Morphological Characterization, Variability and Diversity among Vegetable Soybean (Glycine max L.) Genotypes
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Experimental Details
4.2. Biochemical Analysis
4.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shanmugasundaram, S.; Yan, M.R. Vegetable soybeans for nutritional quality income generation and soil sustainability. In Proceedings of the World Soybean Research Conference VI, Chicago, NY, USA, 4–7 August 1999; p. 450. [Google Scholar]
- Mebrahtu, A.T.; Devine, T.E. Combining ability analysis for selected green pod yield components of vegetable soybean genotypes (Glycine max L.). N. Z. J. Crop Hort. Sci. 2010, 36, 97–105. [Google Scholar] [CrossRef]
- Keatinge, J.D.H.; Easdown, W.J.; Yang, R.Y.; Chadha, M.L.; Shanmugasundaram, S. Overcoming chronic malnutrition in a future warming world: The key importance ofmungbean and vegetable soybean. Euphytica 2011, 180, 129–141. [Google Scholar] [CrossRef]
- Fehr, W.R.; Caviness, C.E. Stages of soybean development. In Iowa State University Cooperative Extension Service, Special Report; Iowa State University: Ames, IA, USA, 1977; p. 80. [Google Scholar]
- Masuda, R. Quality requirement and improvement of vegetable soybean.Vegetable Soybean: Research Needs for Production and Quality Improvement. In Proceedings of the a Workshop, Kenting, Taiwan, 29 April–2 May 1991; p. 92. [Google Scholar]
- Mathur, S. Soybean wonder legume. Beverage Food World 2004, 31, 61–62. [Google Scholar]
- Shanmugasundaram, S.; Yan, M.R. Global expansion of high value vegetable soybean. In Proceedings of the 7th World Soybean Research Conference, Foz do Iguassu, Brazil, 29 February–5 March 2004; pp. 915–920. [Google Scholar]
- Mishra, S.; Pancheshwar, D.K.; Singh, P.; Jha, A. study of genetic variability in recently evolved genotypes of soybean (Glycine max (L.) Merill). Trends Biosci. 2014, 8, 5390–5393. [Google Scholar]
- Baraskar, V.V.; Kacchadia, H.V.; Vacchan, J.H.; Barad, H.R.; Patel, M.B.; Darwankar, M.S. Genetic variability, heritability and genetic advance in soybean [Glycine max (L.) Merrill]. Electron. J. Plant Breed. 2014, 5, 802–806. [Google Scholar]
- Kumar, A.; Pandey, A.; Aochen, C.; Pattanayak, A. Evaluation of genetic diversity and interrelationships of agromorphological characters in soybean (Glycine max L.) genotypes. Proc. Natl. Acad. Sci. India B 2015, 85, 397–405. [Google Scholar]
- IBPGR. Disriptors for Soybean. International Board for Plant Genetic Resources; IBPGR Secretariat: Rome, Italy, 1984; pp. 19–38. [Google Scholar]
- Boerma, H.R.; Specht, J.E.; Carlson, J.B.; Lersten, N.R. Reproductive morphology. In Soybeans: Improvement, Production, and Uses, 2nd ed.; Boerma, H.R., Specht, J.E., Eds.; Agronomy Monograph No. 16; American Society of Agronomy/Crop Science Society of America/Soil Science Society of America: Madison, WI, USA, 1987; pp. 95–134. [Google Scholar]
- Lockhart, J.; Candat, A.; Paszkiewicz, G.; Neveu, M.; Gautier, R.; Logan, D.C.; Avelange-Macherel, M.-H.; Macherel, D. Finding Dt2, the Dominant Gene That Specifies the Semideterminate Growth Habit in Soybean. Plant Cell 2014, 26, 2725. [Google Scholar] [CrossRef]
- Kato, S.; Sayama, T.; Taguchi-Shiobara, F.; Kikuchi, A.; Ishimoto, M.; Cober, E. Effect of change from a determinate to a semi-determinate growth habit on the yield and lodging resistance of soybeans in the northeast region of Japan. Breed. Sci. 2019, 69, 151–159. [Google Scholar] [CrossRef]
- Rao, C.R. Advanced Statistical Methods in Biometrical Research; John Wiley and Sons Ltd.: London, UK, 1952; p. 301. [Google Scholar]
- Basavaraja, G.T.; Naidu, G.K.; Salimath, P.M. Evaluation of vegetable soybean genotypes for yield and component traits. Kar. J. Agric. Sci. 2005, 18, 27–31. [Google Scholar]
- Reni, Y.P.; Raob, Y.K. Genetic variability in soybean (Glycine max (L.) Merrill). Inter. J. Plant Anim. Environ. Sci. 2013, 4, 35–38. [Google Scholar]
- Pagde, L.; Abubakkar, D.; Ingole, G.; Dhuppe, M.V. Study of genetic variability for yield and yield contributing traits in soybean (Glycine max (L.) Merrill). Bioinfolet 2015, 12, 256–258. [Google Scholar]
- Mebrahtu, T.; Mohamed, A. Genetic variation for green pod yield and quality among vegetable soybean genotypes. J. Crop Improv. 2006, 16, 113–130. [Google Scholar] [CrossRef]
- Poornima, R.; Koti, R.V.; Nair, R.N. Physiological basis of yield variation in vegetable soybean and organoleptic test for acceptance. Plant Arch. 2014, 14, 51–54. [Google Scholar]
- Ramya, V.; Mummigatti, U.V. Characterization of vegetable soybean genotypes for phenological, physiological and yield attributing traits. Kar. J. Agric. Sci. 2015, 28, 500–503. [Google Scholar]
- Haruna, M.K.; Turaki, Z.G.S.; Bibinu, A.T.S.; Wali, A.S. Soybean varietal evaluation in Northern Guinea Savanna. J. Bio. Agric. Healthcare 2015, 5, 139–142. [Google Scholar]
- Sureshrao, S.S.; Singh, V.J.; Gampala, S.; Rangare, N.R. Assessment of genetic variability of the main yield related characters in soybean. Int. J. Food Agric. Veterin. Sci. 2014, 4, 69–74. [Google Scholar]
- Mahbub, M.M.; Rahman, M.M.; Hossain, S.; Mahmud, F.; Mir, M.M. Genetic variability, correlation and path analysis for yield and yield components in soybean. Am.-Eur. J. Agric. Environ. Sci. 2015, 15, 231–236. [Google Scholar]
- Sharma, B.K.; Kushwah, S.S.; Verma, K.S.; Singh, O.P. Studies on french bean (Phaseolus vulgaris L.) varieties under different N, P, K and S levels for growth, yield and economics. J. Hort. Sci. 2013, 8, 268–270. [Google Scholar]
- Njoroge, J.N.; Owouche, J.O.; Oyoo, M.E. Evaluation of soybean (Glycine max (L.) Merrill) genotypes for agronomic and quality traits in Kenya. Afr. J. Agric. Res. 2015, 10, 1474–1479. [Google Scholar]
- Kuswantoro, H. Genetic variability and heritability of acid-adaptive soybean promising lines. Biodiversitas 2017, 18, 378–382. [Google Scholar] [CrossRef]
- Gohil, V.N.; Pandya, H.M.; Mehta, D.R. Genetic variability for seed yield and its component traits in soybean. Agric. Sci. Digest. 2006, 26, 73–74. [Google Scholar]
- Karnwal, M.K.; Singh, K. Studies on genetic variability, character association and path coefficient for seed yield and its contributing traits in soybean (Glycine max (L.) Merrill). Legume Res. 2009, 32, 70–73. [Google Scholar]
- Dilnesaw, Z.; Abadi, S.; Getahun, A. Genetic variability and heritability of soybean (Glycine max (L.) Merrill) genotypes in Pawe district, Metekelzone, Benishangule-Gumuz regional state, north western Ethiopia. WudpeckerJ. Agric. Res. 2013, 2, 240–245. [Google Scholar]
- Ekka, P.K.; Lal, G.M. Study on genetic variability and character association in soybean (Glycine max (L.) Merrill) germplasm at Vindhyan zone of Uttar Pradesh. Agric. Sci. Digest. 2016, 36, 69–71. [Google Scholar] [CrossRef]
- Manav, A.R.N. Genetic variability studies for yield and seedling traits in soybean (Glycine max (L.) Merrill). Indian Res. J. Genetic Biotech. 2017, 9, 78–110. [Google Scholar]
- Thakur, D.K.; Gendley, T.K.; Tigga, K.; Sharma, A.C. Study on genetic variability, heritability and genetic advance for seed yield and its attributing traits in soybean [Glycine max (L.) Merrill]. Trends Biosci. 2015, 8, 1994–1996. [Google Scholar]
- Sood, V.K.; Sood, V.P.; Pathania, A.; Chandel, K. Exploiting genotypic variability in relation to genetic divergence among advanced lines of soybean (Glycine max (L) Merrill). Indian J. Plant Genet. Res. 2006, 19, 66–69. [Google Scholar]
- Patil, S.S.; Naik, M.R.; Patil, A.B.; Ghodke, U.R. Genetic diversity in soybean. Legume Res. 2011, 34, 68–70. [Google Scholar]
- KAU. Package of Practices Recommendation: Crops, 15th ed.; Kerala Agricultural University: Thrissur, India, 2016; p. 200. [Google Scholar]
- Yemm, E.W.; Willis, A.J. The estimation of carbohydrates in plant extracts by anthrone. Biochem. J. 1954, 57, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the folin-phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Sadasivam, S.; Balasubraminan, T. Practical Manual in Biochemistry; Tamil Nadu Agricultural University: Coimbatore, India, 1987; p. 14. [Google Scholar]
- Sadasivam, S.; Manickam, A. Biochemical Methods for Agricultural Sciences; Wiley Eastern Limited: New Delhi, India, 1992. [Google Scholar]
- Jackson, M.L. Soil Chemical Analysis; Prentice Hall of India Pvt. Ltd.: New Delhi, India, 1973; p. 498. [Google Scholar]
- Piper, C.S. Soil and Plant Analysis; Hans Publishers: Bombay, India, 1966; p. 368. [Google Scholar]
- Panse, V.G.; Sukhatme, P.V. Statistical Methods for Agricultural Workers, 2nd ed.; ICAR: New Delhi, India, 1967; p. 361. [Google Scholar]
- Wricke, G.; Weber, W.E. Quantitative Genetics and Selection in Plant Breeding; Walter de Gruyter and, Co.: Berlin, Germany, 1986. [Google Scholar]
- Shivasubramanian, S.; Menon, N. Heterosis and inbreeding depression in rice. Madras Agric. J. 1973, 60, 1139–1144. [Google Scholar]
- Robinson, H.F.; Comstock, R.E.; Harvey, V.H. Estimates of heritability and degree of dominance in corn. Agron. J. 1949, 41, 353–359. [Google Scholar] [CrossRef]
- Burton, C.W.; Devane, E.H. Estimating heritability in tall Fescue (Festuca arundinaceae) from replicated clonal material. Agron. J. 1953, 45, 478–481. [Google Scholar] [CrossRef]
- Johnson, H.W.; Robinson, H.F.; Comstock, R.E. Estimates of genetic and environmental variability in soybeans. Agron. J. 1955, 47, 314–318. [Google Scholar] [CrossRef]

| Genotypes | Leaf Shape | Leaf Color | Flower Color | Growth Habit | Pod Color | Pod Pubescence | Pod Shape |
|---|---|---|---|---|---|---|---|
| GM-1 | Pointed ovate | Green | Purple | Determinate | Green | Absent | Slightly curved |
| GM-2 | Lanceolate | Dark green | Purple | Semi determinate | Green | Absent | Straight |
| GM-3 | Pointed ovate | Green | White | Semi determinate | Dark green | Absent | Straight |
| GM-4 | Lanceolate | Dark green | Purple | Determinate | Green | Absent | Slightly curved |
| GM-5 | Pointed ovate | Green | Purple | Semi determinate | Green | Absent | Curved |
| GM-6 | Pointed ovate | Green | White | Semi determinate | Green | Absent | Slightly curved |
| GM-7 | Lanceolate | Green | Purple | Indeterminate | Green | Absent | Slightly curved |
| GM-8 | Pointed ovate | Dark green | White | Determinate | Dark green | Absent | Slightly curved |
| GM-9 | Pointed ovate | Green | Purple | Indeterminate | Green | Absent | Slightly curved |
| GM-10 | Pointed ovate | Green | Purple | Determinate | Green | Absent | Slightly curved |
| GM-11 | Round ovate | Dark green | Purple | Determinate | Green | Present | Slightly curved |
| GM-12 | Lanceolate | Green | Purple | Indeterminate | Green | Absent | Curved |
| GM-13 | Round ovate | Dark green | White | Determinate | Green | Absent | Straight |
| GM-14 | Lanceolate | Green | Purple | Determinate | Green | Absent | Straight |
| GM-15 | Lanceolate | Dark green | Purple | Semi determinate | Green | Absent | Slightly curved |
| GM-16 | Round ovate | Green | Purple | Determinate | Green | Present | Straight |
| GM-17 | Pointed ovate | Green | Purple | Indeterminate | Green | Absent | Curved |
| GM-18 | Round ovate | Green | Purple | Determinate | Green | Absent | Slightly curved |
| GM-19 | Lanceolate | Green | Purple | Semi determinate | Green | Present | Curved |
| GM-20 | Lanceolate | Green | Purple | Semi determinate | Green | Absent | Straight |
| GM-21 | Pointed ovate | Green | Purple | Indeterminate | Green | Absent | Slightly curved |
| GM-22 | Round ovate | Dark green | Purple | Determinate | Dark green | Absent | Slightly curved |
| GM-23 | Pointed ovate | Green | Purple | Semi determinate | Green | Absent | Slightly curved |
| GM-24 | Round ovate | Green | Purple | Determinate | Green | Absent | Straight |
| GM-25 | Round ovate | Dark green | Purple | Determinate | Green | Present | Slightly curved |
| GM-26 | Pointed ovate | Dark Green | Purple | Indeterminate | Green | Absent | Curved |
| GM-27 | Pointed ovate | Green | Purple | Indeterminate | Green | Present | Slightly curved |
| GM-28 | Pointed ovate | Green | Purple | Indeterminate | Green | Absent | Curved |
| Sl. No. | Character | GV | PV | GCV (%) | PCV (%) | h2 (%) | GA (%) | GAM (%) |
|---|---|---|---|---|---|---|---|---|
| 1. | Plantheight (cm) | 392.73 | 394.78 | 43.21 | 43.33 | 99.46 | 40.71 | 88.77 |
| 2. | Daysto50% flowering | 12.89 | 14.20 | 10.51 | 11.05 | 90.47 | 7.03 | 20.59 |
| 3 | Daystofirst harvest | 19.87 | 22.93 | 9.79 | 10.54 | 86.23 | 8.52 | 18.73 |
| 4 | Daytovegetable maturity | 2.90 | 3.37 | 13.41 | 14.49 | 85.58 | 3.24 | 25.55 |
| 5 | Podlength (cm) | 0.36 | 0.36 | 12.88 | 12.91 | 99.64 | 1.23 | 26.50 |
| 6 | Podwidth (cm) | 0.09 | 0.09 | 14.49 | 14.56 | 99.11 | 0.64 | 29.73 |
| 7 | Pod yieldperplant (g) | 361.27 | 458.52 | 29.94 | 33.87 | 78.11 | 34.55 | 54.51 |
| 8 | Podsperplant | 181.16 | 191.61 | 42.27 | 43.51 | 94.35 | 26.92 | 84.58 |
| 9 | Podweight (g) | 0.52 | 0.56 | 29.40 | 30.67 | 91.92 | 1.42 | 58.08 |
| 10 | Numberofharvests | 0.71 | 0.86 | 19.64 | 20.41 | 92.56 | 1.76 | 38.93 |
| 11 | Starch (g/100 g) | 0.29 | 0.29 | 35.54 | 34.66 | 99.30 | 70.91 | 70.91 |
| 12 | Carbohydrate (g/100 g) | 3.84 | 3.85 | 23.99 | 24.01 | 99.87 | 49.39 | 49.39 |
| 13 | Protein (g/100 g) | 10.52 | 10.57 | 23.61 | 23.67 | 99.51 | 48.53 | 48.53 |
| 14 | Vitamin C (mg/100 g) | 8.06 | 8.09 | 33.21 | 33.27 | 99.66 | 68.30 | 68.30 |
| 15 | Iron (mg/100 g) | 6.29 | 6.30 | 50.19 | 50.24 | 99.80 | 100.00 | 103.29 |
| 16 | Calcium (mg/100 g) | 90.12 | 90.14 | 39.53 | 39.54 | 99.97 | 81.43 | 81.426 |
| 17 | Phosphorous (mg/100 g) | 13060.89 | 13205.34 | 24.87 | 25.01 | 98.86 | 50.94 | 50.94 |
| 18 | Polyphenols (g/100 g) | 7.71 | 7.84 | 43.32 | 43.45 | 99.38 | 88.97 | 88.97 |
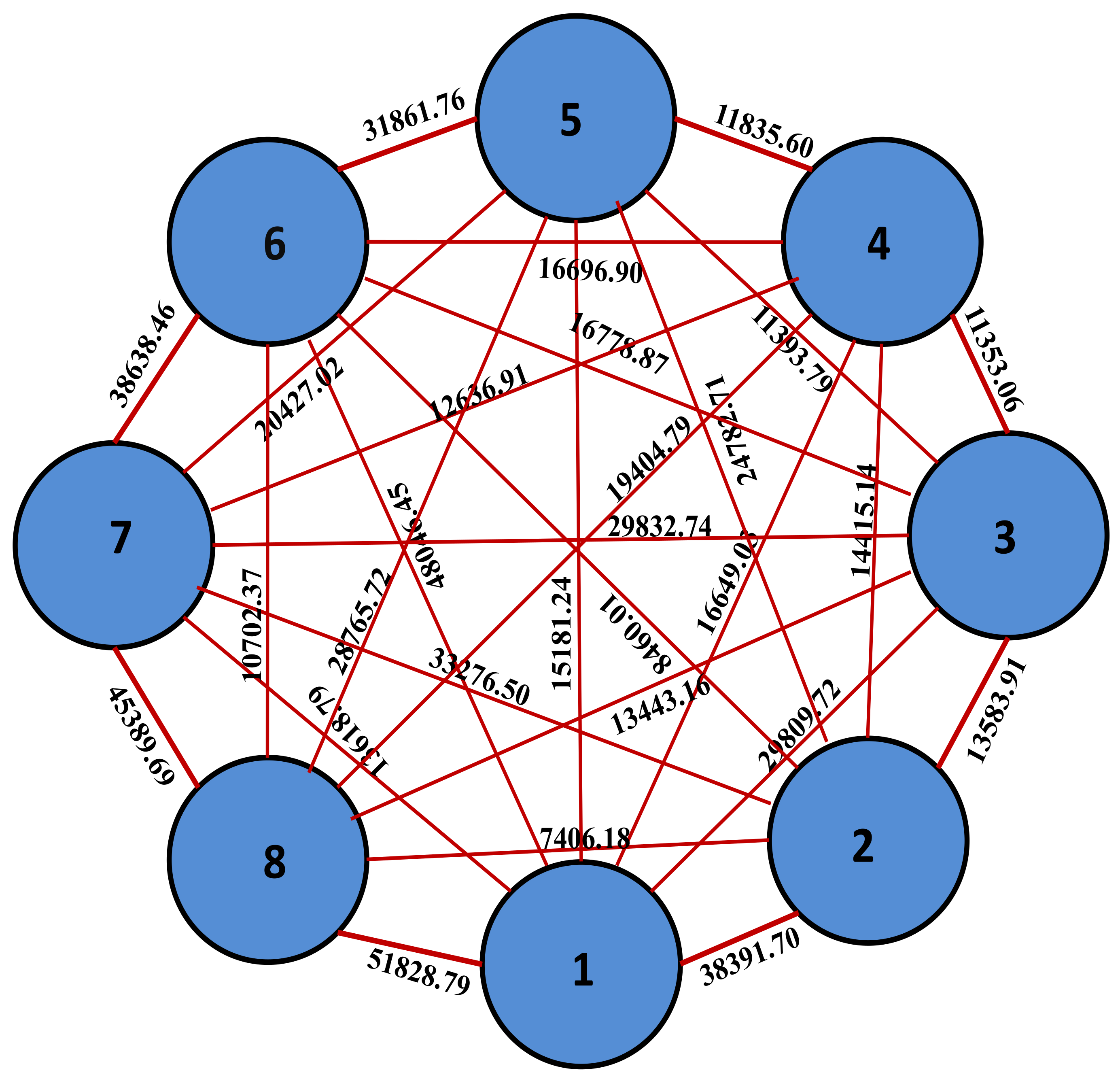
| Cluster | Genotypes Per Cluster | Names of the Genotypes |
|---|---|---|
| I | 4 | GM-10, GM-18, GM-20, GM-25 |
| II | 8 | GM-2, GM-3, GM-12, GM-8, GM-9, GM-13, GM-28, GM-14 |
| III | 3 | GM-7, GM-21, GM-15 |
| IV | 3 | GM-5, GM-19, GM-11 |
| V | 4 | GM-24, GM-22, GM-26, GM-23 |
| VI | 2 | GM-1, GM4 |
| VII | 2 | GM-16, GM-24 |
| VIII | 2 | GM-6, GM-27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shilpashree, N.; Devi, S.N.; Manjunathagowda, D.C.; Muddappa, A.; Abdelmohsen, S.A.M.; Tamam, N.; Elansary, H.O.; El-Abedin, T.K.Z.; Abdelbacki, A.M.M.; Janhavi, V. Morphological Characterization, Variability and Diversity among Vegetable Soybean (Glycine max L.) Genotypes. Plants 2021, 10, 671. https://doi.org/10.3390/plants10040671
Shilpashree N, Devi SN, Manjunathagowda DC, Muddappa A, Abdelmohsen SAM, Tamam N, Elansary HO, El-Abedin TKZ, Abdelbacki AMM, Janhavi V. Morphological Characterization, Variability and Diversity among Vegetable Soybean (Glycine max L.) Genotypes. Plants. 2021; 10(4):671. https://doi.org/10.3390/plants10040671
Chicago/Turabian StyleShilpashree, Nagaraju, Sarojinikunjamma Nirmala Devi, Dalasanuru Chandregowda Manjunathagowda, Anjanappa Muddappa, Shaimaa A. M. Abdelmohsen, Nissren Tamam, Hosam O. Elansary, Tarek K. Zin El-Abedin, Ashraf M. M. Abdelbacki, and Veerabhadregowda Janhavi. 2021. "Morphological Characterization, Variability and Diversity among Vegetable Soybean (Glycine max L.) Genotypes" Plants 10, no. 4: 671. https://doi.org/10.3390/plants10040671
APA StyleShilpashree, N., Devi, S. N., Manjunathagowda, D. C., Muddappa, A., Abdelmohsen, S. A. M., Tamam, N., Elansary, H. O., El-Abedin, T. K. Z., Abdelbacki, A. M. M., & Janhavi, V. (2021). Morphological Characterization, Variability and Diversity among Vegetable Soybean (Glycine max L.) Genotypes. Plants, 10(4), 671. https://doi.org/10.3390/plants10040671







