.; Leger, A.; Roby, D. Very long chain fatty acid and lipid signaling in the response of plants to pathogens. Plant Signal. Behav. 2009, 4, 94–99. [Google Scholar] [CrossRef]
- Reiner, T.; Hoefle, C.; Hückelhoven, R. A barley SKP1-like protein controls abundance of the susceptibility factor RACB and influences the interaction of barley with the barley powdery mildew fungus. Mol. Plant Pathol. 2016, 17, 184–195. [Google Scholar] [CrossRef]
- Figueiredo, J.; Sousa Silva, M.; Figueiredo, A. Subtilisin-like proteases in plant defence: The past, the present and beyond. Mol. Plant Pathol. 2018, 19, 1017–1028. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.; Monteiro, F.; Sebastiana, M. Subtilisin-like proteases in plant–pathogen recognition and immune priming: A perspective. Front. Plant Sci. 2014, 5, 1–4. [Google Scholar] [CrossRef]
- Cabot, C.; Martos, S.; Llugany, M.; Gallego, B.; Tolrà, R.; Poschenrieder, C. A Role for Zinc in Plant Defense against Pathogens and Herbivores. Front. Plant Sci. 2019, 10, 1171. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-K.; Park, J.M.; Joung, Y.H.; Lee, S.; Chung, E.; Kim, S.Y.; Yu, S.H.; Choi, D. A plant EPF-type zinc-finger protein, CaPIF1, involved in defense against pathogens. Mol. Plant Pathol. 2005, 6, 269–285. [Google Scholar] [CrossRef]
- Mutlu, N.; Miklas, P.; Reiser, J.; Coyne, D.; Miklas, P.; Coyne, D. Backcross breeding for improved resistance to common bacterial blight in pinto bean (Phaseolus vulgaris L.). Plant Breed. 2005, 124, 282–287. [Google Scholar] [CrossRef]
- Mutlu, N.; Miklas, P.; Steadman, J.; Vidaver, A.; Lindgren, D.; Reiser, J.; Coyne, D.; Pastor-Corrales, M. Registration of Common Bacterial Blight Resistant Pinto Bean Germplasm Line ABCP-8. Crop. Sci. 2005, 45, 806–807. [Google Scholar] [CrossRef]
- Yu, K.; Shi, C.; Zhang, B. Development and Application of Molecular Markers to Breed Common Bean (Phaseolus vulgaris L.) for Resistance to Common Bacterial Blight (CBB)—Current Status and Future Directions. Appl. Photosynth. 2012. [Google Scholar] [CrossRef][Green Version]
- Miklas, P.N.; Smith, J.R.; Singh, S.P. Registration of Common Bacterial Blight Resistant Dark Red Kidney Bean Germplasm Line USDK-CBB-15. Crop. Sci. 2006, 46, 1005. [Google Scholar] [CrossRef]
- Miklas, P.N.; Smith, J.R.; Riley, R.; Grafton, K.F.; Singh, S.P.; Jung, G.; Coyne, D. Marker -assisted breeding for pyramided resistance to common bacterial blight in common bean. BIC Annu. Rep. 2000, 43, 39–40. [Google Scholar]
- Miklas, P.N.; Zapata, M.; Beaver, J.S.; Grafton, K.F. Registration of Four Dry Bean Germplasms Resistant to Common Bacterial Blight: ICB-3, ICB-6, ICB-8, and ICB-10. Crop. Sci. 1999, 39, 594. [Google Scholar] [CrossRef]
- Porch, T.; Beaver, J.; Debouck, D.; Jackson, S.; Kelly, J.; Dempewolf, H. Use of Wild Relatives and Closely Related Species to Adapt Common Bean to Climate Change. Agronomy 2013, 3, 433–461. [Google Scholar] [CrossRef]
- Mutlu, N.; Urrea, C.A.; Miklas, P.N.; Pastor-Corrales, M.A.; Steadman, J.R.; Lindgren, D.T.; Reiser, J.; Vidaver, A.K.; Coyne, D.P. Registration of Common Bacterial Blight, Rust and Bean Common Mosaic Resistant Great Northern Common Bean Germplasm Line ABC-Weihing. J. Plant Regist. 2008, 2, 53–55. [Google Scholar] [CrossRef]
- Boersma, J.G.; Conner, R.L.; Balasubramanian, P.M.; Navabi, A.; Yu, K.; Hou, A. Combining resistance to common bacterial blight, anthracnose, and bean common mosaic virus into Manitoba-adapted dry bean (Phaseolus vulgaris L.) cultivars. Can. J. Plant Sci. 2014, 94, 405–415. [Google Scholar] [CrossRef]
- Ariyarathne, H.; Coyne, D.; Jung, G.; Skroch, P.; Vidaver, A.; Steadman, J.; Miklas, P.; Bassett, M. Molecular Mapping of Disease Resistance Genes for Halo Blight, Common Bacterial Blight, and Bean Common Mosaic Virus in a Segregating Population of Common Bean. J. Am. Soc. Hortic. Sci. 1999, 124, 654–662. [Google Scholar] [CrossRef]
- Miklas, P.N.; Fourie, D.; Trapp, J.; Davis, J.; Myers, J.R. New loci including Pse-6 conferring resistance to halo bacterial blight on chromosome Pv04 in common bean. Crop. Sci. 2014, 54, 2099–2108. [Google Scholar] [CrossRef]
- Osorno, J.M.; Vander Wal, A.J.; Posch, J.; Simons, K.; Grafton, K.F.; Pasche, J.S.; Valentini, G.; Pastor-Corrales, M. A new black bean with resistance to bean rust: Registration of ‘ND Twilight’. J. Plant Regist. 2021, 15, 28–36. [Google Scholar] [CrossRef]
- Oladzad, A.; Porch, T.; Rosas, J.C.; Moghaddam, S.M.; Beaver, J.; Beebe, S.E.; Burridge, J.; Jochua, C.N.; Miguel, M.A.; Miklas, P.N.; et al. Single and Multi-trait GWAS Identify Genetic Factors Associated with Production Traits in Common Bean Under Abiotic Stress Environments. G3 Genes Genomes Genet. 2019, 9, 1881–1892. [Google Scholar]
- Coyne, D.; Gonzales, J.; Reiser, J.; Sutton, L.; Nuland, D.; Carlson, C.; Lindgren, D.; Steadman, J.; Smith, D.; Schild, J.; et al. ‘Weihing’ Great Northern Disease-resistant Dry Bean. HortScience 2000, 35, 310–312. [Google Scholar] [CrossRef]
- Michaels, T.E.; Smith, T.H.; Larsen, J.; Beattie, A.D.; Pauls, K.P. OAC Rex common bean. Can. J. Plant Sci. 2006, 86, 733–736. [Google Scholar] [CrossRef]
- Federer, W.T.; Crossa, J.J.; Lu, S.I. 4 screening experimental designs for quantitative trait loci, association mapping, genotype-by environment interaction, and other investigations. Front. Physiol. 2012, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Castellamos, G.; Jara, C.E.; Mosquera Cifuentes, G.M. Bean Pathogens: Practical Guide for Lab and Greenhouse Work; Centro Internacional de Agricultura Tropical (CIAT): Cali, Colombia, 2016. [Google Scholar]
- Aritua, V.; Harrison, J.; Sapp, M.; Buruchara, R.; Smith, J.; Studholme, D.J. Genome sequencing reveals a new lineage associated with lablab bean and genetic exchange between Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans. Front. Microbiol. 2015, 6, 1080. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Gupta, V.K. A note on augmented designs. Indian JPl Genet. Resour. 2000, 13, 53–58. [Google Scholar]
- Revelle, W.R. Psych: Procedures for Personality and Psychological Research; Northwestern University: Evanston, IL, USA, 2020. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; p. 2. [Google Scholar]
- Kassambara, A. easyGgplot2: Perform and Customize Easily a Plot with ggplot2, R package version 1.0.9000; 2014. Available online: http://www.sthda.com (accessed on 31 March 2021).
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinform. Appl. 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Z. GAPIT Version 3: An Interactive Analytical Tool for Genomic Association and Prediction. bioRxiv 2021. [Google Scholar] [CrossRef]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Clustering Method: Clustering Criterion and Agglomerative Algorithm. arXiv 2011, arXiv:1111.6285. [Google Scholar]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Xavier, A.; Xu, S.; Muir, W.M.; Rainey, K.M. Genetics and population analysis NAM: Association studies in multiple populations. Bioinformatics 2015, 31, 3862–3864. [Google Scholar]
- Wickham, H.; François, R.; Henry, L.; Müller, K. dplyr: A Grammar of Data Manipulation, R Package Version 0.3.0.2; 2020. Available online: https://dplyr.tidyverse.org/reference/dplyr-package.html (accessed on 31 March 2021).
- Jombart, T.; Ahmed, I.; Bateman, A. Adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- Galili, T. Dendextend: An R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Genome analysis circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Yin, L. Package ‘CMplot’. 2020, pp. 1–8. Available online: https://github.com/YinLiLin/CMplot (accessed on 31 March 2021).
- Gao, X.; Starmer, J.; Martin, E.R. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol. 2008, 32, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; E McClean, P.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2011, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]


| Source | df | CBB | CR | Seed Yield |
|---|---|---|---|---|
| Block | 9 | 0.78 | 0.02 | 1744.29 |
| Entries | 211 | 6.64 ** | 0.11 ** | 310,800.47 ** |
| Test genotypes | 207 | 5.34 ** | 0.10 ** | 307,463.3 ** |
| Controls | 3 | 96.69 ** | 0.52 ** | 134,277.37 ** |
| Test vs Controls | 1 | 5.69 * | 0.31 | 1,262,515.75 ** |
| Error | 27 | 0.83 | 0.05 | 819.96 |
| Corrected Total | 247 |
| TRAIT | YEAR | Source of Variance (df) | Mean Square | Error | Variance Component | Percentage of Total Variance |
|---|---|---|---|---|---|---|
| Seed Yield (kg ha−1) | Year_1 | RACE (1) | 52,806 | CLASS | −9960.07 | 0 |
| CLASS (8) | 570,449 ** | Error | 24,191 | 12.69 | ||
| Error (201) | 166,757 | 166,757 | 87.33 | |||
| Year_2 | RACE (1) | 5,097,230.7 | CLASS | −50,466 | 0 | |
| CLASS (8) | 4,840,309.78 ** | Error | 253,244 | 29.19 | ||
| Error (201) | 614,247.5 | 614,248 | 70.81 | |||
| Year_3 | RACE (1) | 1,545,948.98 | CLASS | −26,236 | 0 | |
| CLASS (8) | 1,994,374.83 ** | Error | 105,248 | 30.66 | ||
| Error (133) | 238,027.7 | 238,028 | 69.34 | |||
| CBB (1–9 scale) | Year_1 | RACE (1) | 82.11 | CLASS | 0.29 | 5.87 |
| CLASS (8) | 24.24 ** | Error | 1.25 | 25.6 | ||
| Error (201) | 3.35 | 3.35 | 68.53 | |||
| Year_2 | RACE (1) | 72.14 | CLASS | 0.05 | 21.36 | |
| CLASS (8) | 29.75 ** | Error | 1.68 | 27.95 | ||
| Error (201) | 1.78 | 1.78 | 50.68 | |||
| Year_3 | RACE (1) | 132.61 | CLASS | 0.51 | 8.1 | |
| CLASS (8) | 36.04 ** | Error | 1.93 | 30.41 | ||
| Error (201) | 3.89 | 3.9 | 61.49 | |||
| CR (1–9) | Year_3 | RACE (1) | 5.66 * | CLASS | 0.04 | 30.77 |
| CLASS (6) | 0.31 ** | Error | 0.01 | 7.69 | ||
| Error (266) | 0.08 | 0.08 | 61.54 |
| Trait | Exp. | Variance Source (df) | Mean Square | Error | Variance Component | % of Total |
|---|---|---|---|---|---|---|
| REP (2) | 12.13 | ERROR | 0.09 | 5.17 | ||
| Common Bacterial Blight (CBB) | Greenhouse | RACE (1) | 4.25 | CLASS | −0.12 | 0.00 |
| CLASS (RACE) (4) | 16.92 ** | GEN | 0.31 | 17.82 | ||
| GEN(RACE*CLASS) (114) | 2.44 ** | ERROR | 0.56 | 32.18 | ||
| Error (238) | 0.08 | 0.78 | 44.83 | |||
| Field | YEAR (2) | 25.69 ** | ERROR | 0.11 | 0.42 | |
| RACE (1) | 48.69 | CLASS | 0.53 | 13.23 | ||
| CLASS(RACE) (8) | 65.56 ** | GEN | 0.27 | 11.61 | ||
| GEN (RACE*CLASS) (201) | 8.36 ** | ERROR | 2.87 | 41.34 | ||
| ERROR (420) | 1.43 | 1.43 | 33.40 | |||
| Seed Yield per Hectare | Field | YEAR (2) | 25,461,147.1 * | ERROR | 119,825.7 | 21.1 |
| RACE (1) | 5,899,456.6 | CLASS | −12,323.3 | 0.00 | ||
| CLASS (RACE) (8) | 5,983,223.5 ** | GEN | 66,726.9 | 11.75 | ||
| GEN (RACE*CLASS) (201) | 710,453.7 ** | ERROR | 203,314.1 | 35.81 | ||
| ERROR (420) | 177,933.9 | 177,933.9 | 31.34 |
| SNP Name | Chr. | Position | p-Value | R2 | Effect | MAF |
|---|---|---|---|---|---|---|
| S10_1437174 | 10 | 1,437,174 | 1.7 × 10−5 | 0.034 | −0.06 | A (0.08) |
| S11_25981923 | 11 | 25,981,923 | 3.4 × 10−5 | 0.031 | 1.38 | T (0.48) |
| S02_8541089 | 2 | 8,541,089 | 4.2 × 10−5 | 0.031 | −0.35 | G (0.49) |
| S08_24383486 | 8 | 24,383,486 | 4.4 × 10−5 | 0.031 | −0.53 | T (0.37) |
| S08_55730740 | 8 | 55,730,740 | 4.6 × 10−5 | 0.031 | 0.91 | T (0.42) |
| S08_8738278 | 8 | 8,738,278 | 7.4 × 10−5 | 0.029 | −2.55 | A (0.50) |
| S04_29756136 | 4 | 29,756,136 | 7.9 × 10−5 | 0.029 | 0.85 | A (0.25) |
| S05_23846983 | 5 | 23,846,983 | 8.3 × 10−5 | 0.029 | 1.39 | G (0.20) |
| S05_25451953 | 5 | 25,451,953 | 8.3 × 10−5 | 0.029 | 0.83 | G (0.10) |
| S11_25981955 | 11 | 25,981,955 | 8.4 × 10−5 | 0.029 | −0.63 | G (0.48) |
| S10_3398159 | 10 | 3,398,159 | 8.5 × 10−5 | 0.029 | 1.33 | C (0.38) |
| S01_35716145 | 1 | 35,716,145 | 8.7 × 10−5 | 0.029 | 0.75 | T (0.26) |
| S08_55730703 | 8 | 55,730,703 | 9.5 × 10−5 | 0.028 | 2.57 | A (0.420 |
| S11_25982002 | 11 | 25,982,002 | 9.7 × 10−5 | 0.028 | −0.58 | A (0.47) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ambachew, D.; Joshua, J.; Mmbaga, M.T.; Blair, M.W. Sources of Resistance to Common Bacterial Blight and Charcoal Rot Disease for the Production of Mesoamerican Common Beans in the Southern United States. Plants 2021, 10, 998. https://doi.org/10.3390/plants10050998
Ambachew D, Joshua J, Mmbaga MT, Blair MW. Sources of Resistance to Common Bacterial Blight and Charcoal Rot Disease for the Production of Mesoamerican Common Beans in the Southern United States. Plants. 2021; 10(5):998. https://doi.org/10.3390/plants10050998
Chicago/Turabian StyleAmbachew, Daniel, Jacqueline Joshua, Margaret T. Mmbaga, and Matthew W. Blair. 2021. "Sources of Resistance to Common Bacterial Blight and Charcoal Rot Disease for the Production of Mesoamerican Common Beans in the Southern United States" Plants 10, no. 5: 998. https://doi.org/10.3390/plants10050998
APA StyleAmbachew, D., Joshua, J., Mmbaga, M. T., & Blair, M. W. (2021). Sources of Resistance to Common Bacterial Blight and Charcoal Rot Disease for the Production of Mesoamerican Common Beans in the Southern United States. Plants, 10(5), 998. https://doi.org/10.3390/plants10050998






