Pan-Genome miRNomics in Brachypodium
Abstract
1. Introduction
2. Results
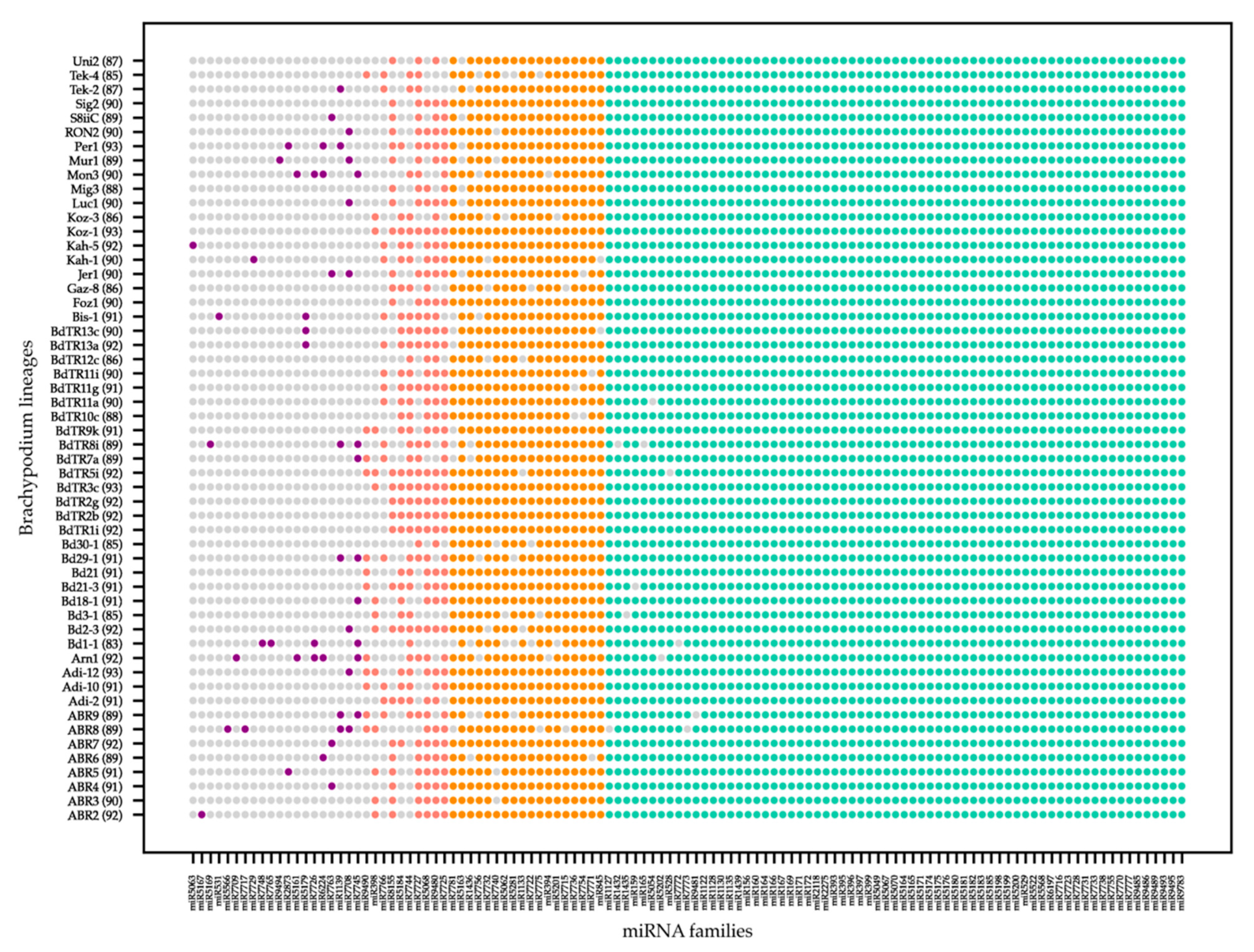
2.1. miRNA Identification Resulted in an Average of 90 miRNA Families in Each Lineage
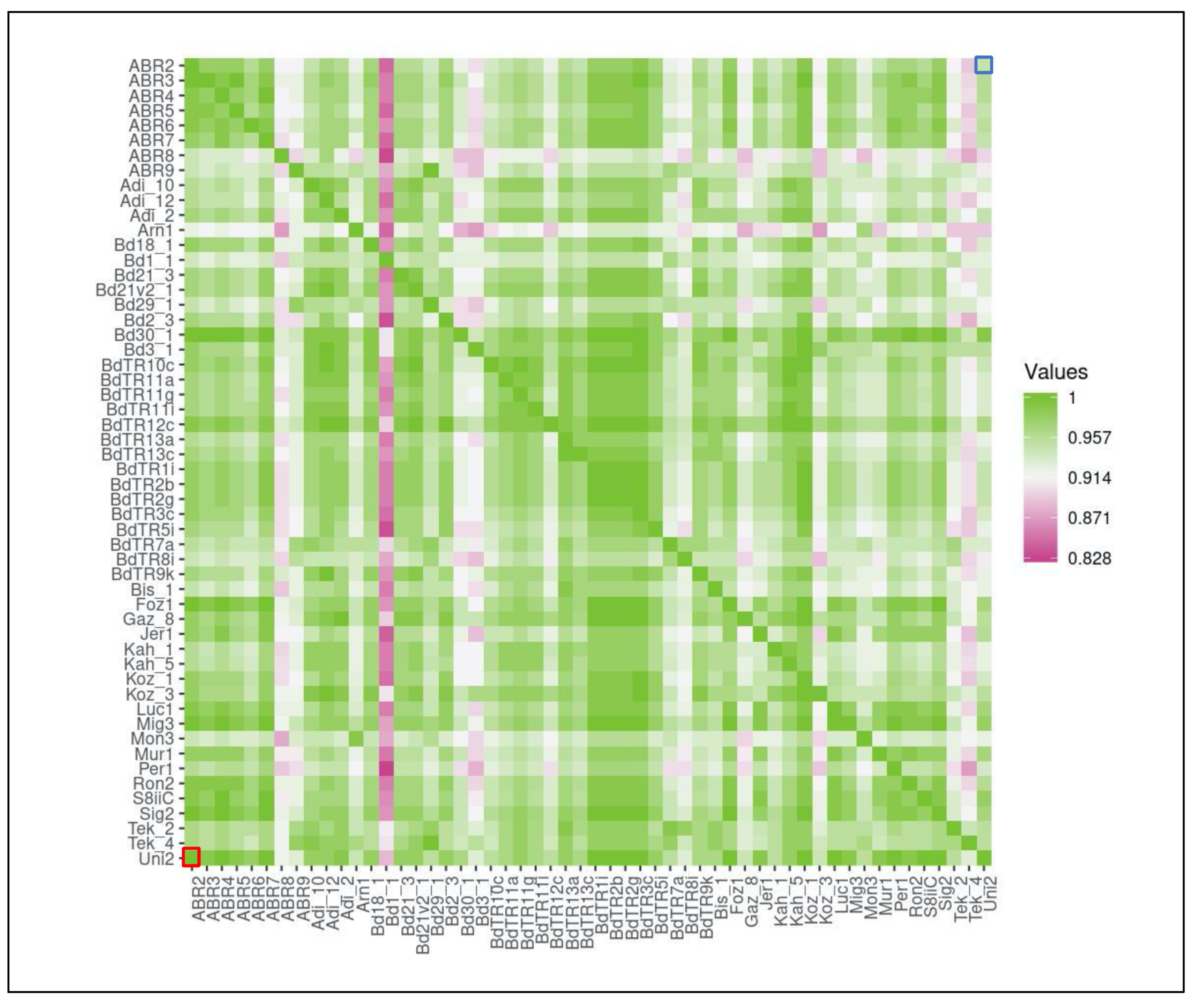
2.2. Pairwise Comparison of Common microRNA Families between Lineages Indicates High Conservation
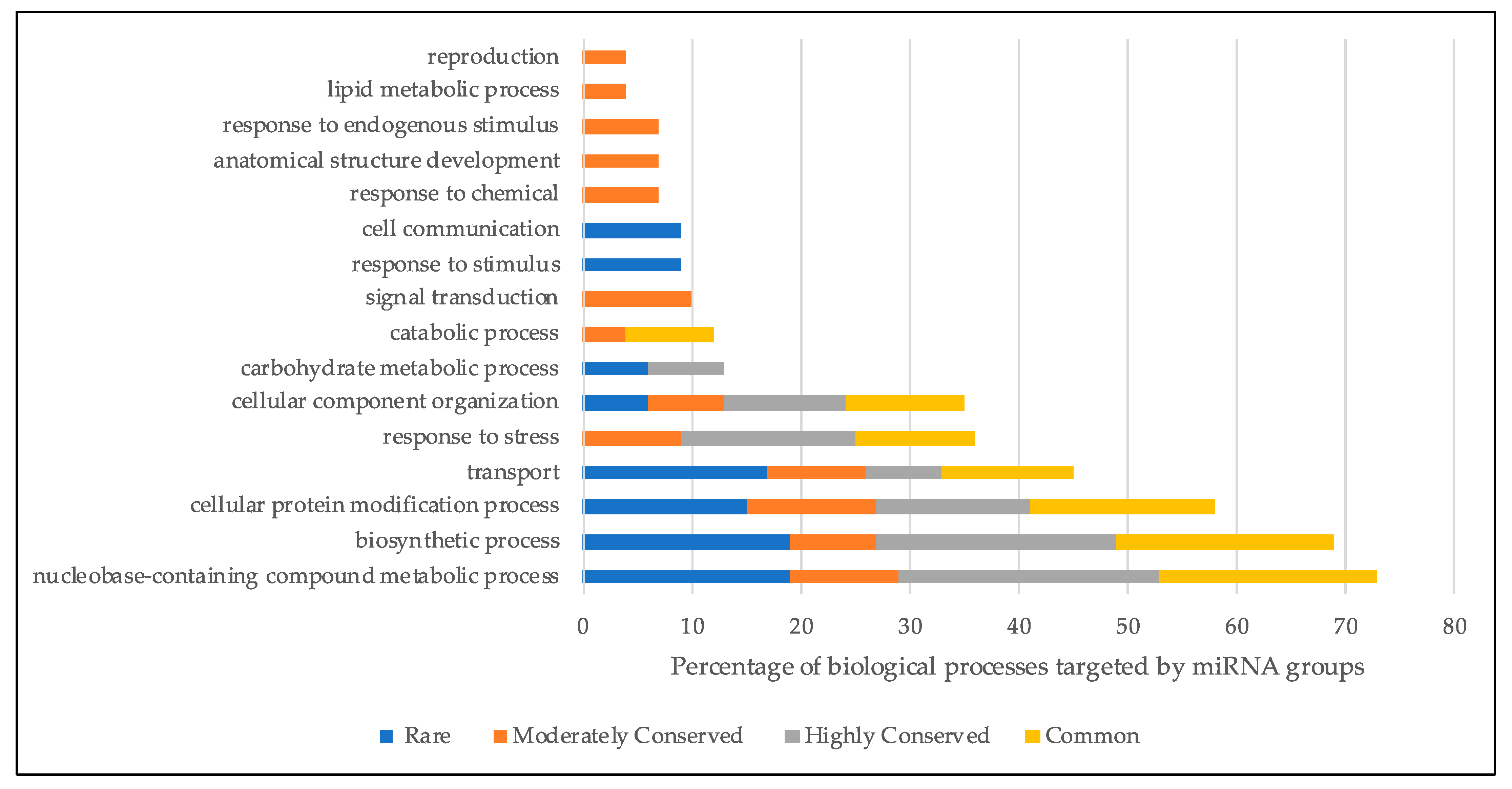
2.3. mRNA Target Analysis Determined the Potential Biological Processes Targeted by Each miRNA Groups
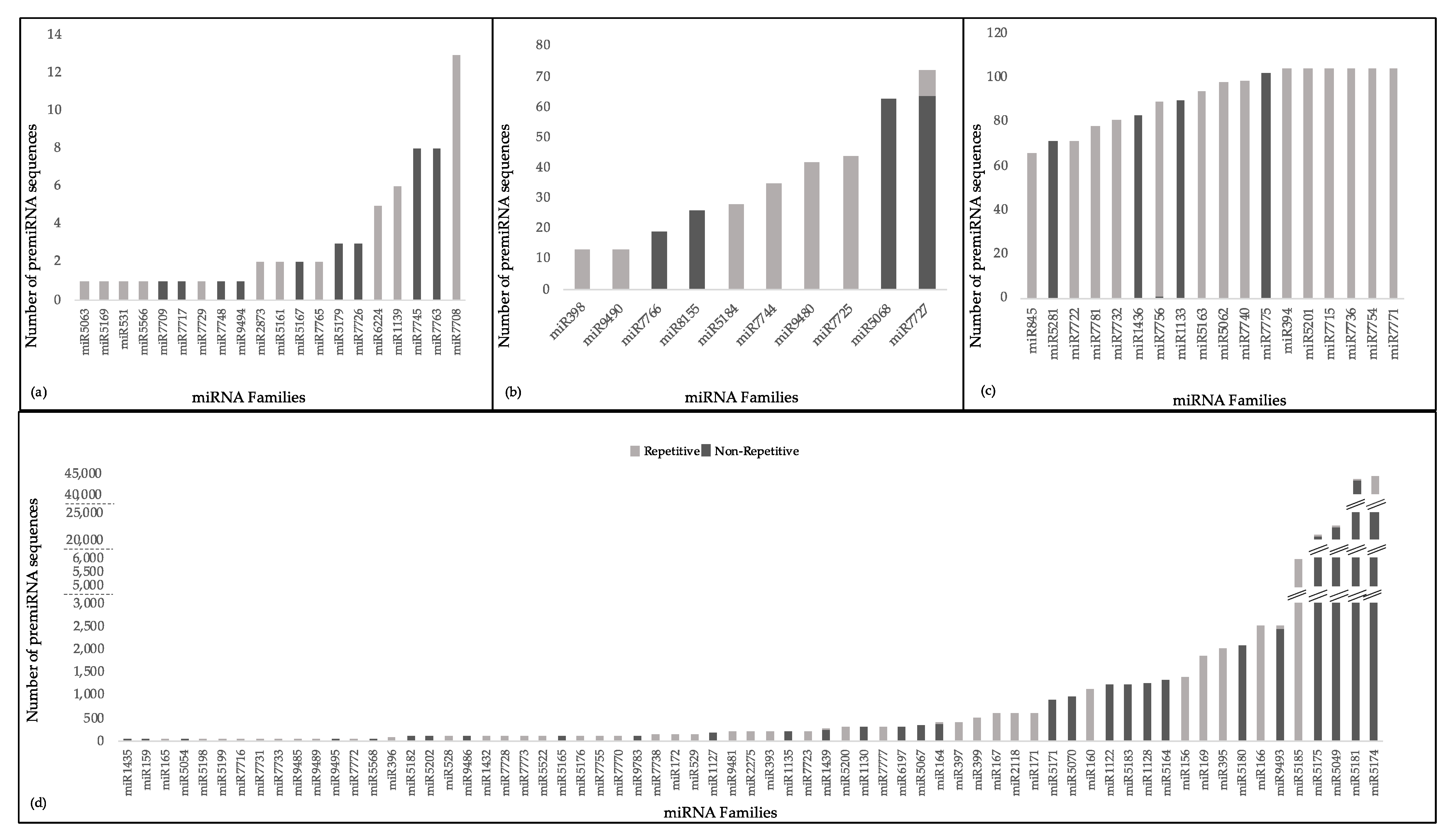
2.4. miRNAs Mostly Target Multiple mRNA Sequences but a Transcript Is Rarely Targeted by Multiple miRNAs
3. Discussion
4. Materials and Methods
4.1. Datasets Used in This Study
4.2. miRNA Identification
4.3. Potential mRNA Target Analysis of Identified miRNAs
4.4. Construction of the Heatmap
4.5. Identification of Repetitive Elements and Conservation among Precursor miRNA Sequences
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Sanger, F.; Coulson, A.R. A Rapid Method for Determining Sequences in DNA by Primed Synthesis with DNA Polymerase. J. Mol. Biol. 1975, 94, 441–448. [Google Scholar] [CrossRef]
- Mardis, E.R. DNA Sequencing Technologies: 2006–2016. Nat. Protoc. 2017, 12, 213–218. [Google Scholar] [CrossRef]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next Generation Sequencing Technologies (and Bioinformatics) in Cancer. Mol. Biol. 2018, 122, 1–15. [Google Scholar] [CrossRef]
- Shendure, J.; Balasubramanian, S.; Church, G.M.; Gilbert, W.; Rogers, J.; Schloss, J.A.; Waterston, R.H. DNA Sequencing at 40: Past, Present and Future. Nature 2017, 550, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Tettelin, H.; Riley, D.; Cattuto, C.; Medini, D. Comparative Genomics: The Bacterial Pan-Genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef]
- Salk, J.J.; Schmitt, M.W.; Loeb, L.A. Enhancing the Accuracy of Next-Generation Sequencing for Detecting Rare and Subclonal Mutations. Nat. Rev. Genet. 2018, 19, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Hurgobin, B.; Edwards, D. SNP Discovery Using a Pangenome: Has the Single Reference Approach Become Obsolete? Biology 2017, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.P.; Contreras-Moreira, B.; Woods, D.P.; des Marais, D.L.; Burgess, D.; Shu, S.; Stritt, C.; Roulin, A.C.; Schackwitz, W.; Tyler, L.; et al. Extensive Gene Content Variation in the Brachypodium Distachyon Pan-Genome Correlates with Population Structure. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Zhao, Q.; Feng, Q.; Lu, H.; Li, Y.; Wang, A.; Tian, Q.; Zhan, Q.; Lu, Y.; Zhang, L.; Huang, T.; et al. Pan-Genome Analysis Highlights the Extent of Genomic Variation in Cultivated and Wild Rice. Nat. Genet. 2018, 50, 278–284. [Google Scholar] [CrossRef]
- Golicz, A.A.; Bayer, P.E.; Barker, G.C.; Edger, P.P.; Kim, H.R.; Martinez, P.A.; Chan, C.K.K.; Severn-Ellis, A.; McCombie, W.R.; Parkin, I.A.P.; et al. The Pangenome of an Agronomically Important Crop Plant Brassica oleracea. Nat. Commun. 2016, 7, 1–8. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome Analysis of Multiple Pathogenic Isolates of Streptococcus Agalactiae: Implications for the Microbial “Pan-Genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef]
- Akpinar, B.A.; Avsar, B.; Lucas, S.J.; Budak, H. Plant Abiotic Stress Signaling. Plant Signal. Behav. 2012, 7, 1450–1455. [Google Scholar] [CrossRef]
- Rasool, S.; Ahmad, P.; Rehman, M.U.; Arif, A.; Anjum, N.A. Achieving Crop Stress Tolerance and Improvement—An Overview of Genomic Techniques. Appl. Biochem. Biotechnol. 2015, 177, 1395–1408. [Google Scholar] [CrossRef] [PubMed]
- Waititu, J.K.; Zhang, C.; Liu, J.; Wang, H. Plant Non-Coding Rnas: Origin, Biogenesis, Mode of Action and Their Roles in Abiotic Stress. Int. J. Mol. Sci. 2020, 21, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Akpinar, B.A. Plant MiRNAs: Biogenesis, Organization and Origins. Funct. Integr. Genom. 2015, 15, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific Effects of MicroRNAs on the Plant Transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef]
- Sakaguchi, J.; Watanabe, Y. MiR165/166 and the Development of Land Plants. Dev. Growth Differ. 2012, 54, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Akpinar, B.A.; Yuce, M.; Lucas, S.; Vrána, J.; Burešová, V.; Doležel, J.; Budak, H. Molecular Organization and Comparative Analysis of Chromosome 5B of the Wild Wheat Ancestor Triticum dicoccoides. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef]
- Cheah, B.H.; Nadarajah, K.; Divate, M.D.; Wickneswari, R. Identification of Four Functionally Important MicroRNA Families with Contrasting Differential Expression Profiles between Drought-Tolerant and Susceptible Rice Leaf at Vegetative Stage. BMC Genom. 2015, 16, 1–18. [Google Scholar] [CrossRef]
- Akpinar, B.A.; Budak, H. Dissecting MiRNAs in Wheat D Genome Progenitor, Aegilops tauschii. Front. Plant Sci. 2016, 7, 1–17. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, Z.; Duan, C.; Chen, Y.; Meng, Q.; Wu, J.; Hao, Z.; Wang, Z.; Li, M.; Yong, H.; et al. Dual Transcriptome Analysis Reveals Insights into the Response to Rice Black-Streaked Dwarf Virus in Maize. J. Exp. Bot. 2016, 67, 4593–4609. [Google Scholar] [CrossRef] [PubMed]
- Franke, K.R.; Schmidt, S.A.; Park, S.; Jeong, D.H.; Accerbi, M.; Green, P.J. Analysis of Brachypodium MiRNA Targets: Evidence for Diverse Control during Stress and Conservation in Bioenergy Crops. BMC Genom. 2018, 19, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.H.; Schmidt, S.A.; Rymarquis, L.A.; Park, S.; Ganssmann, M.; German, M.A.; Accerbi, M.; Zhai, J.; Fahlgren, N.; Fox, S.E.; et al. Parallel Analysis of Rna Ends Enhances Global Investigation of Micrornas and Target RNAs of Brachypodium distachyon. Genome Biol. 2013, 14, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. PsRNATarget: A Plant Small RNA Target Analysis Server (2017 Release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-Throughput Functional Annotation and Data Mining with the Blast2GO Suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Manavella, P.A.; Yang, S.W.; Palatnik, J. Keep Calm and Carry on: MiRNA Biogenesis under Stress. Plant J. 2019, 99, 832–843. [Google Scholar] [CrossRef]
- Unver, T.; Budak, H. Conserved Micrornas and Their Targets in Model Grass Species Brachypodium distachyon. Planta 2009, 230, 659–669. [Google Scholar] [CrossRef]
- Baev, V.; Milev, I.; Naydenov, M.; Apostolova, E.; Minkov, G.; Minkov, I.; Yahubyan, G. Implementation of a de novo Genome-Wide Computational Approach for Updating Brachypodium MiRNAs. Genom. 2011, 97, 282–293. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Integrating MicroRNA Annotation and Deep-Sequencing Data. Nucleic Acids Res. 2011, 39 (Suppl. 1), 152–157. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced MiRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Zhang, L.; Zheng, Y.; Jagadeeswaran, G.; Li, Y.; Gowdu, K.; Sunkar, R. Identification and Temporal Expression Analysis of Conserved and Novel MicroRNAs in Sorghum. Genomics 2011, 98, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Kurtoglu, K.Y.; Kantar, M.; Budak, H. New Wheat MicroRNA Using Whole-Genome Sequence. Funct. Integr. Genom. 2014, 14, 363–379. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Meyers, B.C. Revisiting Criteria for Plant MicroRNA Annotation in the Era of Big Data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Narjala, A.; Nair, A.; Tirumalai, V.; Vivek Hari Sundar, G.; Shivaprasad, P.V. A Conserved Sequence Signature Is Essential for Robust Plant MiRNA Biogenesis. Nucleic Acids Res. 2020, 48, 3103–3118. [Google Scholar] [CrossRef]
- Fahlgren, N.; Howell, M.D.; Kasschau, K.D.; Chapman, E.J.; Sullivan, C.M.; Cumbie, J.S.; Givan, S.A.; Law, T.F.; Grant, S.R.; Dangl, J.L.; et al. High-Throughput Sequencing of Arabidopsis MicroRNAs: Evidence for Frequent Birth and Death of MIRNA Genes. PLoS ONE 2007, 2, e219. [Google Scholar] [CrossRef]
- Morea, E.G.O.; da Silva, E.M.; e Silva, G.F.F.; Valente, G.T.; Barrera Rojas, C.H.; Vincentz, M.; Nogueira, F.T.S. Functional and Evolutionary Analyses of the MiR156 and MiR529 Families in Land Plants. BMC Plant Biol. 2016, 16, 1–13. [Google Scholar] [CrossRef]
- Zhang, S.D.; Ling, L.Z.; Zhang, Q.F.; Xu, J.D.; Cheng, L. Evolutionary Comparison of Two Combinatorial Regulators of SBP-Box Genes, MiR156 and MiR529, in Plants. PLoS ONE 2015, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, Q.; Zhang, Y.; Li, Z.; Guo, J.; Chen, X.; Zhang, G. Genome-Wide Identification, Characterization, and Expression Patterns Analysis of the SBP-Box Gene Family in Wheat (Triticum aestivum L.). Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Bartel, D.P. Antiquity of MicroRNAs and Their Targets in Land Plants. Plant Cell 2005, 17, 1658–1673. [Google Scholar] [CrossRef] [PubMed]
- Wójcik, A.M.; Nodine, M.D.; Gaj, M.D. MiR160 and MiR166/165 Contribute to the LEC2-Mediated Auxin Response Involved in the Somatic Embryogenesis Induction in Arabidopsis. Front. Plant Sci. 2017, 8, 1–17. [Google Scholar] [CrossRef]
- Mussig, C.; Biesgen, C.; Lisso, J.; Uwer, U.; Weiler, E.W.; Altmann, T. A Novel Stress-Inducible 12-Oxophytodienoate Reductase from Arabidopsis thaliana Provides a Potential Link between Brassinosteroid-Action and Jasmonic-Acid Synthesis. J. Plant Physiol. 2000, 157, 143–152. [Google Scholar] [CrossRef]
- Wilson, P.B.; Streich, J.C.; Murray, K.D.; Eichten, S.R.; Cheng, R.; Aitken, N.C.; Spokas, K.; Warthmann, N.; Gordon, S.P.; Vogel, J.P.; et al. Global Diversity of the Brachypodium Species Complex as a Resource for Genome-Wide Association Studies Demonstrated for Agronomic Traits in Response to Climate. Genetics 2019, 211, 317–331. [Google Scholar] [CrossRef]
- Filiz, E.; Ozdemir, B.S.; Budak, F.; Vogel, J.P.; Tuna, M.; Budak, H. Molecular, Morphological, and Cytological Analysis of Diverse Brachypodium Distachyon Inbred Lines. Genome 2009, 52, 876–890. [Google Scholar] [CrossRef]
- Gordon, S.P.; Priest, H.; Des Marais, D.L.; Schackwitz, W.; Figueroa, M.; Martin, J.; Bragg, J.N.; Tyler, L.; Lee, C.R.; Bryant, D.; et al. Genome Diversity in Brachypodium Distachyon: Deep Sequencing of Highly Diverse Inbred Lines. Plant J. 2014, 79, 361–374. [Google Scholar] [CrossRef]
- Wu, L.; Liu, D.; Wu, J.; Zhang, R.; Qin, Z.; Liu, D.; Li, A.; Fu, D.; Zhai, W.; Mao, L. Regulation of FLOWERING LOCUS T by a MicroRNA in Brachypodium distachyon. Plant Cell 2013, 25, 4363–4377. [Google Scholar] [CrossRef]
- Koroban, N.V.; Kudryavtseva, A.V.; Krasnov, G.S.; Sadritdinova, A.F.; Fedorova, M.S.; Snezhkina, A.V.; Bolsheva, N.L.; Muravenko, O.V.; Dmitriev, A.A.; Melnikova, N.V. The Role of MicroRNA in Abiotic Stress Response in Plants. Mol. Biol. 2016, 50, 337–343. [Google Scholar] [CrossRef]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, A.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The Genome Portal of the Department of Energy Joint Genome Institute: 2014 Updates. Nucleic Acids Res. 2014, 42, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Kantar, M.; Akpınar, B.A.; Valárik, M.; Lucas, S.J.; Doležel, J.; Hernández, P.; Budak, H. Subgenomic Analysis of MicroRNAs in Polyploid Wheat. Funct. Integr. Genom. 2012, 12, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Kurtoglu, K.Y.; Kantar, M.; Lucas, S.J.; Budak, H. Unique and Conserved MicroRNAs in Wheat Chromosome 5D Revealed by Next-Generation Sequencing. PLoS ONE 2013, 8, e69801. [Google Scholar] [CrossRef] [PubMed]
- Alptekin, B.; Budak, H. Wheat MiRNA Ancestors: Evident by Transcriptome Analysis of A, B, and D Genome Donors. Funct. Integr. Genom. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Akpinar, B.A.; Kantar, M.; Budak, H. Root Precursors of MicroRNAs in Wild Emmer and Modern Wheats Show Major Differences in Response to Drought Stress. Funct. Integr. Genom. 2015, 15, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Markham, N.; Zuker, M. UNAFold: Software for Nucleic Acid Folding and Hybridization. Methods Mol. Biol. 2008, 453, 3–31. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Sen, T.Z.; Budak, H. mirMachine: A One-Stop Shop for Plant miRNA Annotation. J. Vis. Exp. 2021, 171, e62430. [Google Scholar] [CrossRef]
- Lucas, S.J.; Budak, H. Sorting the Wheat from the Chaff: Identifying MiRNAs in Genomic Survey Sequences of Triticum aestivum Chromosome 1AL. PLoS ONE 2012, 7, e40859. [Google Scholar] [CrossRef]
- Dai, X.; Zhao, P.X. PsRNATarget: A Plant Small RNA Target Analysis Server. Nucleic Acids Res. 2011, 39 (Suppl. 2), 155–159. [Google Scholar] [CrossRef]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A Web Server for Clustering and Comparing Biological Sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-Enabled Heat Mapping for All. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. Available online: http://www.repeatmasker.org (accessed on 9 April 2021).
- MAFFT Version 7. Available online: http://mafft.cbrc.jp (accessed on 10 April 2021).

| Magnoliophyta | Coniferophyta | Embryophyta | ||
|---|---|---|---|---|
| Rare | miR1139, miR2873, miR5063, miR5161, miR5167, miR5169, miR5179, miR531, miR5566, miR6224, miR7708, miR7709, miR7717, miR7726, miR7729, miR7745, miR7748, miR7763, miR7765, miR9494 | √ (m) | ||
| Mod. Cons. | miR398 | √ | √ | |
| miR5068, miR5184, miR7725, miR7727, miR7744, miR7766, miR9480, miR9490 | √ (m) | |||
| miR8155 | √ (e) | |||
| Highly Cons. | miR394 | √ | ||
| miR1133, miR1436, miR5062, miR5163, miR5201, miR7715, miR7722, miR7732, miR7736, miR7740, miR7754, miR7756, miR7771, miR7775, miR7781 | √ (m) | |||
| miR5281, miR845 | √ * | |||
| Common | miR1122, miR1127, miR1128, miR1130, miR1135, miR1432, miR1435, miR1439, miR5049, miR5054, miR5067, miR5070, miR5164, miR5165, miR5171, miR5174, miR5175, miR5176, miR5180, miR5181, miR5182, miR5183, miR5185, miR5198, miR5199, miR5200, miR5202, miR528, miR5522, miR5568, miR6197, miR7716, miR7723, miR7728, miR7731, miR7733, miR7738, miR7755, miR7770, miR7772, miR7773, miR7777, miR9481, miR9485, miR9486, miR9489, miR9493, miR9495, miR9783 | √ (m) | ||
| miR156, miR159, miR160, miR166, miR171, miR395, miR396 | √ | √ | √ | |
| miR164, miR169, miR397 | √ | √ | ||
| miR165 | √ (e) | |||
| miR167 | √ | √ | ||
| miR172, miR393 | √ | |||
| miR2118, miR2275, miR399 | √ * | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muslu, T.; Biyiklioglu-Kaya, S.; Akpinar, B.A.; Yuce, M.; Budak, H. Pan-Genome miRNomics in Brachypodium. Plants 2021, 10, 991. https://doi.org/10.3390/plants10050991
Muslu T, Biyiklioglu-Kaya S, Akpinar BA, Yuce M, Budak H. Pan-Genome miRNomics in Brachypodium. Plants. 2021; 10(5):991. https://doi.org/10.3390/plants10050991
Chicago/Turabian StyleMuslu, Tugdem, Sezgi Biyiklioglu-Kaya, Bala Ani Akpinar, Meral Yuce, and Hikmet Budak. 2021. "Pan-Genome miRNomics in Brachypodium" Plants 10, no. 5: 991. https://doi.org/10.3390/plants10050991
APA StyleMuslu, T., Biyiklioglu-Kaya, S., Akpinar, B. A., Yuce, M., & Budak, H. (2021). Pan-Genome miRNomics in Brachypodium. Plants, 10(5), 991. https://doi.org/10.3390/plants10050991







