Species Diversity of Pin Nematodes (Paratylenchus spp.) from Potato Growing Regions of Southern Alberta, Canada
Abstract
1. Introduction
2. Results
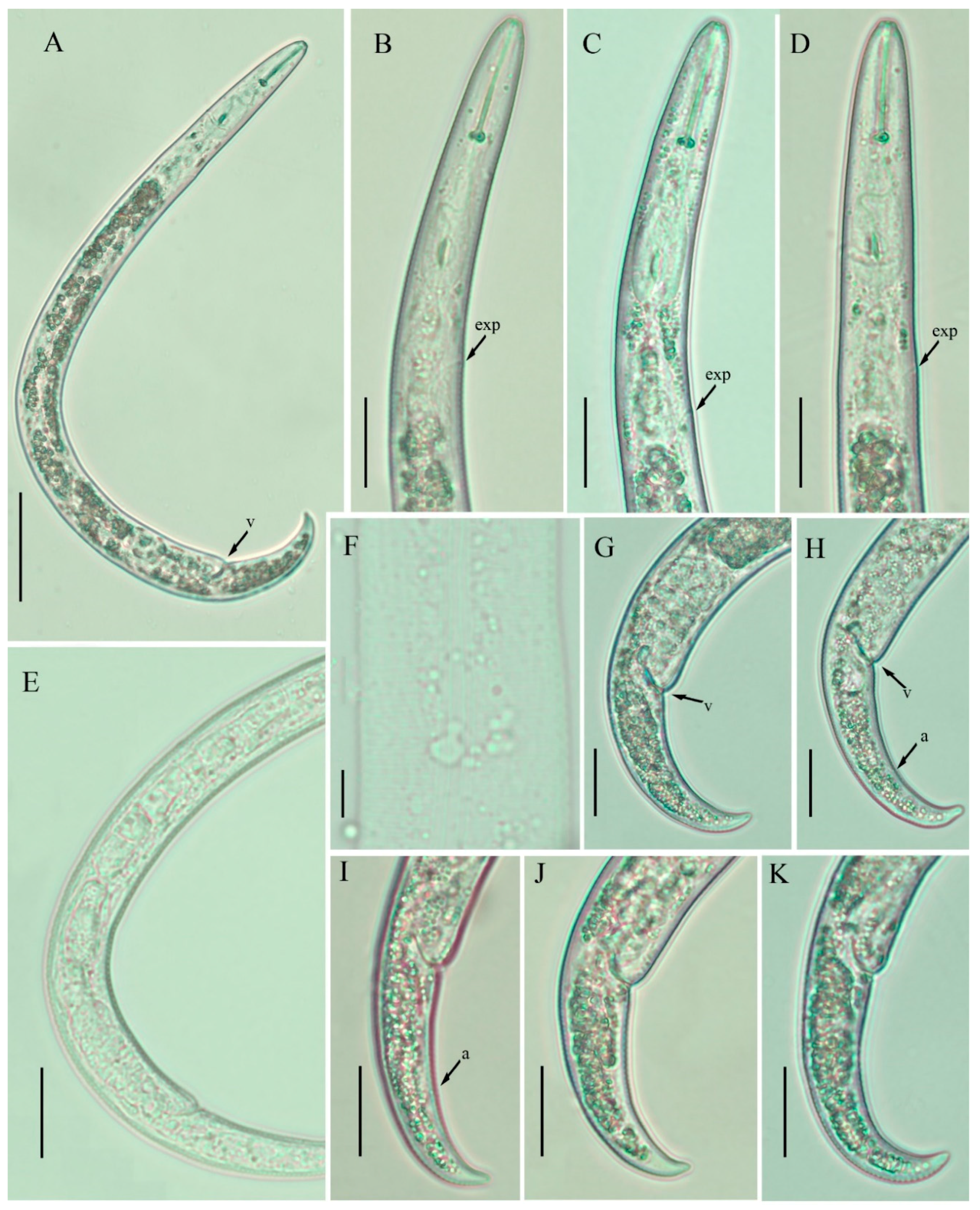
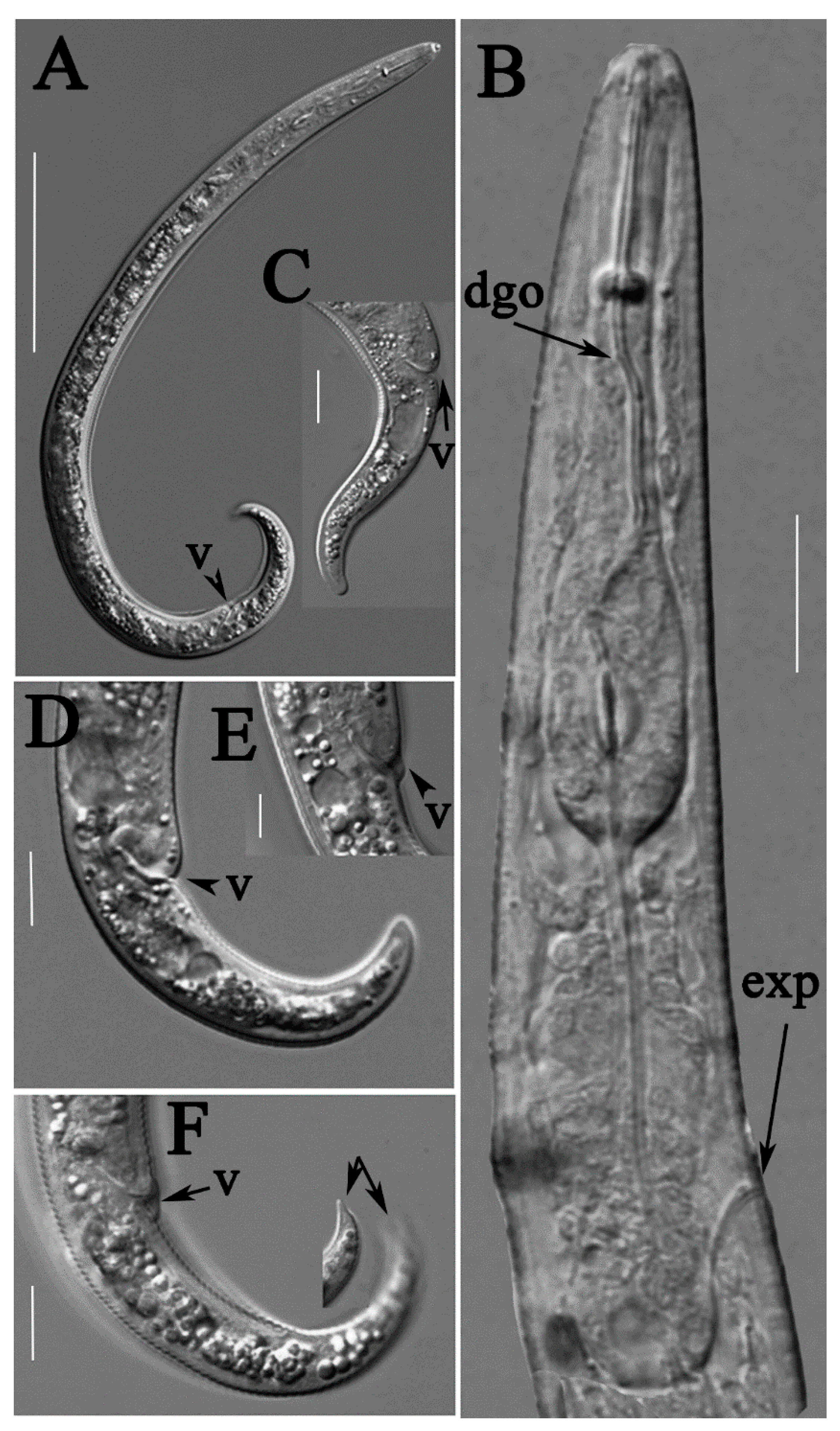
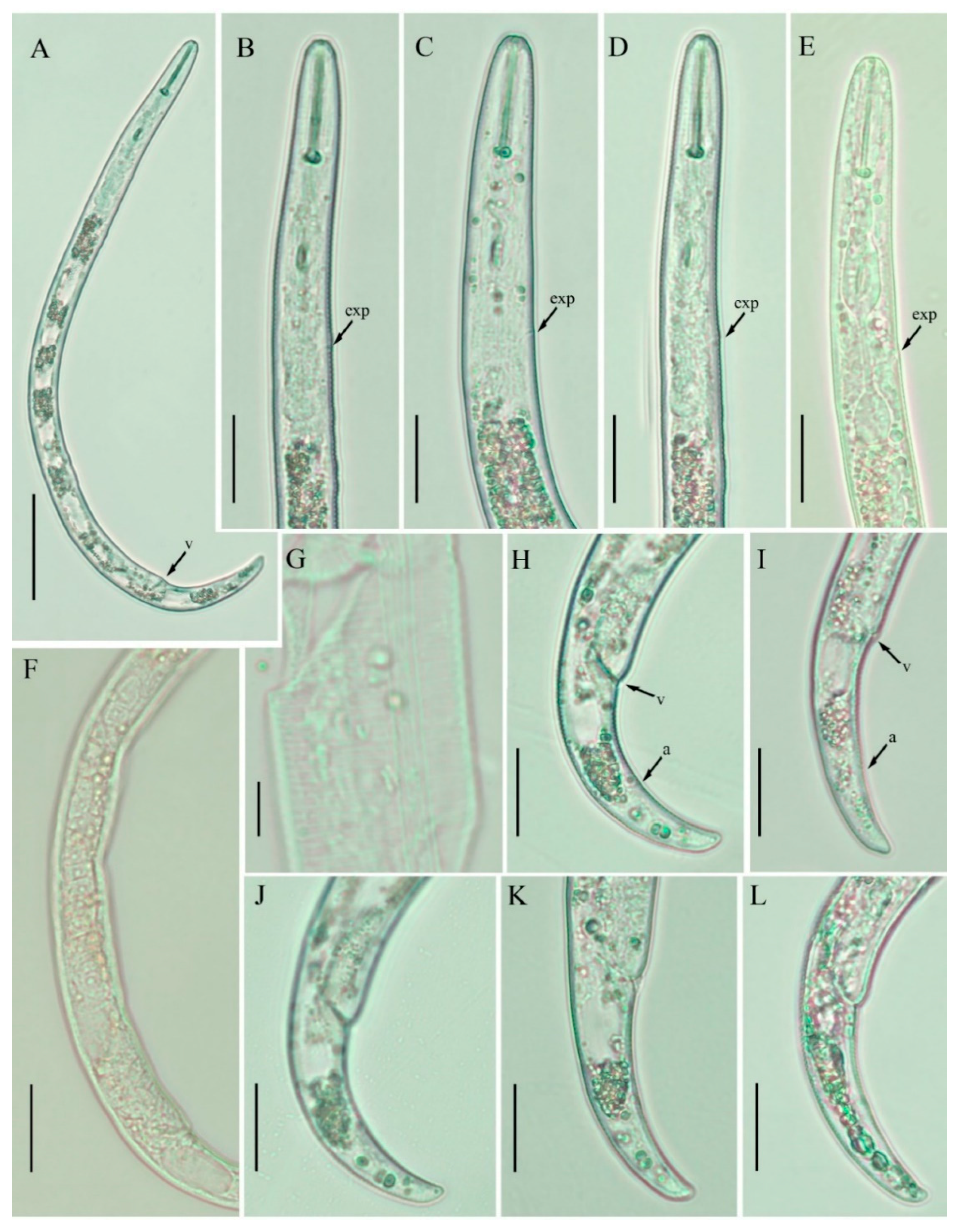
2.1. Description of Female Paratylenchus neoprojectus Wu and Hawn
2.1.1. Juveniles
2.1.2. Remarks
2.1.3. Habitat and Locality
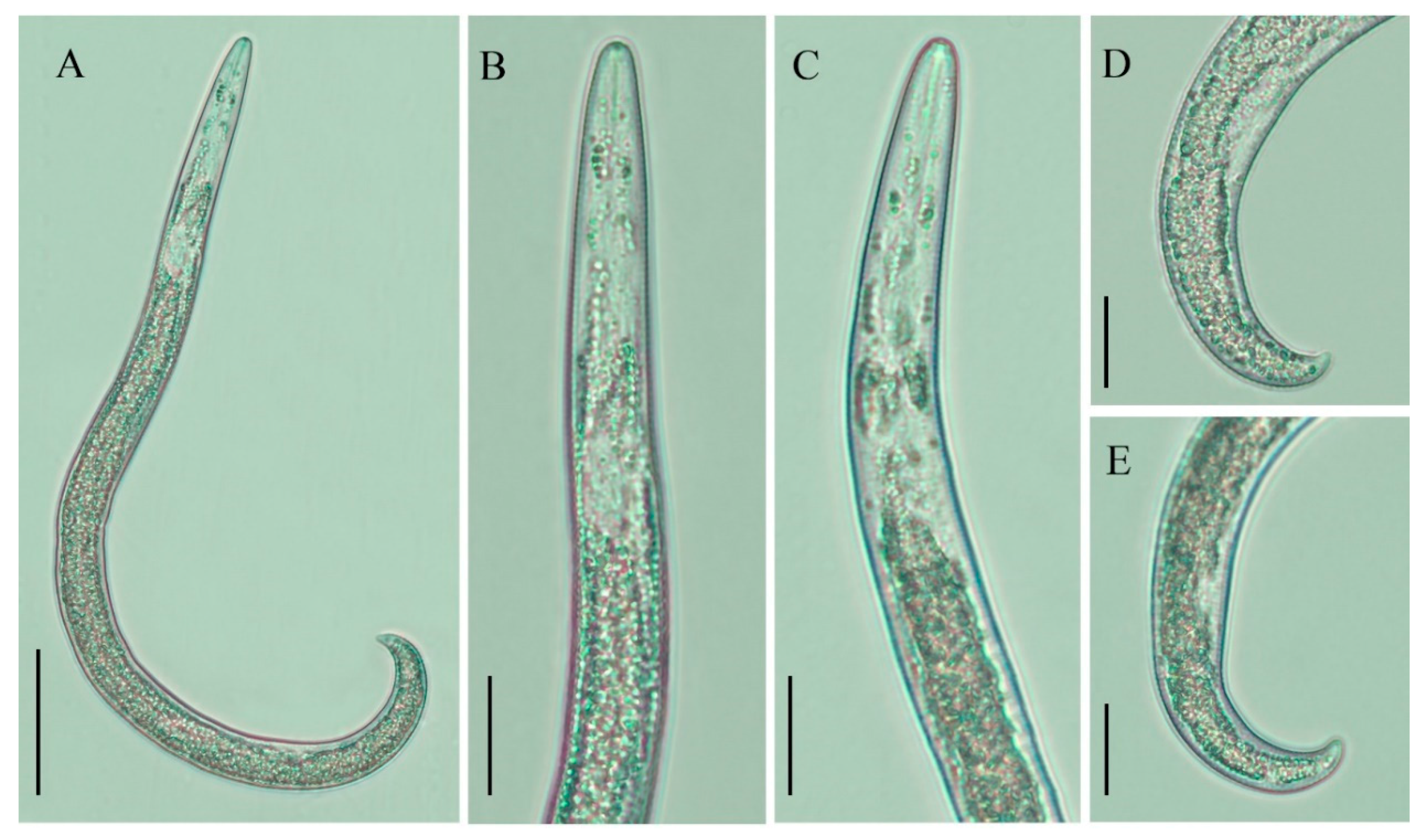
2.2. Description of Female Paratylenchus tateae Wu and Townshend
2.2.1. Juveniles
2.2.2. Remarks
2.2.3. Habitat and Locality
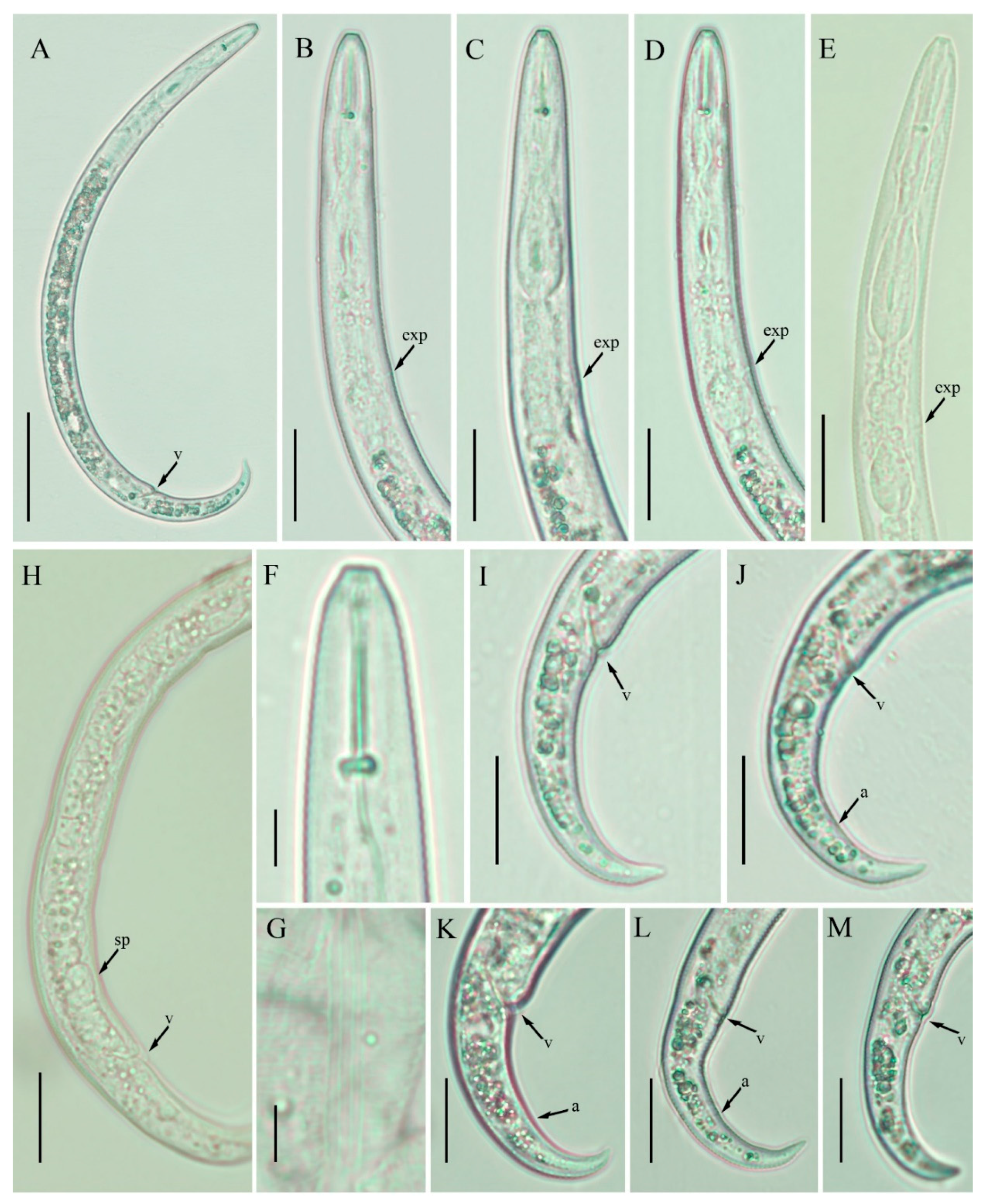
2.3. Description of Female Paratylenchus enigmaticus sp. nov.
2.3.1. Juvenile
2.3.2. Diagnosis and Relationship
2.3.3. Remarks
2.3.4. Type Habitat and Locality
2.3.5. Etymology
2.3.6. Type Material
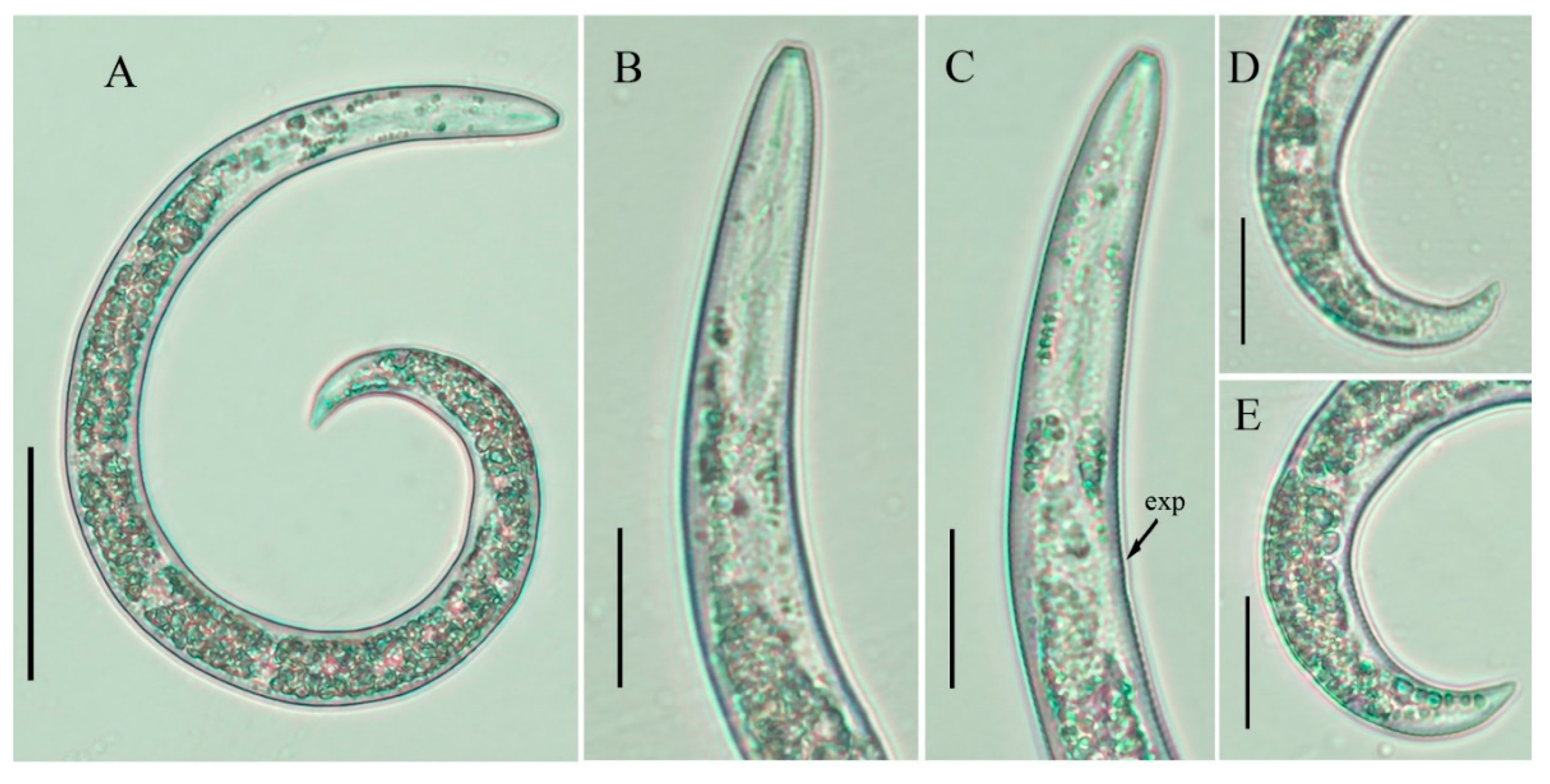
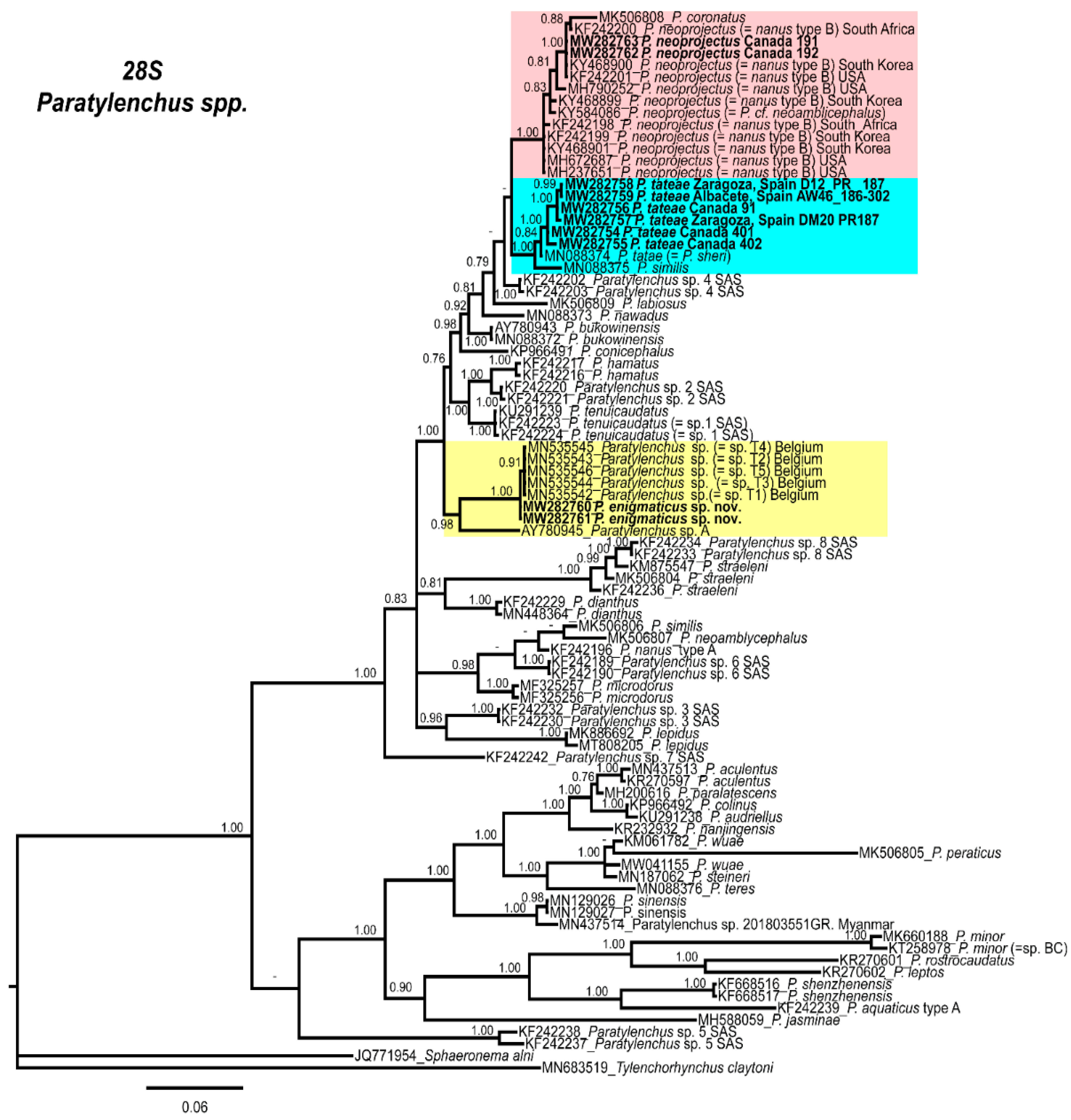
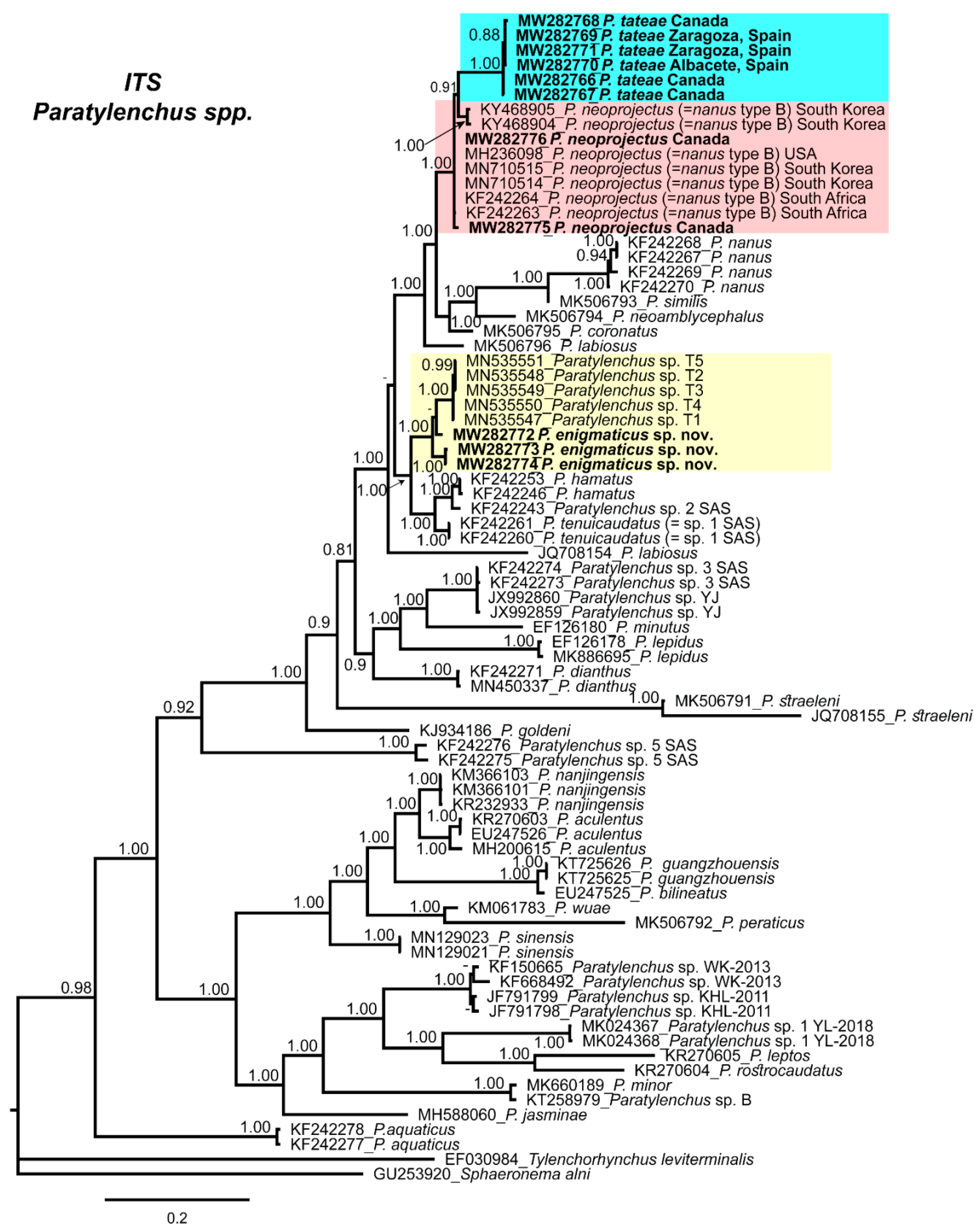
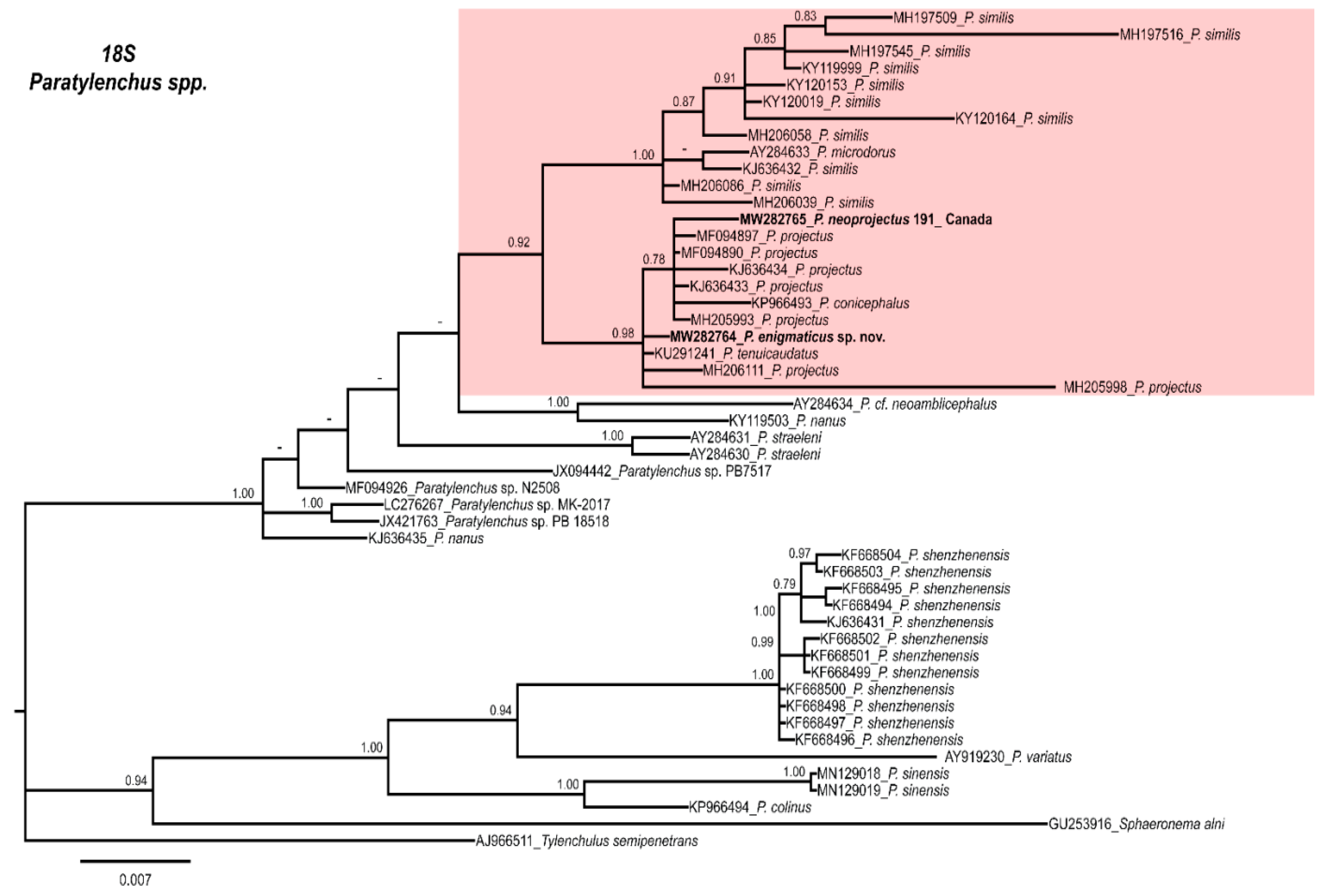
2.4. Molecular Characterization and Phylogenetic Analysis of Paratylenchus Populations from Canada and Spain
3. Discussion
4. Materials and Methods
4.1. Isolation and Morphological/Morphometrical Studies
4.2. DNA Extraction, PCR and Sequencing
4.3. Phylogenetic Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Statistics Canada. Table 32-10-0358-01 Area, production and farm value of potatoes. Stat. Can. 2020. [Google Scholar] [CrossRef]
- Potter, J.; McKeown, A. Fluctuations of populations of the pin nematode Paratylenchus projectus under selected potato management practices. Phytoprotection 2002, 83, 147–155. [Google Scholar] [CrossRef][Green Version]
- Mahran, A.; Tenuta, M.; Shinners-Carenelly, T.; Mundo-Ocampo, M.; Daayf, F. Prevalence and species identification of Pratylenchus spp. in Manitoba potato fields and host suitability of ‘Russet Burbank’. Can. J. Plant Pathol. 2010, 32, 272–282. [Google Scholar] [CrossRef]
- Forge, T.A.; Larney, F.J.; Kawchuk, L.M.; Pearson, D.C.; Koch, C.; Blackshaw, R.E. Crop rotation effects on Pratylenchus neglectus populations in the root zone of irrigated potatoes in southern Alberta. Can. J. Plant Pathol. 2015, 37, 363–368. [Google Scholar] [CrossRef]
- Loof, P.A.A. Paratylenchus projectus, CI.H., Descriptions of Plant-Parasitic Nematodes; Set 5, No. 71; Commonwealth Agricultural Bureau: St Albans, UK, 1975. [Google Scholar]
- Siddiqi, M.R. Tylenchida: Parasites of Plants and Insects, 2nd ed.; CABI Publishing: Wallingford, UK, 2000; p. 833. [Google Scholar]
- Wang, K.; Li, Y.; Xie, H.; Wu, W.J.; Xu, C.L. Pin nematode slow decline of Anthurium andreanum, a new disease caused by the pin nematode Paratylenchus shenzhenensis. Plant Dis. 2016, 100, 940–945. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pereira, G.F. Survey of Plant-Parasitic Nematodes in Pulse Crop Fields of the Canadian Prairies. Master’s Thesis, University of Manitoba, Winnipeg, MB, Canada, 2018. [Google Scholar]
- Townshend, J.L.; Eggens, J.L.; McCollum, N.K. Occurrence and population densities of nematodes associated with forage crops in eastern Canada. Can. Plant Dis. Surv. 1973, 53, 131–136. [Google Scholar]
- Townshend, J.L.; Potter, J.W.; Marks, C.F.; Loughton, A. The pin nematode, Paratylenchus projectus, in rhubarb in Ontario. Can. J. Plant Sci. 1973, 53, 377–381. [Google Scholar] [CrossRef]
- Townshend, J.L.; Potter, J.W. Evaluation of forage legumes, grasses, and cereals as hosts of forage nematodes. Nematologica 1976, 22, 196–201. [Google Scholar] [CrossRef]
- Townshend, J.L.; Cline, R.A.; Driks, V.A.; Marks, C.F. Assessment of turfgrasses for the management of Pratylenchus penetrans and Paratylenchus projectus in orchards. Can. J. Plant Sci. 1984, 64, 355–360. [Google Scholar] [CrossRef]
- Kimpinski, J. Nematodes associated with potato in Prince Edward Island and New Brunswick. Ann. App. Nematol. 1987, 1, 17–19. [Google Scholar]
- Jenkins, W.R. Paratylenchus projectus, new species (Nematoda: Criconematidae), with a key to the species of Paratylenchus. J. Wash. Acad. Sci. 1956, 46, 296–298. [Google Scholar]
- Wood, F.H. Biology and host range of Paratylenchus projectus Jenkins, 1956 (Nematoda: Criconematidae) from a sub-alpine tussock grassland. N. Z. J. Agric. Res. 1973, 16, 381–384. [Google Scholar] [CrossRef]
- Micoletzky, H. Die Freilebenden Erd-Nematoden. Archiv für Naturgeschichte Berlin A 1921, 87, 1–650. [Google Scholar]
- Jenkins, W.R.; Taylor, D.P. Paratylenchus dianthus, n. sp. (Nematoda, Criconematidae), a parasite of carnation. Proc. Helminthol. Soc. Wash. 1956, 23, 124–127. [Google Scholar]
- Thorne, G.; Allen, M.W. Paratylenchus hamatus n. sp. and Xiphinema index n. sp., two nematodes associated with fig roots, with a note on Paratylenchus anceps Cobb. Proc. Helminthol. Soc. Wash. 1950, 17, 27–35. [Google Scholar]
- Andrássy, I. Neue und wenig bekannte nematoden aus Jugoslawien. Ann. Hist.-Nat. Musei Natl. Hung. 1959, 51, 259–275. [Google Scholar]
- Geraert, E. The genus Paratylenchus. Nematologica 1965, 11, 301–334. [Google Scholar] [CrossRef]
- Wang, K.; Xie, H.; Li, Y.; Xu, C.L.; Yu, L.; Wang, D.W. Paratylenchus shenzhenensis n. sp. (Nematoda: Paratylenchinae) from the rhizosphere soil of Anthurium andreanum in China. Zootaxa 2013, 3750, 167–175. [Google Scholar] [CrossRef]
- Ghaderi, R. The damage potential of pin nematodes, Paratylenchus Micoletzky, 1922 sensu lato spp. (Nematoda: Tylenchulidae). J. Crop Prot. 2019, 8, 243–257. [Google Scholar]
- Ghaderi, R.; Geraert, E.; Karegar, A. The Tylenchulidae of the World; Identification of the Family Tylenchulidae (Nematoda: Tylenchida), 2nd ed.; Academia Press: Ghent, Belgium, 2016; p. 453. [Google Scholar]
- Yu, Q.; Ye, W.; Powers, T. Morphological and Molecular Characterization of Gracilacus wuae n. sp. (Nematoda: Criconematoidea) associated with Cow Parsnip (Heracleum maximum) in Ontario, Canada. J. Nematol. 2016, 48, 203–213. [Google Scholar]
- Ghaderi, R.; Kashi Nahanji, L.; Karegar, A. Contribution to the study of the genus Paratylenchus Micoletzky, 1922 sensu lato (Nematoda: Tylenchulidae). Zootaxa 2014, 3841, 151–187. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.Y.; Hawn, E.J. Paratylenchus neoprojectus n. sp. (Paratylenchinae: Nematoda) from alfalfa fields in Alberta, Canada. Can. J. Zool. 1975, 53, 1841–1843. [Google Scholar] [CrossRef]
- Wu, Y.L.; Townshend, J.L. Paratylenchus tateae n. sp. (Paratylenchinae, Nematoda). Can. J. Zool. 1973, 51, 109–111. [Google Scholar] [CrossRef]
- Van den Berg, E.; Tiedt, L.R.; Subbotin, S.A. Morphological and molecular characterisation of several Paratylenchus Micoletzky, 1922 (Tylenchida: Paratylenchidae) species from South Africa and USA, together with some taxonomic notes. Nematologica 2014, 16, 323–358. [Google Scholar] [CrossRef]
- Bajaj, H.K. On the species of Paratylenchus Micoletzky (Nematoda: Criconematina) from Haryana, India. Indian J. Nematol. 1987, 17, 318–326. [Google Scholar]
- Ghaderi, R.; Karegar, A. Some species of Paratylenchus (Nematoda: Tylenchulidae) from Iran. Iran. J. Plant Pathol. 2013, 49, 137–156, (In Persian with English Abstract). [Google Scholar]
- Raski, D.J. Revision of the genus Paratylenchus Micoletzky, 1922 and descriptions of new species. Part II of three parts. J. Nematol. 1975, 7, 274–295. [Google Scholar]
- Anderson, R.V.; Kimpinski, J. Paratylenchus labiosus n. sp. (Nematoda: Paratylenchidae) from Canada. Can. J. Zool. 1977, 55, 1992–1996. [Google Scholar] [CrossRef]
- Wu, L.Y. Paratylenchus brevihastus n. sp. (Nematoda: Criconematidae). Can. J. Zool. 1962, 40, 391–393. [Google Scholar] [CrossRef]
- Claerbout, J.; Vandevelde, I.; Venneman, S.; Kigozi, A.; de Sutter, N.; Neukermans, J.; Bleyaert, P.; Bert, W.; Hofte, M.; Viaene, N. A thorough study of a Paratylenchus sp. in glasshouse-grown lettuce: Characterisation, population dynamics, host plants and damage threshold as keys to its integrated management. Ann. Appl. Biol. 2020, 178, 62–79. [Google Scholar] [CrossRef]
- Cob, N.A. Notes on Paratylenchus, a genus of nemas. J. Wash. Acad. Sci. 1923, 13, 251–257. [Google Scholar]
- Wu, L.Y. Paratylenchus tenuicaudatus n. sp. (Nematoda: Criconematidae). Can. J. Zool. 1961, 39, 163–165. [Google Scholar] [CrossRef]
- Raski, D.J. Paratylenchoides gen. n. and two new species (Nematoda: Paratylenchidae). Proc. Helminthol. Soc. Wash. 1973, 40, 230–233. [Google Scholar]
- Khan, E.; Prasad, S.K.; Mathur, V.K. Two new species of the genus Paratylenchus Micoletzky, 1922 (Nematoda: Criconematidae) from India. Nematologica 1967, 13, 79–84. [Google Scholar] [CrossRef]
- Colbran, R.C. Studies of plant and soil nematodes. 10. Paratylenchus coronatus n. sp. (Nematoda: Criconematidae), a pin nematode associated with citrus. Qld. J. Agric. Anim. Sci. 1965, 22, 277–279. [Google Scholar]
- Mirbabaei, H.; Eskandari, A.; Ghaderi, R.; Karegar, A. On the synonymy of Trophotylenchulus asoensis and T. okamotoi with T. arenarius, and intra-generic structure of Paratylenchus (Nematoda: Tylenchulidae). J. Nematol. 2019, 51, 1–14. [Google Scholar] [CrossRef]
- Brzeski, M.W.; Hanel, L. Paratylenchinae: Evaluation of diagnostic morpho-biometrical characters of females in the genus Paratylenchus Micoletzky, 1922 (Nematoda: Tylenchulidae). Nematology 2000, 2, 253–261. [Google Scholar] [CrossRef]
- Akyazi, F.; Felek, A.F.; Cermak, V.; Cudejkova, M.; Foit, J.; Yildiz, S.; Hanel, L. Description of Paratylenchus (Gracilacus) straeleni (De Coninck, 1931) Oostenbrink, 1960 (Nematoda: Criconematoidea, Tylenchulidae) from hazelnut in Turkey and its comparison with other world populations. Helminthologia 2015, 52, 270–279. [Google Scholar] [CrossRef]
- Brzeski, M.W. Paratylenchinae: Morphology of some known species and descriptions of Gracilacus bilineata sp. n. and G. vera sp. n. (Nematoda: Tylenchulidae). Nematologica 1995, 41, 535–565. [Google Scholar] [CrossRef]
- Bahmani, J.; Barooti, S.; Ghaderi, R. On occurrence of Paratylenchus labiosus Anderson & Kimpinski, 1977 (Nematoda: Tylenchulidae) in Iran, with discussion on the validity of the species. J. Crop Prot. 2014, 3, 273–281. [Google Scholar]
- Maria, M.; Gu, J.; Fang, Y.; He, J.; Castillo, P.; Li, H. Radopholoides japonicus n. sp. (Nematoda: Pratylenchidae) found in rhizosphere soil associated with Podocarpus macrophyllus from Japan. Nematologica 2017, 19, 1095–1105. [Google Scholar] [CrossRef]
- Fang, E.; Li, H.; Maria, M.; Bert, W. Description of Pseudaphelenchus zhoushanensis n. sp. (Tylenchina: Aphelenchoididae) found in the wood of Pinus thunbergii at Zhoushan Islands, Zhejiang Province, China. Nematologica 2016, 18, 1151–1164. [Google Scholar] [CrossRef]
- Pedram, M.; Pourhashemi, M.; Hosseinzadeh, J.; Koolivand, D. Comments on taxonomic status and host association of some Laimaphelenchus spp. (Rhabditida: Aphelenchoidea). Nematologica 2018, 20, 483–489. [Google Scholar] [CrossRef]
- Powers, T.; Harris, T.; Higgins, R.; Mullin, P.; Sutton, L.; Powers, K. MOTUs, morphology and biodiversity estimation. A case study using nematodes of the suborder Criconematina and a conserved 18S DNA barcode. J. Nematologica 2011, 43, 35–48. [Google Scholar]
- Powers, T.; Bernard, E.C.; Harris, T.; Higgins, R.; Olson, M.; Lodema, M.; Mullin, P.; Sutton, L.; Powers, K.S. COI haplotype groups in Mesocriconema (Nematoda: Criconematidae) and their morphospecies associations. Zootaxa 2014, 3827, 101–146. [Google Scholar] [CrossRef]
- Powers, T.; Bernard, E.; Harris, T.; Higgins, R.; Olson, M.; Olson, S.; Lodema, M.; Matczyszyn, J.; Mullin, P.; Sutton, L.; et al. Species discovery and diversity in Lobocriconema (Criconematidae: Nematoda) and related plant-parasitic nematodes from North American ecoregions. Zootaxa 2016, 4085, 301–344. [Google Scholar] [CrossRef]
- Powers, T.; Harris, T.; Higgins, R.; Mullin, P.; Powers, K. An 18S rDNA Perspective on the Classification of Criconematoidea. J. Nematologica 2017, 49, 236–244. [Google Scholar] [CrossRef]
- Gutiérrez-Gutiérrez, C.; Palomares-Rius, J.E.; Cantalapiedra-Navarrete, C.; Landa, B.B.; Esmenjaud, D.; Castillo, P. Molecular analysis and comparative morphology to resolve a complex of cryptic Xiphinema species. Zool. Scr. 2010, 39, 483–498. [Google Scholar] [CrossRef]
- Gutiérrez-Gutiérrez, C.; Cantalapiedra-Navarrete, C.; Decraemer, W.; Vovlas, N.; Prior, T.; Palomares-Rius, J.E.; Castillo, P. Phylogeny, diversity, and species delimitation in some species of the Xiphinema americanum-group complex (Nematoda: Longidoridae), as inferred from nuclear and mitochondrial DNA sequences and morphology. Eur. J. Plant Pathol. 2012, 134, 561–597. [Google Scholar] [CrossRef]
- Palomares-Rius, J.E.; Cantalapiedra-Navarrete, C.; Castillo, P. Cryptic species in plant-parasitic nematodes. Nematologica 2014, 16, 1105–1118. [Google Scholar] [CrossRef]
- Čermák, V.; Renčo, M. The family Paratylenchidae Thorne, 1949 in the rhizosphere of grass and woody species in Europe: A review of the literature. Helminthol. 2010, 47, 139–146. [Google Scholar] [CrossRef]
- Esmaeili, M.; Heydari, R.; Castillo, P.; Ziaie Bidhendi, M.; Palomares-Rius, J.E. Molecular characterization of two known species of Paratylenchus Micoletzky, 1922 from Iran with notes on the validity of Paratylenchus audriellus Brown, 1959. Nematologica 2016, 18, 591–604. [Google Scholar] [CrossRef]
- Maria., M.; Miao, W.; Castillo, P.; Zheng, J. A new pin nematode, Paratylenchus sinensis n. sp. (Nematoda: Paratylenchinae) in the rhizosphere of white mulberry from Zhejiang Province, China. Eur. J. Plant Pathol. 2020, 156, 1023–1039. [Google Scholar] [CrossRef]
- Rhoades, H.L.; Linford, M.B. Molting of pre-adult nematodes of the genus Paratylenchus stimulated by root diffusates. Science 1961, 130, 1476–1477. [Google Scholar] [CrossRef]
- Fisher, J.M. Effect of temperature and host on Paratylenchus neoamblycephalus and effect of the nematode on the host. Aust. J. Agric. Res. 1967, 18, 921–929. [Google Scholar] [CrossRef]
- Wu, Y.L. Paratylenchus projectus (Paratylenchinae: Nematoda) and some closely related species. Can. J. Zool. 1975, 53, 1875–1881. [Google Scholar] [CrossRef]
- Kimpinski, J.; Smith, E.M. Nematodes in potato soils in New Brunswick. Can. Plant Dis. Surv. 1988, 68, 147–148. [Google Scholar]
- Jenkins, W.R. A rapid centrifugal-flotation technique for separating nematodes from soil. Plant Dis. Rep. 1964, 48, 692. [Google Scholar]
- Seinhorst, J.W. A rapid method for the transfer of nematodes from fixative to anhydrous glycerin. Nematologica 1959, 4, 67–69. [Google Scholar] [CrossRef]
- De Grisse, A.T. Redescription ou modifications de quelques techniques utilisées dans l’étude des nématodes phytoparasitaires. Mededelingen van de Faculteit Landbouwwetenschappen Rijksuniversiteit. Gent 1969, 34, 351–369. [Google Scholar]
- Maria, M.; Powers, T.O.; Tian, Z.; Zheng, J. Distribution and description of criconematids from Hangzhou, Zhejiang Province, China. J. Nematol. 2018, 50, 183–206. [Google Scholar]
- Holterman, M.; van der Wurff, A.; van den Elsen, S.; van Megen, H.; Holovachov, O.; Bakker, J.; Helder, J. Phylum wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol. Biol. Evol. 2006, 23, 1792–1800. [Google Scholar] [CrossRef]
- Holterman, M.; Rybarczyk, K.; van Den Elsen, S.; van Megen, H.; Mooyman, P.; Peña- Santiago, R.; Bongers, T.; Bakker, J.; Helder, J. A ribosomal DNA-based framework for the detection and quantification of stress-sensitive nematode families in terrestrial habitats. Mol. Ecol. Resour. 2008, 8, 23–34. [Google Scholar] [CrossRef]
- Ferris, V.R.; Ferns, J.M.; Faghihi, J. Variation in spacer ribosomal DNA in some cyst-forming species of plant-parasitic nematodes. Fundam. Appl. Nematol. 1993, 16, 177–184. [Google Scholar]
- Curran, J.; Driver, F.; Ballard, J.W.O.; Milner, R.J. Phylogeny of Metarhizium: Analysis of ribosomal DNA sequence data. Mycol. Res. 1994, 98, 547–552. [Google Scholar] [CrossRef]
- Maroteaux, L.; Herzog, M.; Soye-Gobillard, M.O. Molecular organization of dinogflagellate ribosomal DNA: Evolutionary implications of the deduced 5.8 S rRNA secondary structure. Biosystems 1985, 18, 307–319. [Google Scholar] [CrossRef]
- Carta, L.; Li, S. Improved 18S small subunit rDNA primers for problematic nematode amplification. J. Nematol. 2018, 50, 533–542. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]


| Present Study | Wu & Hawn [26] | * Van den Berg et al. [28] | |||
|---|---|---|---|---|---|
| Characters | Females | Juveniles | Females | Females | Juveniles |
| n | 11 | 4 | 76 | 17 | 4 |
| Body length | 383.5 ± 36.7 (330.0–434.0) | 342.0 ± 19.6 (322.0–365.0) | 327–405 | 359 (300–415) | 339.5 (299–390) |
| a | 24.0 ± 1.7 (21.0–26.0) | 22.3 ± 1.9 (20.5–24.3) | 18–26 | 22.1 (19.5–24.6) | 20.4 (17.7–22.9) |
| b | 3.8 ± 0.3 (3.3–4.3) | 3.9 ± 0.3 (3.5–4.1) | 3.8–4.6 | 3.9 (3.5–4.4) | 4.1 (3.7–4.7) |
| c | 14.6 ± 1.8 (12.1–18.5) | 12.8 ± 1.6 (11.0–15.0) | 14–16 | 15.3 (14–18.5) | 13.8 (12.3–18.9) |
| c’ | 2.7 ± 0.2 (2.3–3.0) | 2.3 ± 0.3 (1.9–2.6) | - | 2.4 (2.0–2.8) | 2.2 (1.7–2.5) |
| V | 84.4 ± 1.3 (82.0–85.8) | - | 82–85.7 | 84 (82.5–85) | - |
| Stylet percentage | 7.0 ± 0.8 (5.8–8.3) | - | - | 8 (6.8–9.3) | - |
| Lip height | 3.3 ± 0.4 (3.0–4.0) | - | - | 3.5 (3–4) | - |
| Lip width | 6.4 ± 0.4 (6.0–7.0) | - | - | 7 (6.5–7.5) | - |
| Stylet length | 25.3 ± 1.3 (25.0–29) | 13.3 ± 1.0 (12.0–14.0) | 28–31 | 28.5 (26–31) | 10 (3.5–14.5) |
| Median bulb length | 23.4 ± 1.6 (21.0–25.0) | - | - | - | - |
| Median bulb width | 9.3 ± 0.8 (8.0–11.0) | - | - | - | - |
| Anterior end to excretory pore | 79.1 ± 4.8 (70.0–85.0) | 75.0 ± 5.2 (70.0–80.0) | - | 77.5 (71–85) | 71 (65–78.5) |
| Pharynx length | 99.0 ± 4.2 (92.0–106.0) | 89.0 ± 6.2 (80.0–93.0) | 82–94 | 92 (85–110) | 83.5 (72.5–94.5) |
| Maximum body width | 16.0 ± 1.4 (13.5–18.0) | 15.4 ± 0.4 (15.0–15.8) | - | 16 (13–20) | - |
| Vulva body width | 13.6 ± 1.3 (12.0–15.0) | - | - | - | - |
| Anal body width | 9.7 ± 0.9 (8.0–11.2) | 11.7 ± 0.7 (10.7–12.4) | - | - | - |
| Distance from vulva to anus | 33.5 ± 5.8 (28.0–44.0) | - | 29–44 | 33.5 (26–44) | - |
| Distance from vulva to tail terminus | 60.0 ± 7.3 (50.0–72.0) | - | - | - | - |
| Tail length | 26.0 ± 2.9 (22.0–30.0) | 27.0 ± 3.5 (22.0–30.0) | 23–27 | 23.5 (17.5–29.5) | 23 (20.5–29.5) |
| Canadian Populations | Spanish Populations | Wu & Townshend [27] | ||||
|---|---|---|---|---|---|---|
| Characters | Females | Juveniles | Females | Females | ||
| Populations | 091 | 041 | 091 | Ariza, Zaragoza | Alpera, Albacete | type population |
| n | 18 | 18 | 6 | 20 | 8 | 43 |
| Body length | 333.6 ± 33.7 (269.0–380.0) | 349.5 ± 25.4 (314.0–388.0) | 315.5 ± 26.4 (267.0–342.0) | 346.2 ± 25.8 (310.0–425.0) | 334.4 ± 14.3 (310.0–353.0) | 315–401 |
| a | 23.4 ± 1.4 (21.4–26.2) | 23.9 ± 1.9 (20.4–27.0) | 22.0 ± 1.9 (18.8–24.0) | 21.8 ± 1.6 (17.4–24.3) | 21.7 ± 1.7 (19.1–23.5) | 19–26 |
| b | 3.6 ± 0.3 (3.2–4.0) | 3.9 ± 0.3 (3.3–4.7) | 3.9 ± 0.2 (3.5–4.1) | 3.7 ± 0.2 (3.3–4.2) | 3.6 ± 0.2 (3.3–4.1) | 3.8–5.9 |
| c | 11.9 ± 1.1 (10.0–13.8) | 13.5 ± 1.4 (11.6–16.9) | 15.4 ± 1.9 (13.5–18.9) | 13.9 ± 1.9 (10.5–17.7) | 13.2 ± 1.8 (11.3–15.3) | 11.7–15.8 |
| c’ | 3.5 ± 0.4 (3.0–4.5) | 3.3 ± 0.3 (2.8–3.9) | 2.6 ± 0.1 (2.4–2.8) | 2.9 ± 0.4 (2.5–3.8) | 2.9 ± 0.2 (2.6–3.1) | - |
| V | 82.3 ± 1.2 (80.0–84.3) | 82.9 ± 0.9 (80.8–84.1) | - | 82.9 ± 1.4 (80.2–85.6) | 82.6 ± 1.4 (81.3–85.0) | 80.5–84.7 |
| Lip height | 2.6 ± 0.2 (2.0–3.0) | 2.8 ± 0.3 (2.0–3.0) | - | - | - | - |
| Lip width | 5.5 ± 0.2 (5.0–6.0) | 5.6 ± 0.4 (5.0–6.0) | - | 5.2 ± 0.4 (4.5–6.0) | 5.2 ± 0.4 (4.5–6.0) | - |
| Stylet length | 17.3 ± 0.9 (15.0–19.0) | 16.5 ± 0.9 (14.5–18.0) | 12.0 ± 1.1 (10.0–13.0) | 15.5 ± 0.4 (14.5–16.0) | 15.4 ± 0.4 (15.0–16.0) | 15–16.8 |
| Median bulb length | 21.9 ± 1.5 (19.4–24.2) | 20.6 ± 2.3 (16.0–24.0) | - | 18.2 ± 1.7 (15.5–22.0) | 17.4 ± 1.2 (16.0–19.0) | - |
| Median bulb width | 8.1 ± 0.6 (7.2–9.0) | 8.2 ± 0.8 (7.2–10.0) | - | 8.9 ± 0.6 (8.0–10.0) | 8.6 ± 0.5 (8.0–9.5) | - |
| Anterior end to excretory pore | 73.9 ± 3.8 (64.0–81.0) | 73.4 ± 5.3 (63.0–84.0) | 66.8 ± 4.8 (60.0–71.0) | 78.2 ± 6.0 (70.5–93.0) | 77.4 ± 3.8 (72.5–84.0) | 68–81 |
| Pharynx length | 91.7 ± 3.4 (83.0–98.0) | 90.2 ± 4.9 (82.0–98.0) | 80.3 ± 2.9 (76.0–83.0) | 93.1 ± 5.0 (85.5–103.0) | 92.1 ± 5.6 (85.5–102.0) | 77–89 |
| Maximum body width | 14.2 ± 1.2 (12.0–16.0) | 14.6 ± 0.8 (13.0–16.0) | 14.3 ± 0.6 (13.0–15.0) | 16.0 ± 1.9 (14.5–21.5) | 15.5 ± 1.3 (14.5–18.5) | - |
| Anal body width | 8.1 ± 0.8 (6.0–9.0) | 7.9 ± 0.6 (7.0–9.0) | 7.9 ± 0.2 (7.5–8.0) | 8.7 ± 0.4 (8.0–9.5) | 8.9 ± 0.9 (8.0–11.0) | - |
| Distance from vulva to anus | 30.6 ± 3.6 (26.0–39.0) | 33.5 ± 4.2 (27.0–43.0) | - | - | - | 28–41 |
| Distance from vulva to tail terminus | 58.8 ± 5.6 (52.0–70.6) | 59.6 ± 4.8 (51.0–67.0) | - | - | - | - |
| Tail length | 28.2 ± 3.0 (24.0–35.0) | 26.1 ± 2.5 (21.0–30.0) | 20.7 ± 1.7 (18.0–23.0) | 25.3 ± 3.3 (21.5–32.5) | 25.6 ± 2.9 (22.5–30.0) | 22–33 |
| Canadian Population | * Belgian Population Claerbout et al. [34] | |||||||
|---|---|---|---|---|---|---|---|---|
| Holotype | Paratype | |||||||
| Characters | Female | Females | Juveniles | T1 | T2 | T3 | T4 | T5 |
| n | 11 | 5 | 10 | 10 | 10 | 10 | 10 | |
| Body length | 372 | 382.7 ± 30.9 (343.0–431.0) | 344.3 ± 9.5 (331.0–357.0) | 365 ± 40 (308–465) | 335 ± 20 (302–360) | 365 ± 39 (313–422) | 358 ± 43 (300–411) | 328 ± 31 (293–368) |
| a | 24.6 | 25.7 ± 2.1 (21.7–28.7) | 23.8 ± 0.4 (23.1–24.4) | 24.2 ± 3.8 (14.9–27.6) | 24.3 ± 3.4 (19.3–27.2) | 26.7 ± 2.3 (22–29) | 23.7 ± 2.6 (18.5–27.5) | 23.2 ± 3.3 (18.1–28.1) |
| b | 3.9 | 4.1 ± 0.3 (3.7–4.7) | 4.2 ± 0.2 (3.9–4.4) | 3.7 ± 0.7 (2.7–4.6) | - | 3.4 ± 0.7 (2.5–4.9) | 3.2 ± 0.5 (2.8–4.2) | - |
| c | 15.7 | 15.4 ± 1.3 (12.9–17.5) | 14.9 ± 0.5 (14.4–15.7) | 15.0 ± 1.5 (12.3–17.2) | 14.9 ± 1.5 (13.2–17) | 14.9 ± 1.9 (12.7–17.8) | 14.8 ± 2.3 (13.7–19.8) | 13.0 ± 1.5 (10.1–15.7) |
| c′ | 2.5 | 2.6 ± 0.3 (2.3–3.1) | 2.3 ± 0.3 (1.9–2.6) | - | - | - | - | - |
| V | 84.1 | 85 ± 0.9 (83.0–86.3) | - | 83.2 ± 2.1 (80.4–87.8) | 83.2 ± 2.1 (80–87) | 83.0 ± 1.5 (80–84) | 83.5 ± 0.9 (82.8–84.9) | 83.1 ± 2.1 (80.1–88) |
| Lip height | 3.1 | 3.0 ± 0.3 (2.6–3.6) | - | - | - | - | - | - |
| Lip width | 7.5 | 7.1 ± 0.4 (6.5–7.7) | - | - | - | - | - | - |
| Stylet length | 28.9 | 28.8 ± 1.1 (27.3–30.8) | 12.5 ± 0.9 (11.2–13.5) | 27.3 ± 1.3 (23.5–28.4) | 25.5 ± 1.6 (22.3–26.5) | 26.6 ± 1.5 (25.2–30.5) | 26.8 ± 1.3 (24.6–27.9) | 27.0 ± 1.5 (24.6–28.6) |
| Stylet percentage | 7.7 | 7.6 ± 0.5 (6.8–8.2) | - | 7.5 ± 0.9 (6.0–8.8) | 7.6 ± 0.7 (7.2–8.8) | 7.3 ± 0.7 (6.2–7.9) | 7.6 ± 0.8 (6.6–8.4) | 8.3 ± 0.5 (7.3–8.9) |
| Median bulb length | 21.2 | 20.4 ± 1.0 (18.5–21.3) | - | - | - | - | - | - |
| Median bulb width | 9.8 | 9.6 ± 1.1 (8.0–11.4) | - | - | - | - | - | - |
| Anterior end to excretory pore | 79 | 76.0 ± 4.2 (70.0–82.0) | 65.2 ± 2.8 (63.0–70.0) | - | - | - | - | - |
| Pharynx length | 95 | 93.8 ± 5.2 (83.0–100.0) | 81.6 ± 4.3 (76.0–88.0) | 100.7 ± 19.7 (75.2–137.7) | 88.0 ± 23.3 (42.9–105.8) | 109.9 ± 16.9 (83.3–123.5) | 114.7 ± 18.4 (84.6–125.7) | 120.4 ± 14.6 (95.0–144.0) |
| Maximum body width | 15.1 | 15.0 ± 1.2 (12.6–16.4) | 14.4 ± 0.3 (14.2–14.9) | - | - | - | - | - |
| Vulva body width | 12.7 | 13.1 ± 1.0 (11.4–14.7) | - | - | - | - | - | - |
| Anal body width | 9.5 | 9.7 ± 0.9 (7.7–10.6) | 10.2 ± 1.2 (8.8–11.7) | - | - | - | - | - |
| Distance from vulva to anus | 36 | 33.3 ± 4.0 (26.0–37.0) | - | - | - | - | - | - |
| Distance from vulva to tail terminus | 59.6 | 59.9 ± 3.1 (53.4–65.0) | - | - | - | - | - | - |
| Tail length | 23.6 | 24.9 ± 2.1 (22.0–29.0) | 23.2 ± 0.8 (22.0–24.0) | 24.4 ± 3.1 (21.7–30.8) | 22.6 ± 1.6 (20.3–26.2) | 24.6 ± 1.8 (21.0–26.1) | 24.5 ± 3.3 (21.2–23.7) | 25.4 ± 2.6 (22.0–30.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munawar, M.; Yevtushenko, D.P.; Palomares-Rius, J.E.; Castillo, P. Species Diversity of Pin Nematodes (Paratylenchus spp.) from Potato Growing Regions of Southern Alberta, Canada. Plants 2021, 10, 188. https://doi.org/10.3390/plants10020188
Munawar M, Yevtushenko DP, Palomares-Rius JE, Castillo P. Species Diversity of Pin Nematodes (Paratylenchus spp.) from Potato Growing Regions of Southern Alberta, Canada. Plants. 2021; 10(2):188. https://doi.org/10.3390/plants10020188
Chicago/Turabian StyleMunawar, Maria, Dmytro P. Yevtushenko, Juan E. Palomares-Rius, and Pablo Castillo. 2021. "Species Diversity of Pin Nematodes (Paratylenchus spp.) from Potato Growing Regions of Southern Alberta, Canada" Plants 10, no. 2: 188. https://doi.org/10.3390/plants10020188
APA StyleMunawar, M., Yevtushenko, D. P., Palomares-Rius, J. E., & Castillo, P. (2021). Species Diversity of Pin Nematodes (Paratylenchus spp.) from Potato Growing Regions of Southern Alberta, Canada. Plants, 10(2), 188. https://doi.org/10.3390/plants10020188






