Morphological Characterization and DNA Barcoding of Duckweed Species in Saudi Arabia
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Morphological Characteristics
4.3. Molecular Markers Analysis: DNA Barcoding
4.4. Phylogenetic Analysis Based on DNA Sequence
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gupta, C.; Prakash, D. Duckweed: An effective tool for phytoremediation. Toxicol. Environ. Chem. 2013, 95, 1256–1266. [Google Scholar] [CrossRef]
- Cheng, J.J.; Stomp, A.M. Growing Duckweed to recover nutrients from Wastewaters and for production of fuel ethanol and animal feed. Clean Soil Air Water 2009, 37, 17–26. [Google Scholar] [CrossRef]
- Sońta, M.; Rekiel, A.; Batorska, M. Use of Duckweed (Lemna L.) in sustainable livestock production and aquaculture—A review. Ann. Anim. Sci. 2019, 19, 257–271. [Google Scholar] [CrossRef]
- Les, D.H.; Landolt, E.; Crawford, D.J. Systematics of the Lemnaceae (duckweeds): Inferences from micromolecular and morphological data. Plant Syst. Evol. 1997, 204, 161–177. [Google Scholar] [CrossRef]
- Crawford, D.J.; Landolt, E.; Les, D.H. An allozyme study of two sibling species of Lemna (Lemnaceae) with comments on their morphology, ecology and distribution. Bull. Torrey Bot. Club 1996, 123, 1–6. [Google Scholar] [CrossRef]
- Crawford, D.J.; Landolt, E.; Les, D.H.; Archibald, J.K.; Kimball, R.T. Allozyme variation within and divergence between Lemna gibba and L. disperma: Systematic and biogeographic implications. Aquat. Bot. 2005, 83, 119–128. [Google Scholar] [CrossRef]
- An, D.; Li, C.; Zhou, Y.; Wu, Y.; Wang, W. Genomes and transcriptomes of duckweeds. Front. Chem. 2018, 6, 230. [Google Scholar] [CrossRef]
- Wang, W.; Wu, Y.; Yan, Y.; Ermakova, M.; Kerstetter, R.; Messing, J. DNA barcoding of the Lemnaceae, a family of aquatic monocots. BMC Plant Biol. 2010, 10, 205. [Google Scholar] [CrossRef]
- Bog, M.; Appenroth, K.-J.; Sree, K.S. Duckweed (Lemnaceae): Its molecular taxonomy. Front. Sustain. Food Syst. 2019, 3, 117. [Google Scholar] [CrossRef]
- Masrahi, Y.S.; Al-Turki, T.A.; Sayed, O.H. Wolffiella hyalina (Delile) Monod (Lemnaceae)—A new record for the flora of Saudi Arabia. Feddes Rep. 2010, 121, 189–193. [Google Scholar] [CrossRef]
- Landolt, E. Key to the Determination of Taxa within the Family of Lemnaceae; Geobotanischen Institutes der Eidg. Technische Hochschule, Stiftung Rübel: Zurich, Switzerland, 1980; pp. 13–21. [Google Scholar]
- Les, D.H.; Crawford, D.J.; Landolt, E.; Gabel, J.D.; Kimball, R.T. Phylogeny and systematics of Lemnaceae, the duckweed family. Syst. Bot. 2002, 27, 221–240. [Google Scholar]
- Azer, S. Taxonomic revision of genus Lemna L. (lemnaceae gray) in Egypt. Ann. Agric. Sci. 2013, 58, 257–263. [Google Scholar] [CrossRef][Green Version]
- Bog, M.; Baumbach, H.; Schween, U.; Hellwig, F.; Landolt, E.; Appenroth, K.J. Genetic structure of the genus Lemna L. (Lemnaceae) as revealed by amplified fragment length polymorphism. Planta 2010, 232, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Klaus, J.; Nikolai, B.; Eric, L. Telling duckweed apart: Genotyping technologies for the Lemnaceae. Chin. J. Appl. Environ. Biol. 2013, 19, 1–10. [Google Scholar] [CrossRef]
- Bog, M.; Schneider, P.; Hellwig, F.; Sachse, S.; Kochieva, E.Z.; Martyrosian, E.; Landolt, E.; Appenroth, K.-J. Genetic characterization and barcoding of taxa in the genus Wolffia Horkel ex Schleid. (Lemnaceae) as revealed by two plastid markers and amplified fragment length polymorphism (AFLP). Planta 2013, 237, 1–13. [Google Scholar] [CrossRef]
- Zhang, J.; Azizullah, A. Genetic Diversity and DNA Barcoding in the Duckweed Family. In The Duckweed Genomes; Springer: Cham, Switzerland, 2020; pp. 59–65. [Google Scholar]
- Xue, H.; Xiao, Y.; Jin, Y.; Li, X.; Fang, Y.; Zhao, H.; Zhao, Y.; Guan, J. Genetic diversity and geographic differentiation analysis of duckweed using inter-simple sequence repeat markers. Mol. Biol. Rep. 2012, 39, 547–554. [Google Scholar] [CrossRef]
- Sieben, E. The status and distribution of vascular plants (Magnoliophyta, Lycophyta, Pteridophyta). In The Status and Distribution of Freshwater Biodiversity in Southern Africa; Darwall, W., Smith, K.G., Tweddle, D., Skelton, P., Eds.; IUCN: Gland, Switzerland, 2009; pp. 83–98. [Google Scholar]
- Ding, Y.; Fang, Y.; Guo, L.; Li, Z.; He, K.; Zhao, Y.; Zhao, H. Phylogenic study of Lemnoideae (duckweeds) through complete chloroplast genomes for eight accessions. PeerJ 2017, 5, e4186. [Google Scholar] [CrossRef]
- Tippery, N.; Les, D.; Crawford, D. Evaluation of phylogenetic relationships in Lemnaceae using nuclear ribosomal data. Plant Biol. 2015, 17, 50–58. [Google Scholar] [CrossRef]
- Borisjuk, N.; Chu, P.; Gutierrez, R.; Zhang, H.; Acosta, K.; Friesen, N.; Sree, K.; Garcia, C.; Appenroth, K.; Lam, E. Assessment, validation and deployment strategy of a two-barcode protocol for facile genotyping of duckweed species. Plant Biol. 2015, 17, 42–49. [Google Scholar] [CrossRef]
- Landolt, E. Biosystematic investigations in the family of duckweeds (Lemnaceae). The family of Lemnaceae a monographic study. Veroff Geobot. Inst. ETH 1986, 71, 1–563. [Google Scholar]
- Xu, Y.; Ma, S.; Huang, M.; Peng, M.; Bog, M.; Sree, K.S.; Appenroth, K.-J.; Zhang, J. Species distribution, genetic diversity and barcoding in the duckweed family (Lemnaceae). Hydrobiologia 2015, 743, 75–87. [Google Scholar] [CrossRef]
- Halder, S.; Venu, P. Lemna landoltii sp. nov. (Lemnaceae) from India. Taiwania 2012, 58, 12–14. [Google Scholar]
- Les, D.H.; Crawford, D.J. Landoltia (Lemnaceae), a new genus of duckweeds. Novon 1999, 9, 530–533. [Google Scholar] [CrossRef]
- Wang, W.; Haberer, G.; Gundlach, H.; Gläßer, C.; Nussbaumer, T.; Luo, M.; Lomsadze, A.; Borodovsky, M.; Kerstetter, R.; Shanklin, J. The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat. Commun. 2014, 5, 3311. [Google Scholar] [CrossRef]
- Jordan, W.C.; Courtney, M.W.; Neigel, J.E. Low levels of intraspecific genetic variation at a rapidly evolving chloroplast DNA locus in North American duckweeds (Lemnaceae). Am. J. Bot. 1996, 83, 430–439. [Google Scholar] [CrossRef]
- Li, T. Annual Variation of Meteorological Radiation in Hainan Island. Meteorol. Mon. 2002, 28, 45–47. [Google Scholar]
- Ceschin, S.; Leacche, I.; Pascucci, S.; Abati, S. Morphological study of Lemna minuta Kunth, an alien species often mistaken for the native L. minor L. (Araceae). Aquat. Bot. 2016, 131, 51–56. [Google Scholar] [CrossRef]
- CBOL Plant Working Group; Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar]
- Lahaye, R.; Savolainen, V.; Duthoit, S.; Maurin, O.; Van der Bank, M. A test of psbK-psbI and atpF-atpH as potential plant DNA barcodes using the flora of the Kruger National Park (South Africa) as a model system. Nat. Preced. 2008. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 1, 2–3. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Ragan, M.A. Phylogenetic inference based on matrix representation of trees. Mol. Phylogenetics Evol. 1992, 1, 53–58. [Google Scholar] [CrossRef]
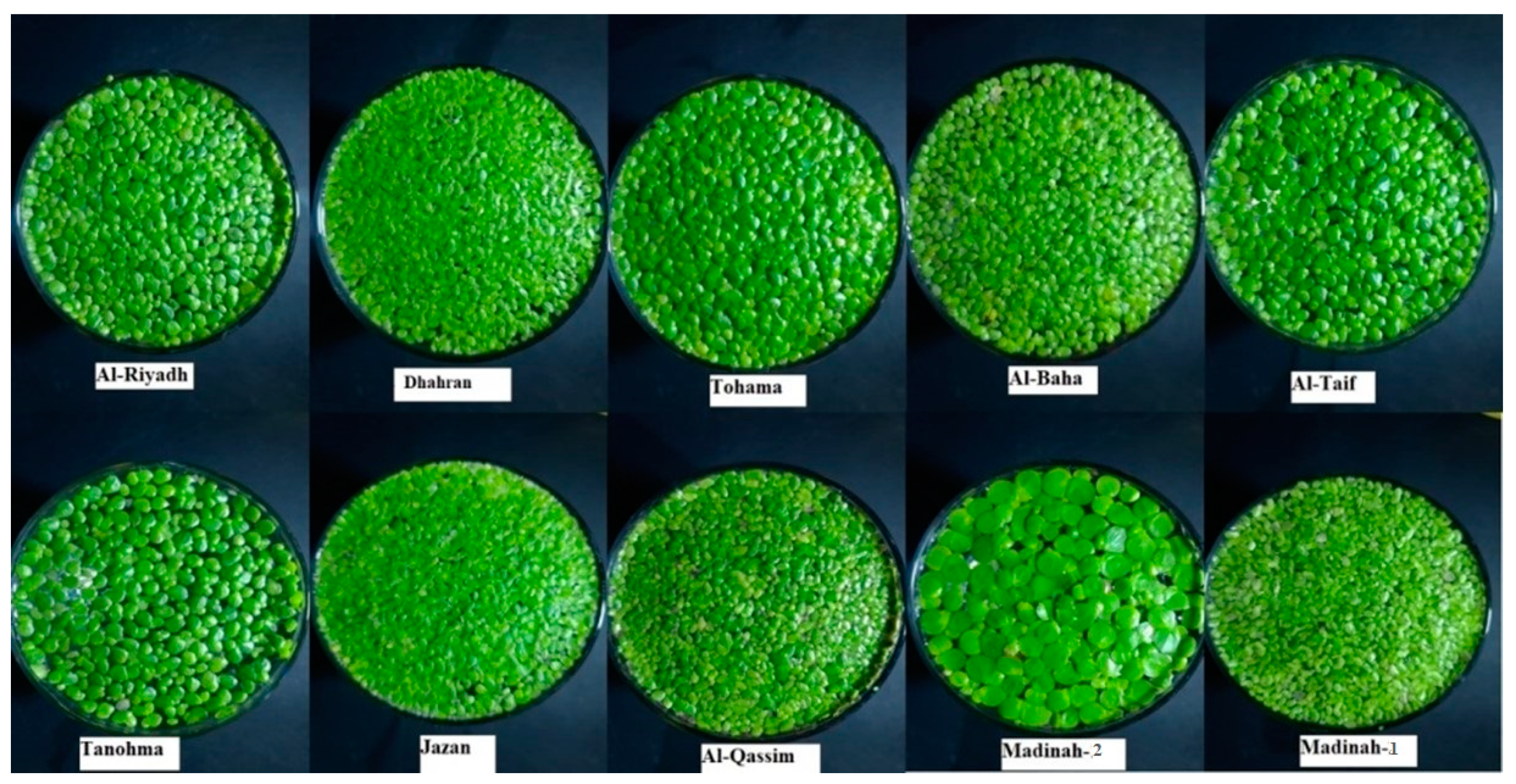


| FSh | FS | FC | FA | T | |
|---|---|---|---|---|---|
| Riyadh | Obovate | Asymmetry | Dark Green | Rounded | Present |
| Dhahran | Obovate | Symmetry | Light Green | Rounded | Present |
| Tohama | Obovate | Asymmetry | Dark Green | Rounded | Present |
| Al-Baha | Obovate | Symmetry | Dark Green | Rounded | Present |
| Al-Taif | Obovate | Asymmetry | Light Green | Rounded | Present |
| Tanomah | Obovate | Asymmetry | Light Green | Rounded | Present |
| Jazan | Obovate | Symmetry | Light Green | Rounded | Present |
| Al-Qassim | Obovate | Asymmetry | Dark Green | Rounded | Present |
| Madinah-1 | Obovate | symmetry | Light Green | Rounded | Present |
| Madinah-2 | Obovate | Asymmetry | Dark Green | Rounded | Present |
| Number of Roots | Frond Width (mm) | Frond Thickness (mm) | Frond Length (mm) | NCF Number of Contiguous Fronds | Root Length (mm) | Number of Fronds | |
|---|---|---|---|---|---|---|---|
| Riyadh | 2.00 ± 0.01 b | 5.7 ± 0.12 b | 2.3 ± 0.17 c | 7.0 ± 0.02 a | 4.0 ± 0.01 b | 2.35 ± 0.13 b | 4.00 ± 0.01 b |
| Al-Taif | 2.00 ± 0.01 b | 5.8 ± 0.06 b | 3.2 ± 0.06 a | 6.5 ± 0.06 b | 5.0 ± 0.01 a | 2.02 ± 0.32 b | 3.67 ± 0.50 b |
| Tohama | 3.00 ± 0.01 b | 5.8 ± 0.06 b | 3.0 ± 0.00 b | 6.5 ± 0.04 b | 3.0 ± 0.01 c | 2.29 ± 0.37 b | 4.00 ± 0.01 b |
| Jazan | 2.44 ± 0.53 b | 3.5 ± 0.10 d | 0.5 ± 0.06 f | 4.6 ± 0.04 e | 3.0 ± 0.01 c | 0.84 ± 0.05 c | 3.67 ± 0.50 b |
| Tanomah | 1.89 ± 0.33 b | 3.4 ± 0.06 d | 0.5 ± 0.0 f | 4.5 ± 0.09 e | 4.0 ± 0.01 b | 3.76 ± 0.49 a | 3.56 ± 0.53 b |
| Al-Baha | 1.67 ± 0.50 b | 3.5 ± 0.06 d | 1.0 ± 0.0 d | 4.7 ± 0.09 e | 4.0 ± 0.01 b | 1.98 ± 0.45 b | 3.00 ± 0.01 bc |
| Dhahran | 2.00 ± 0.01 b | 3.0 ± 0.01 e | 0.4 ± 0.0 f | 5.0 ± 0.07 d | 3.0 ± 0.01 c | 1.54 ± 0.28 b | 3.22 ± 0.44 bc |
| Al-Qassim | 8.11 ± 0.78 a | 4.0 ± 0.01 c | 0.7 ± 0.0 e | 6.0 ± 0.06 c | 5.0 ± 0.01 a | 2.14 ± 0.09 b | 6.00 ± 0.01 a |
| Madinah-1 | 2.00 ± 0.01 b | 8.5 ± 0.01 a | 0.9 ± 0.0 d | 1.0 ± 0.06 g | 4.0 ± 0.01 b | 0.84 ± 0.09 c | 2.89 ± 0.33 bc |
| Madinah-2 | 9.22 ± 2.39 a | 4.0 ± 0.06 c | 0.4 ± 0.0 f | 4.0 ± 0.05 f | 3.0 ± 0.01 c | 0.74 ± 0.09 c | 2.44 ± 0.53 bc |
| Tukey HSD | 1.60 | 0.38 | 0.18 | 0.45 | 0.42 | 0.60 | 1.10 |
| Region | psbK-psbI | trnH-psbA | matK | atpF-atpH | rpoC1 | rbcL |
|---|---|---|---|---|---|---|
| Riyadh | L. japonica (OK546023) | L. gibba (OK103562) | L. gibba (OK095301) | L. gibba (OK095300) | L. minor (OK376426) | L. gibba (OK571367) |
| Dhahran | L. japonica (OK571365) | L. gibba (OK383787) | L. gibba (OK375248) | L.gibba (OK598953) | L. gibba (Ok546019) | L. gibba (OK598946) |
| Tohama | L. japonica (OK350360) | L. gibba (OK383788) | L. gibba (OK546020) | L. gibba (OK598959) | L. minor (OK376427) | L. gibba (OK598947) |
| Al-Baha | L. japonica (OK350359) | L. gibba (OK103563) | L. gibba (OK571364) | ----- | L. minor (OK493446) | L. gibba (OK598949) |
| Jazan | L. perpusilla (OK350358) | L. aequinoctialis (OK383786) | L. aequinoctialis (OK375249) | L. aequinoctialis (OK598956) | L. aequinoctialis (OK493447) | L. minor (OK598948) |
| Al-Taif | L. japonica (OK546024) | L. gibba (OK383789) | L. gibba (OK546021) | L. gibba (OK598958) | L. gibba (OK493448) | L. gibba (OK598951) |
| Tanomah | L. japonica (OK350361) | L. gibba (OK383790) | L. gibba (OK546026) | L. gibba (OK598957) | L. minor (OK493449) | L. gibba (OK598952) |
| Al-Qassim | L. punctata (OK350363) | L. punctata (OK247674) | L. punctata (OK375250) | L. punctata (OK598955) | L. punctata (OK493450) | L. punctata (OK571368) |
| Madinah-1 | L. perpusilla (OK350362) | L.aequinoctialis (OK571366) | L.aequinoctialis (OK546022) | L. aequinoctialis (OK598954) | L. aequinoctialis (OK493451) | L. minor (OK571369) |
| Madinah-2 | S. polyryiza (OK546025) | S. polyryiza (OK247675) | ----- | S. polyryiza (OK247676) | S. polyryiza (OK493452) | S. polyryiza (OK598950) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Dakhil, M.; Alghamdi, S.; Migdadi, H.; Afzal, M.; Ali, A.A. Morphological Characterization and DNA Barcoding of Duckweed Species in Saudi Arabia. Plants 2021, 10, 2438. https://doi.org/10.3390/plants10112438
Al-Dakhil M, Alghamdi S, Migdadi H, Afzal M, Ali AA. Morphological Characterization and DNA Barcoding of Duckweed Species in Saudi Arabia. Plants. 2021; 10(11):2438. https://doi.org/10.3390/plants10112438
Chicago/Turabian StyleAl-Dakhil, Mohammed, Salem Alghamdi, Hussein Migdadi, Muhammad Afzal, and Ahmed Abdelrahim Ali. 2021. "Morphological Characterization and DNA Barcoding of Duckweed Species in Saudi Arabia" Plants 10, no. 11: 2438. https://doi.org/10.3390/plants10112438
APA StyleAl-Dakhil, M., Alghamdi, S., Migdadi, H., Afzal, M., & Ali, A. A. (2021). Morphological Characterization and DNA Barcoding of Duckweed Species in Saudi Arabia. Plants, 10(11), 2438. https://doi.org/10.3390/plants10112438






