RNA Sequencing Reveals Rice Genes Involved in Male Reproductive Development under Temperature Alteration
Abstract
1. Introduction
2. Results
2.1. Effects of Temperature on Seed Setting Rate
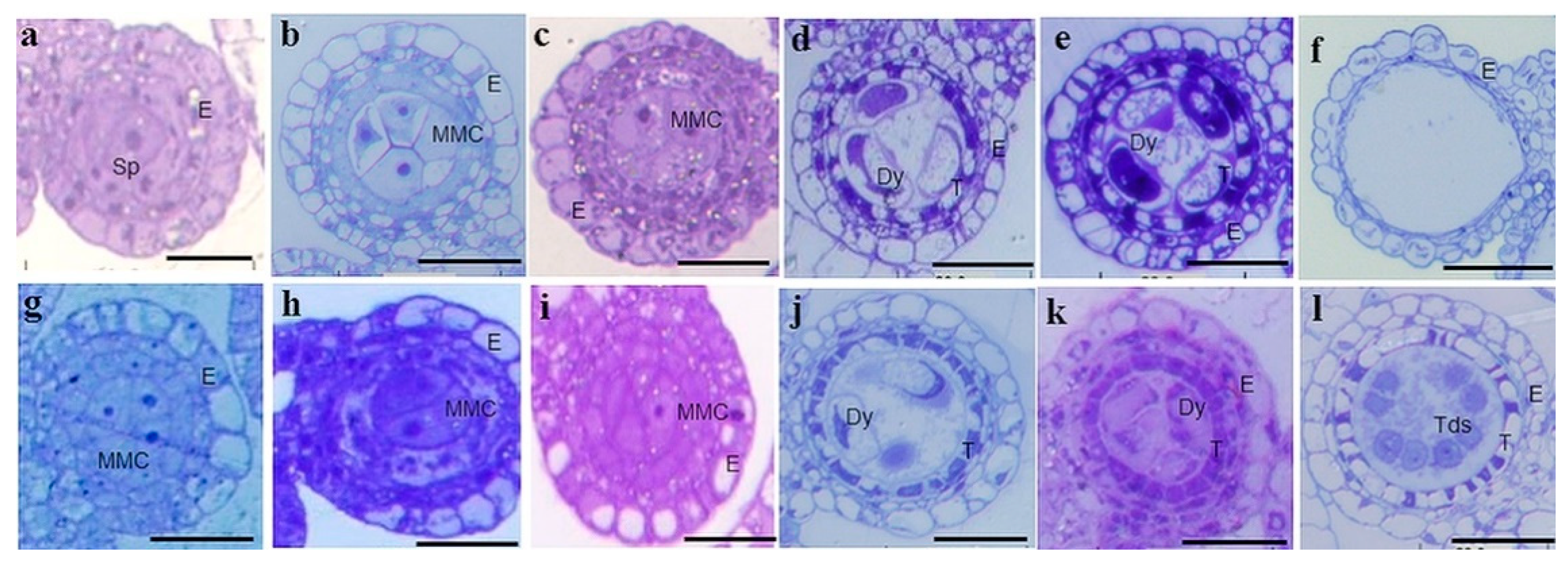
2.2. Cytology Characterization of Anther Development in IR68301S
2.3. Transcriptome Data and Read Assembly
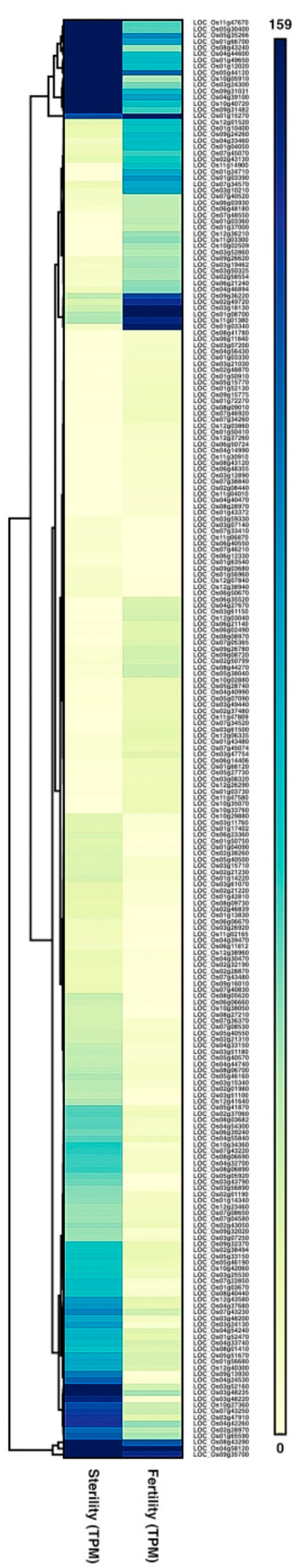
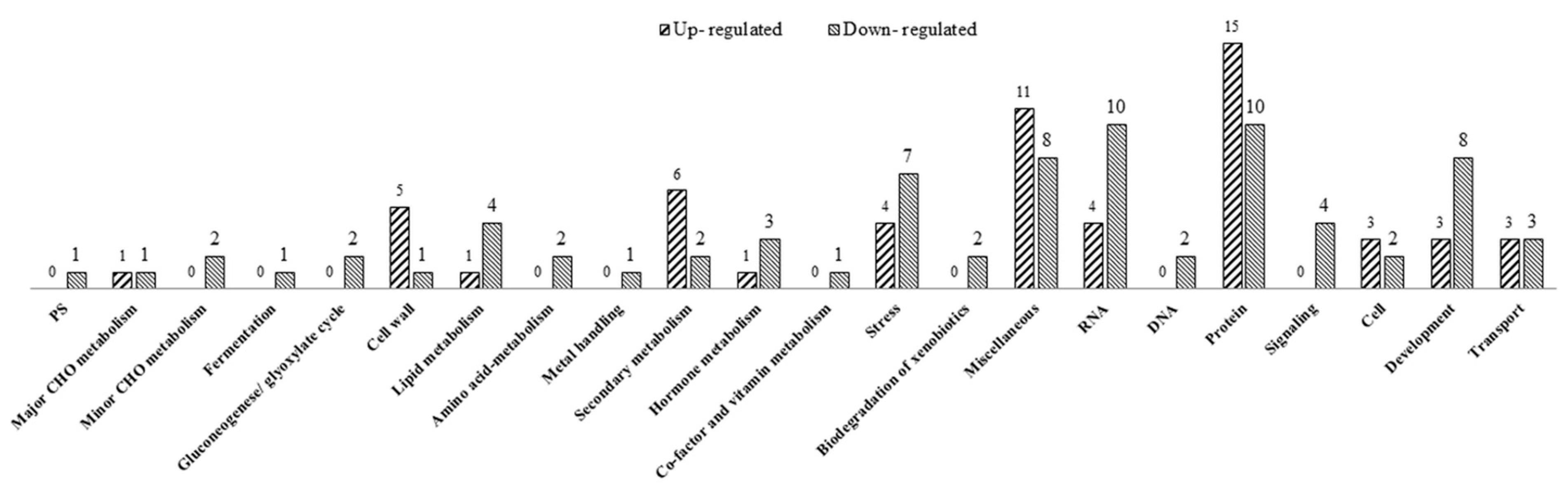
2.4. Differentially Expressed Genes (DEGs) between Fertile and Sterile Conditions
2.5. DEGs Validation
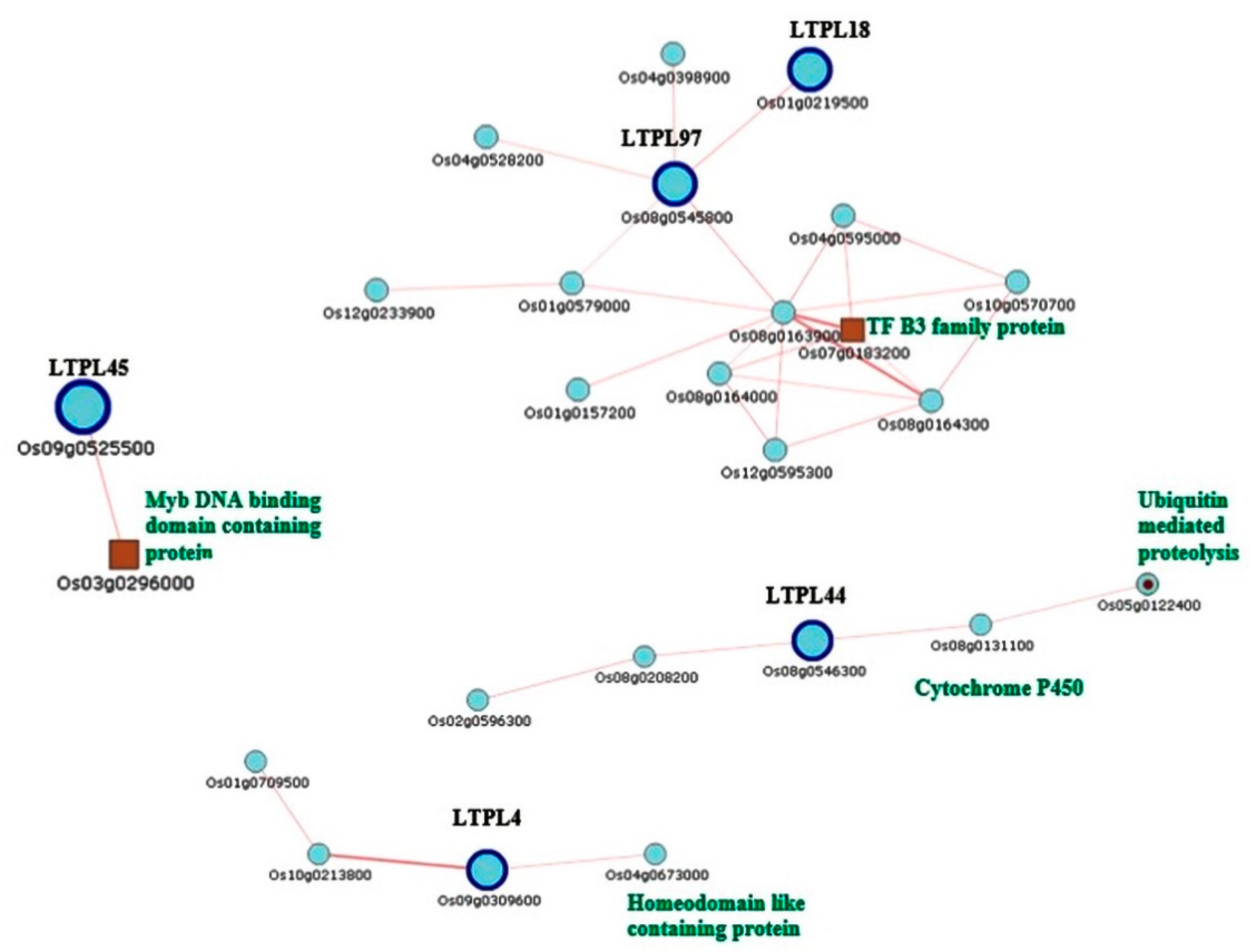
2.6. TGMS-Related Genes and Common Temperature Responsive Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Condition
4.2. Observation of Anther Development by Semithin Sections
4.3. RNA Isolation, Library Construction and Transcriptome Sequencing
4.4. Identification of Differentially Expressed Genes, Functional Annotation and Enrichment Analysis
4.5. Gene Expression Using Quantitative Real-Time RT-PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- USDA. World Agricultural Production. Available online: https://apps.fas.usda.gov/psdonline/circulars/production.pdf (accessed on 27 November 2016).
- Virmani, S.; Sun, Z.; Mou, T.; Ali, A.J.; Mao, C. Two-Line Hybrid Rice Breeding Manual; International Rice Research Institute: Manila, Philippines, 2003; pp. 6–11. [Google Scholar]
- Maruyama, K.; Araki, H.; Kato, H. Thermosensitive genetic male sterility induced by irradiation. In Rice Genetics II: (In 2 Parts); World Scientific: International Rice Research Institute: Manila, Philippines, 1991; pp. 227–232. [Google Scholar]
- Arasakesary, S.; Manonmani, S.; Pushpam, R.; Robin, S. New temperature sensitive genic male sterile lines with better outcrossing ability for production of two-line hybrid rice. Rice Sci. 2015, 22, 49–52. [Google Scholar] [CrossRef]
- Song, L.; Liu, Z.; Tong, J.; Xiao, L.; Ma, H.; Zhang, H. Comparative proteomics analysis reveals the mechanism of fertility alternation of thermosensitive genic male sterile rice lines under low temperature inducement. Proteomics 2015, 15, 1884–1905. [Google Scholar] [CrossRef]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S. Plant drought stress: Effects, mechanisms and management. Sustain. Agric. 2009, 1, 153–188. [Google Scholar]
- Moraes de Freitas, G.P.; Basu, S.; Ramegowda, V.; Thomas, J.; Benitez, L.C.; Braga, E.B.; Pereira, A. Physiological and transcriptional responses to low-temperature stress in rice genotypes at the reproductive stage. Plant Signal. Behav. 2019, 14, e1581557. [Google Scholar] [CrossRef]
- Liu, K.; Deng, J.; Lu, J.; Wang, X.; Lu, B.; Tian, X.; Zhang, Y. High nitrogen levels alleviate yield loss of super hybrid rice caused by high temperatures during the flowering stage. Front. Plant Sci. 2019, 10, 357. [Google Scholar] [CrossRef]
- Dhatt, B.K.; Abshire, N.; Paul, P.; Hasanthika, K.; Sandhu, J.; Zhang, Q.; Obata, T.; Walia, H. Metabolic dynamics of developing rice seeds under high night-time temperature stress. Front. Plant Sci. 2019, 10, 1443. [Google Scholar] [CrossRef]
- Zhang, D.; Luo, X.; Zhu, L. Cytological analysis and genetic control of rice anther development. J. Genet. Genom. 2011, 38, 379–390. [Google Scholar] [CrossRef]
- Guo, J.X.; Liu, Y.G. Molecular Control of Male Reproductive Development and Pollen Fertility in Rice F. J. Integr. Plant Biol. 2012, 54, 967–978. [Google Scholar] [CrossRef]
- Kalaiyarasi, R.; Vaidyanathan, P. Cytological screening of rice TGMS lines. Plant Breed. 2003, 122, 334–338. [Google Scholar] [CrossRef]
- Ding, J.; Lu, Q.; Ouyang, Y.; Mao, H.; Zhang, P.; Yao, J.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. USA 2012, 109, 2654–2659. [Google Scholar] [CrossRef]
- Ding, J.; Shen, J.; Mao, H.; Xie, W.; Li, X.; Zhang, Q. RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol. Plant 2012, 5, 1210–1216. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, Q.; Li, J.; Jiang, D.; Zhou, L.; Wu, P.; Lu, S.; Li, F.; Zhu, L.; Liu, Z. Photoperiod-and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res 2012, 22, 649–660. [Google Scholar] [CrossRef]
- Zhou, H.; Zhou, M.; Yang, Y.; Li, J.; Zhu, L.; Jiang, D.; Dong, J.; Liu, Q.; Gu, L.; Zhou, L. RNase Z S1 processes Ub L40 mRNAs and controls thermosensitive genic male sterility in rice. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Xu, H.; Gao, Y.; Wang, J. Transcriptomic analysis of rice (Oryza sativa) developing embryos using the RNA-Seq technique. PLoS ONE 2012, 7, e30646. [Google Scholar] [CrossRef]
- Hennig, L.; Gruissem, W.; Grossniklaus, U.; Köhler, C. Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol. 2004, 135, 1765–1775. [Google Scholar] [CrossRef]
- Ma, J.; Skibbe, D.S.; Fernandes, J.; Walbot, V. Male reproductive development: Gene expression profiling of maize anther and pollen ontogeny. Genome Biol. 2008, 9, 1–17. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, J.; Liu, J.; Xia, M.; Wang, W.; Shen, F. Transcriptome analysis of early anther development of cotton revealed male sterility genes for major metabolic pathways. J. Plant Growth Regul. 2015, 34, 223–232. [Google Scholar] [CrossRef]
- Ma, J.; Wei, H.; Song, M.; Pang, C.; Liu, J.; Wang, L.; Zhang, J.; Fan, S.; Yu, S. Transcriptome profiling analysis reveals that flavonoid and ascorbate-glutathione cycle are important during anther development in Upland cotton. PLoS ONE 2012, 7, e49244. [Google Scholar] [CrossRef]
- Liu, C.; Liu, Z.; Li, C.; Zhang, Y.; Feng, H. Comparative transcriptome analysis of fertile and sterile buds from a genetically male sterile line of Chinese cabbage. Vitr. Cell. Dev. Biol. Plant 2016, 52, 130–139. [Google Scholar] [CrossRef]
- Li, Z.; Cheng, Y.; Cui, J.; Zhang, P.; Zhao, H.; Hu, S. Comparative transcriptome analysis reveals carbohydrate and lipid metabolism blocks in Brassica napus L. male sterility induced by the chemical hybridization agent monosulfuron ester sodium. BMC Genom. 2015, 16, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.; Fu, F.; Liu, M.; Zhao, H.; Liu, C.; Li, J.; Tang, Z.; Xu, X.; Qiu, X.; Wang, R. Comparative transcriptome analysis of recessive male sterility (RGMS) in sterile and fertile Brassica napus lines. PLoS ONE 2015, 10, e0144118. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.-J.; Seo, M.; Jang, Y.-J.; Cho, S.; Lee, G.P. Transcriptome profiling of differentially expressed genes in floral buds and flowers of male sterile and fertile lines in watermelon. BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Li, Q.; Wang, Z.; Wang, Y.; Ma, R.; Zhu, L.; He, G.; Chen, R. Genes associated with thermosensitive genic male sterility in rice identified by comparative expression profiling. BMC Genom. 2014, 15, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, Z.; Guo, Z.; Song, G.; Cheng, Q.; Jiang, D.; Zhu, Y.; Yang, D. Comparative transcriptomes profiling of photoperiod-sensitive male sterile rice Nongken 58S during the male sterility transition between short-day and long-day. BMC Genom. 2011, 12, 1–10. [Google Scholar] [CrossRef]
- Yang, J.; Chen, X.; Zhu, C.; Peng, X.; He, X.; Fu, J.; Ouyang, L.; Bian, J.; Hu, L.; Sun, X. RNA-seq reveals differentially expressed genes of rice (Oryza sativa) spikelet in response to temperature interacting with nitrogen at meiosis stage. BMC Genom. 2015, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Wangpakapattanawong, P.; Finlayson, R.; Öborn, I.; Roshetko, J.M.; Sinclair, F.; Shono, K.; Borelli, S.; Hillbrand, A.; Conigliaro, M. Agroforestry in Rice-Production Landscapes in Southeast Asia: A Practical Manual; FAO Regional Office for Asia and the Pacific: Bangkok, Thailand, 2017. [Google Scholar]
- Pei, X.; Jing, Z.; Tang, Z.; Zhu, Y. Comparative transcriptome analysis provides insight into differentially expressed genes related to cytoplasmic male sterility in broccoli (Brassica oleracea var. italica). Sci. Hortic. 2017, 217, 234–242. [Google Scholar] [CrossRef]
- Nonomura, K.-I.; Nakano, M.; Fukuda, T.; Eiguchi, M.; Miyao, A.; Hirochika, H.; Kurata, N. The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 2004, 16, 1008–1020. [Google Scholar] [CrossRef]
- Nonomura, K.-I.; Nakano, M.; Eiguchi, M.; Suzuki, T.; Kurata, N. PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 2006, 119, 217–225. [Google Scholar] [CrossRef]
- Yuan, W.; Li, X.; Chang, Y.; Wen, R.; Chen, G.; Zhang, Q.; Wu, C. Mutation of the rice gene PAIR3 results in lack of bivalent formation in meiosis. Plant J. 2009, 59, 303–315. [Google Scholar] [CrossRef]
- Endo, M.; Tsuchiya, T.; Hamada, K.; Kawamura, S.; Yano, K.; Ohshima, M.; Higashitani, A.; Watanabe, M.; Kawagishi-Kobayashi, M. High temperatures cause male sterility in rice plants with transcriptional alterations during pollen development. Plant Cell Physiol. 2009, 50, 1911–1922. [Google Scholar]
- Li, W.; Zhao, F.A.; Fang, W.; Xie, D.; Hou, J.; Yang, X.; Zhao, Y.; Tang, Z.; Nie, L.; Lv, S. Identification of early salt stress responsive proteins in seedling roots of upland cotton (Gossypium hirsutum L.) employing iTRAQ-based proteomic technique. Front. Plant Sci. 2015, 6, 732. [Google Scholar] [CrossRef]
- Kim, H.U.; Yun, C.H.; Cho, W.S.; Kang, S.K.; Chung, T.Y. Nucleotide sequence of rice cDNA that encodes a ubiquitin protein and a 79-amino acid protein. Plant Physiol. 1995, 108, 865. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, D.; Lv, X.; Wang, Y.; Xun, Z.; Liu, Z.; Li, F.; Lu, H. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 2014, 26, 2939–2961. [Google Scholar] [CrossRef]
- Gao, H.; Zhang, Y.; Wang, W.; Zhao, K.; Liu, C.; Bai, L.; Li, R.; Guo, Y. Two membrane-anchored aspartic proteases contribute to pollen and ovule development. Plant Physiol. 2017, 173, 219–239. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bai, X.; Wang, X.; Chu, C. OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 2007, 164, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Wu, J.; Sheng, W.-T.; Zhou, B.; Zhou, L.-J.; Zhuang, W.; Yao, D.-P.; Deng, Q.-Y. Comparative analysis of anther transcriptome profiles of two different rice male sterile lines genotypes under cold stress. Int. J. Mol. Sci. 2015, 16, 11398–11416. [Google Scholar] [CrossRef]
- Zou, C.; Jiang, W.; Yu, D. Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J. Exp. Bot. 2010, 61, 3901–3914. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. WRKY proteins: Signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015, 2015, 807560. [Google Scholar] [CrossRef]
- Pinot, F.; Beisson, F. Cytochrome P450 metabolizing fatty acids in plants: Characterization and physiological roles. FEBS J. 2011, 278, 195–205. [Google Scholar] [CrossRef]
- Morant, M.; Jørgensen, K.; Schaller, H.; Pinot, F.; Møller, B.L.; Werck-Reichhart, D.; Bak, S. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 2007, 19, 1473–1487. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pinot, F.; Sauveplane, V.; Werck-Reichhart, D.; Diehl, P.; Schreiber, L.; Franke, R.; Zhang, P.; Chen, L.; Gao, Y. Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 2010, 22, 173–190. [Google Scholar] [CrossRef] [PubMed]
- Lamb, D.C.; Waterman, M.R. Unusual properties of the cytochrome P450 superfamily. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120434. [Google Scholar] [CrossRef]
- Wang, Y.; Liao, M.; Hoe, N.; Acharya, P.; Deng, C.; Krutchinsky, A.N.; Correia, M.A. A role for protein phosphorylation in cytochrome P450 3A4 ubiquitin-dependent proteasomal degradation. J. Biol. Chem. 2009, 284, 5671–5684. [Google Scholar] [CrossRef]
- Zhang, D.; Liang, W.; Yin, C.; Zong, J.; Gu, F.; Zhang, D. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 2010, 154, 149–162. [Google Scholar] [CrossRef]
- Piffanelli, P.; Ross, J.H.; Murphy, D. Biogenesis and function of the lipidic structures of pollen grains. Sex. Plant Reprod. 1998, 11, 65–80. [Google Scholar] [CrossRef]
- Wang, L.; Wu, N.; Zhu, Y.; Song, W.; Zhao, X.; Li, Y.; Hu, Y. The divergence and positive selection of the plant-specific BURP-containing protein family. Ecol. Evol. 2015, 5, 5394–5412. [Google Scholar] [CrossRef]
- Schrick, K.; Nguyen, D.; Karlowski, W.M.; Mayer, K.F. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004, 5, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Venkata, B.; Schrick, K. START domains in lipid/sterol transfer and signaling in plants. Curr. Adv. Biochem. Cell Biol. Plant Lipids 2006, 57. Available online: https://www.semanticscholar.org/paper/START-domains-in-lipid%2Fsterol-transfer-and-in-Venkata-Schrick/81ab331bcb732324c7aa46c230d8e0c91caa091d (accessed on 27 November 2016).
- Chueasiri, C.; Chunthong, K.; Pitnjam, K.; Chakhonkaen, S.; Sangarwut, N.; Sangsawang, K.; Suksangpanomrung, M.; Michaelson, L.V.; Napier, J.A.; Muangprom, A. Rice ORMDL controls sphingolipid homeostasis affecting fertility resulting from abnormal pollen development. PLoS ONE 2014, 9, e106386. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Kingsford, C. Salmon: Accurate, versatile and ultrafast quantification from RNA-seq data using lightweight-alignment. BioRxiv 2015, 021592. [Google Scholar]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Preced. 2010. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Temperature (°C) | Replicates | Seed Setting RATE (%) | Max. Temp. Mean ± SD | |
|---|---|---|---|---|
| Mean ± SD | (Average) | |||
| 24 | 1 | 78.1 ± 2.1 | 75.4 ± 1.8 | 24.38 ±0.6 |
| 2 | 72.6 ± 1.6 | 24.4 ±0.6 | ||
| 26 | 1 | 61.9 ± 6.6 | 64.3 ± 5.1 | 25.9 ± 1.4 |
| 2 | 66.7 ± 3.7 | 26.1 ± 1.5 | ||
| 28 | 1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 29.4 ± 0.5 |
| 2 | 0.0 ± 0.0 | 28.28 ± 0.1 | ||
| 32 | 1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 32.1 ± 0.4 |
| 2 | 0.0 ± 0.0 | |||
| Map to Genome | IR68301S-Fertility | Percentage | IR68301S-Sterility | Percentage |
|---|---|---|---|---|
| Clean reads | 47,317,390 | 100 | 47,443,806 | 100 |
| Total base pairs | 4,258,565,100 | 100 | 4,269,942,540 | 100 |
| Total mapped reads | 37,921,504 | 80.14 | 39,071,301 | 82.35 |
| Perfect match | 26,041,749 | 55.04 | 26,434,966 | 55.72 |
| Unique match | 37,193,432 | 78.60 | 38,354,275 | 80.84 |
| Multi-position match | 728,072 | 1.54 | 717,026 | 1.51 |
| Total rice loci | 66,153 | 100 | 66,153 | 100 |
| Total expressed loci | 21,149 | 31.96 | 21,201 | 32.04 |
| GO term | Ontology | Description | Number in Input List | Number in BG/Ref | p-Value | FDR |
|---|---|---|---|---|---|---|
| GO:0009653 | P | anatomical structure morphogenesis | 14 | 1141 | 0.00000041 | 0.000025 |
| GO:0048856 | P | anatomical structure development | 17 | 1665 | 0.00000026 | 0.000025 |
| GO:0016043 | P | cellular component organization | 17 | 1935 | 0.0000021 | 0.000082 |
| GO:0007275 | P | multicellular organismal development | 19 | 3543 | 0.00043 | 0.01 |
| GO:0032502 | P | developmental process | 20 | 3791 | 0.00036 | 0.01 |
| GO:0032501 | P | multicellular organismal process | 19 | 3619 | 0.00056 | 0.011 |
| GO:0008289 | F | lipid binding | 5 | 346 | 0.0014 | 0.041 |
| Gene | Description |
|---|---|
| Common Temperature Responsive Genes | |
| LOC_Os01g50410 | STE_MEKK_ste11_MAP3K.6—STE kinases include homologs to sterile 7, sterile 11 and sterile 20 from yeast, expressed |
| LOC_Os11g04010 | ICE-like protease p20 domain containing protein, putative, expressed |
| LOC_Os02g08440 | WRKY71, expressed |
| LOC_Os01g24710 | jacalin-like lectin domain containing protein, expressed |
| LOC_Os01g37000 | carboxyl-terminal peptidase, putative, expressed |
| LOC_Os02g48870 | aspartic proteinase nepenthesin-2 precursor, putative, expressed |
| LOC_Os09g32020 | ubiquitin fusion degradation protein, putative, expressed |
| LOC_Os01g12020 | LTPL18—Protease inhibitor/seed storage/LTP family protein precursor, expressed |
| LOC_Os09g35700 | LTPL45—Protease inhibitor/seed storage/LTP family protein precursor, expressed |
| LOC_Os04g40470 | cytochrome P450, putative, expressed |
| LOC_Os01g72270 | cytochrome P450, putative, expressed |
| LOC_Os08g05620 | cytochrome P450, putative, expressed |
| LOC_Os01g63540 | cytochrome P450, putative, expressed |
| LOC_Os01g50750 | zinc finger, C3HC4 type domain containing protein, expressed |
| TGMS-related gene | |
| LOC_Os09g16010 | BURP domain containing protein, expressed |
| LOC_Os06g50724 | START domain containing protein, expressed |
| LOC_Os07g46210 | LTPL2—Protease inhibitor/seed storage/LTP family protein precursor, expressed |
| LOC_Os03g24300 | LTPL1—Protease inhibitor/seed storage/LTP family protein precursor, expressed |
| LOC_Os09g13930 | LTPL4—Protease inhibitor/seed storage/LTP family protein precursor, expressed |
| LOC_Os04g14990 | BURP domain containing protein, expressed |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khlaimongkhon, S.; Chakhonkaen, S.; Tongmark, K.; Sangarwut, N.; Panyawut, N.; Wasinanon, T.; Sikaewtung, K.; Wanchana, S.; Mongkolsiriwatana, C.; Chunwonges, J.; et al. RNA Sequencing Reveals Rice Genes Involved in Male Reproductive Development under Temperature Alteration. Plants 2021, 10, 663. https://doi.org/10.3390/plants10040663
Khlaimongkhon S, Chakhonkaen S, Tongmark K, Sangarwut N, Panyawut N, Wasinanon T, Sikaewtung K, Wanchana S, Mongkolsiriwatana C, Chunwonges J, et al. RNA Sequencing Reveals Rice Genes Involved in Male Reproductive Development under Temperature Alteration. Plants. 2021; 10(4):663. https://doi.org/10.3390/plants10040663
Chicago/Turabian StyleKhlaimongkhon, Sudthana, Sriprapai Chakhonkaen, Keasinee Tongmark, Numphet Sangarwut, Natjaree Panyawut, Thiwawan Wasinanon, Kannika Sikaewtung, Samart Wanchana, Chareerat Mongkolsiriwatana, Julapark Chunwonges, and et al. 2021. "RNA Sequencing Reveals Rice Genes Involved in Male Reproductive Development under Temperature Alteration" Plants 10, no. 4: 663. https://doi.org/10.3390/plants10040663
APA StyleKhlaimongkhon, S., Chakhonkaen, S., Tongmark, K., Sangarwut, N., Panyawut, N., Wasinanon, T., Sikaewtung, K., Wanchana, S., Mongkolsiriwatana, C., Chunwonges, J., & Muangprom, A. (2021). RNA Sequencing Reveals Rice Genes Involved in Male Reproductive Development under Temperature Alteration. Plants, 10(4), 663. https://doi.org/10.3390/plants10040663





