Plum Pox Virus Strain C Isolates Can Reduce Sour Cherry Productivity
Abstract
:1. Introduction
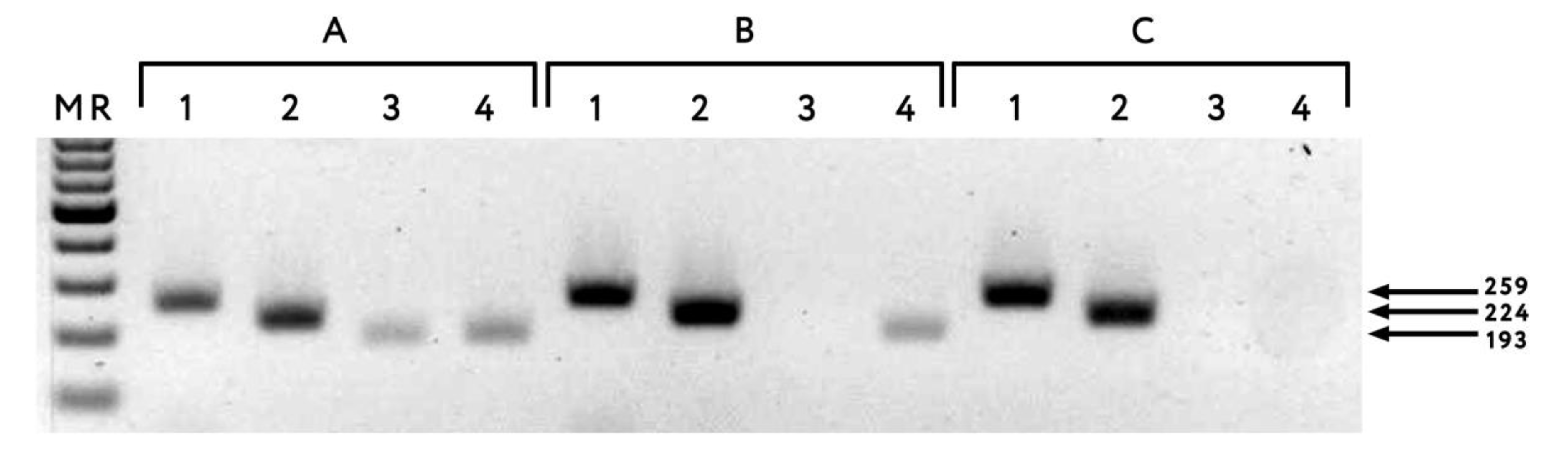
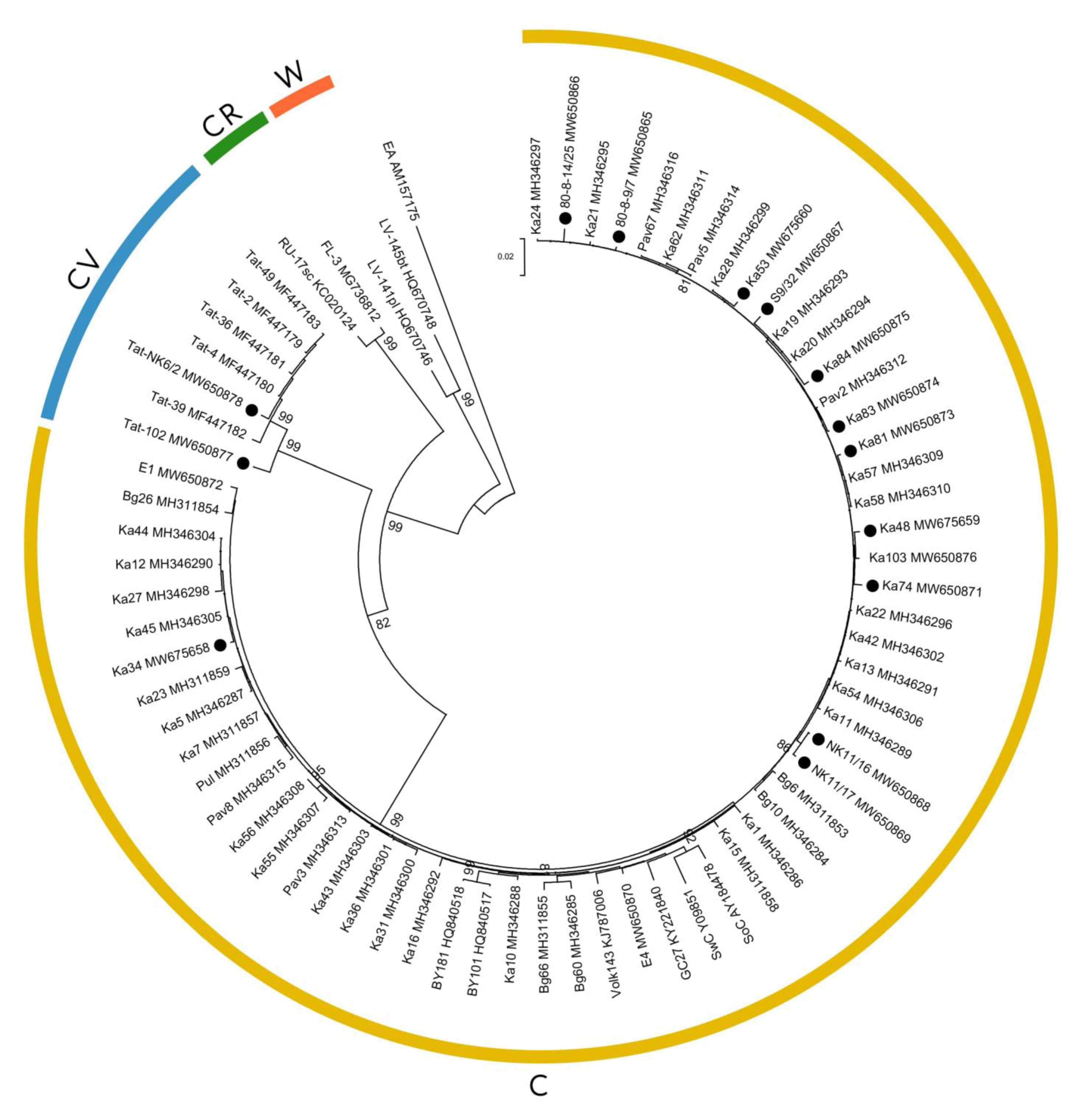
2. Results
3. Discussion
4. Materials and Methods
4.1. Plants
4.2. Sampling
4.3. ELISA
4.4. RT-PCR
4.5. Determination of PPV Strain
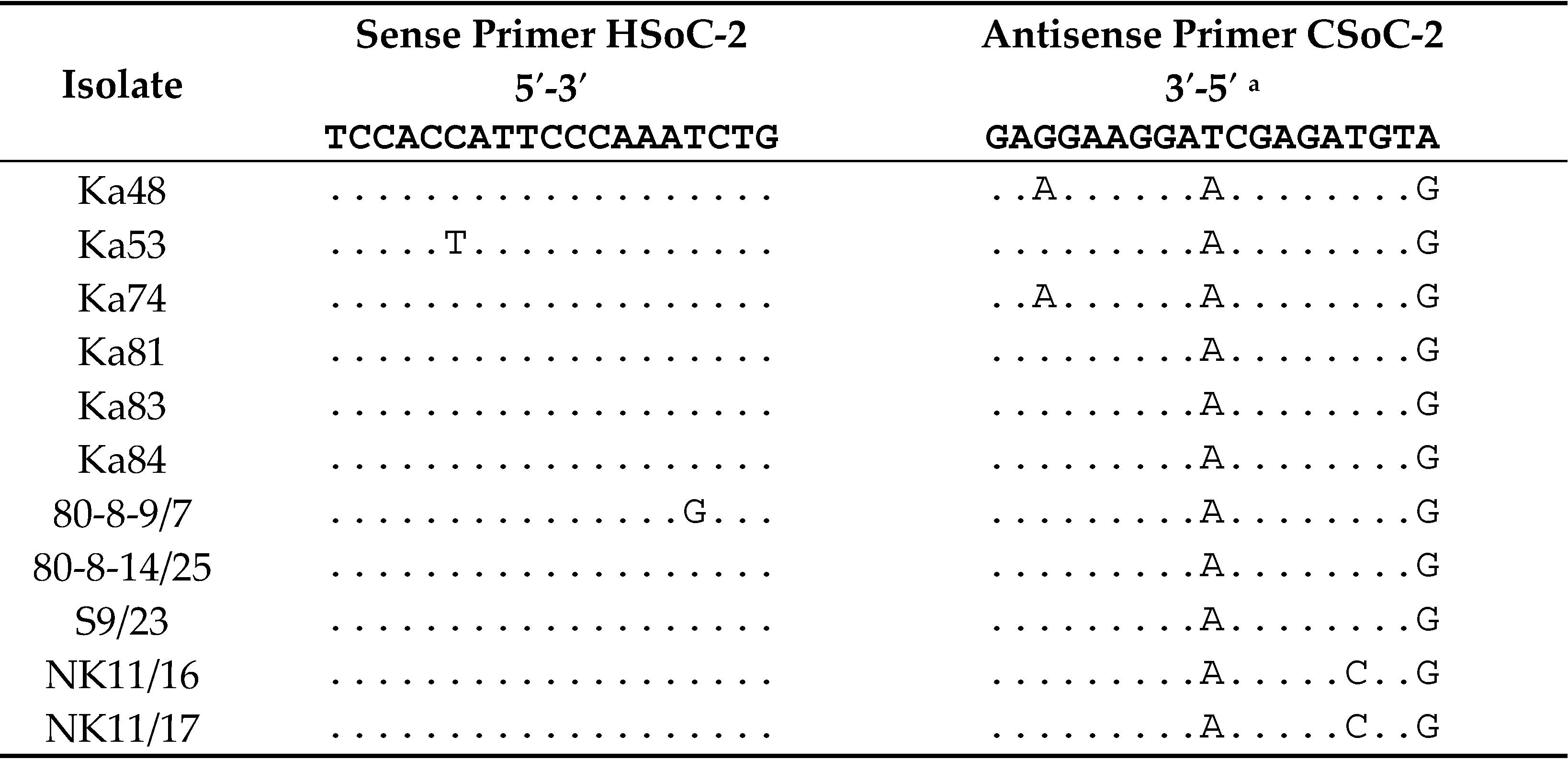
4.6. Sequencing of the 3’-Terminal Genomic Region
4.7. Sequence Analysis
4.8. Study on the Impact of PPV on Cherry Productivity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bujdoso, G.; Hrotko, K. Cherry production. In Cherries: Botany, Production and Uses; Quero-Garcia, J., Iezzoni, A., Pulawska, J., Lang, G., Eds.; CABI: Boston, MA, USA, 2017; pp. 1–13. [Google Scholar]
- Myrta, A.; Savino, V. Virus and virus-like diseases of cherry in the Mediterranean region. Acta Hortic. 2008, 795, 891–896. [Google Scholar] [CrossRef]
- Rubio, M.; Martinez-Gomes, P.; Marais, A.; Sanchez-Navarro, J.A.; Pallas, V.; Candresse, T. Recent advances and prospects in Prunus virology. Ann. Appl. Biol. 2017, 171, 125–138. [Google Scholar] [CrossRef]
- James, D.; Cieslinska, M.; Pallas, V.; Flores, R.; Candresse, T.; Jelkmann, W. Viruses, viroids, phytoplasmas and genetic disorders of cherry. In Cherries: Botany, Production and Uses; Quero-Garcia, J., Iezzoni, A., Pulawska, J., Lang, G., Eds.; CABI: Boston, MA, USA, 2017; pp. 386–419. [Google Scholar]
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Amari, K.; Sanches-Pina, M.A.; Myrta, A.; Sanchez-Navarro, J.A. Ilarviruses in Prunus spp.: A continued concern for fruit trees. Phytopathology 2012, 102, 1108–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Sanchez, R.; Morales-Corts, M.R.; Gomes-Sanchez, M.A. Sour and duke cherry viruses in South-West Europe. Phytopathol. Mediterr. 2017, 56, 62–69. [Google Scholar]
- Garcia, J.A.; Glasa, M.; Cambra, M.; Candresse, T. Plum pox virus and sharka: A model potyvirus and a major disease. Mol. Plant Pathol. 2014, 15, 226–241. [Google Scholar] [CrossRef] [PubMed]
- James, D.; Varga, A.; Sanderson, D. Genetic diversity of Plum pox virus: Strains, diseases and related challenges for control. Can. J. Plant Pathol. 2013, 35, 431–441. [Google Scholar] [CrossRef]
- Chirkov, S.; Sheveleva, A.; Ivanov, P.; Zakubanskiy, A. Analysis of genetic diversity of Russian sour cherry Plum pox virus isolates provides evidence of a new strain. Plant Dis. 2018, 102, 569–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myrta, A.; Potere, O.; Crescenzi, A.; Nuzzaci, M.; Boscia, D. Properties of two monoclonal antibodies specific to the cherry strain of Plum pox virus. J. Plant Pathol. 2000, 82, 95–101. [Google Scholar]
- Nemchinov, L.; Crescenzi, A.; Hadidi, A.; Piazzolla, P.; Verderevskaya, T. Present status of the new cherry subgroup of plum pox virus (PPV-C). In Plant Virus Disease Control; Hadidi, A., Khetarpal, R.K., Kogazenawa, H., Eds.; APS Press: St. Paul, MN, USA, 1998; pp. 629–638. [Google Scholar]
- Szemes, M.; Kalman, M.; Myrta, A.; Boscia, D.; Nemeth, M.; Kolber, M.; Dorgai, L. Integrated RT-PCR/nested PCR diagnosis for differentiating between subgroup of plum pox virus. J. Virol. Methods 2001, 92, 165–175. [Google Scholar] [CrossRef]
- Kalashyan, J.A.; Bilkej, N.D.; Verderevskaya, T.D.; Rubina, E.V. Plum pox virus on sour cherry in Moldova. EPPO Bull. 1994, 24, 645–649. [Google Scholar] [CrossRef]
- Crescenzi, A.; d’Aquino, L.; Comes, S.; Nuzzaci, M.; Piazzolla, P.; Boscia, D.; Hadidi, A. Characterization of the sweet cherry isolate of plum pox potyvirus. Plant Dis. 1997, 81, 711–714. [Google Scholar] [CrossRef] [PubMed]
- Nemchinov, L.; Hadidi, A.; Kolber, M.; Nemeth, M. Molecular evidence for the occurrence of plum pox virus—Cherry subgroup in Hungary. Acta Hortic. 2008, 472, 503–510. [Google Scholar] [CrossRef]
- Malinowski, T.; Sowik, I.; Salavei, A.V.; Kukharchyk, N.V. Partial characterization of biological properties of PPV-C isolates found in Belarus and establishment of in vitro cultures of infected L2 and OWP-C rootstocks. In Proceedings of the 22nd International Conference on Virus and Other Transmissible Diseases of Fruit Crops, Rome, Italy, 3–8 June 2012; p. 152. [Google Scholar]
- Kajić, V.; Černi, S.; Škorić, D. Plum pox virus on sour cherry in Croatia. In Proceedings of the 22nd International Conference on Virus and Other Transmissible Diseases of Fruit Crops, Rome, Italy, 3–8 June 2012; p. 157. [Google Scholar]
- Jelkmann, W.; Sanderson, D.; Berwarth, C.; James, D. First detection and complete genome characterization of a Cherry (C) strain isolate of plum pox virus from sour cherry (Prunus cerasus) in Germany. J. Plant Dis. Prot. 2018, 125, 267–272. [Google Scholar] [CrossRef]
- Glasa, M.; Shneyder, Y.; Predajňa, L.; Zhivaeva, T.; Prikhodko, Y. Characterization of Russian Plum pox virus isolates provides further evidence of a low molecular heterogeneity within the PPV-C strain. J. Plant Pathol. 2014, 96, 597–601. [Google Scholar]
- Sheveleva, A.A.; Ivanov, P.; Gasanova, T.; Osipov, G.; Chirkov, S. Sequence analysis of Plum pox virus strain C isolates from Russia revealed prevalence of the D96E mutation in the universal epitope and interstrain recombination events. Viruses 2018, 10, 450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chirkov, S.; Ivanov, P.; Sheveleva, A. Detection and partial molecular characterization of atypical plum pox virus isolates from naturally infected sour cherry. Arch. Virol. 2013, 158, 1383–1387. [Google Scholar] [CrossRef]
- Glasa, M.; Prikhodko, Y.; Predajna, L.; Nagyova, A.; Shneyder, Y.; Zhivaeva, T.; Subr, Z.; Cambra, M.; Candresse, T. Characterization of sour cherry isolates of Plum pox virus from the Volga basin in Russia reveals a new cherry strain of the virus. Phytopathology 2013, 103, 972–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemeth, M. History and importance of plum pox in stone-fruit production. EPPO Bull. 1994, 24, 525–536. [Google Scholar] [CrossRef]
- Cambra, M.; Capote, N.; Myrta, A.; Llacer, G. Plum pox virus and the estimated costs associated with sharka disease. EPPO Bull. 2006, 36, 202–204. [Google Scholar] [CrossRef]
- Olmos, A.; Cambra, M.; Dasi, M.A.; Candresse, T.; Esteban, O.; Gorris, M.T.; Asensio, M. Simultaneous detection and typing of plum pox potyvirus (PPV) isolates by hemi-nested PCR and PCR-ELISA. J. Virol. Methods 1997, 68, 127–137. [Google Scholar] [CrossRef]
- Glasa, M.; Malinowski, T.; Predajna, L.; Pupola, N.; Dekena, D.; Michalczuk, L.; Candresse, T. Sequence variability, recombination analysis and specific detection of the W strain of Plum pox virus. Phytopathology 2011, 101, 980–985. [Google Scholar] [CrossRef] [PubMed]
- Kamenova, I.; Borisova, A. Molecular variability of the coat protein gene of prunus necrotic ringspot virus on sweet and sour cherry in Bulgaria. J. Plant Pathol. 2021, 103, 97–104. [Google Scholar] [CrossRef]
- IPPC-FAO. International Standards for Phytosanitary Measures: Diagnostic Protocols: Plum pox virus. ISPM 27, Annex 2 (DP2). 2018. Available online: https://assets.ippc.int/static/media/files/publication/en/2019/07/DP_02_2018_En_PlumPox_Rev_2018-09-21.pdf (accessed on 26 October 2021).
- Wetzel, T.; Candresse, T.; Macquaire, G.; Ravelonandro, M.; Dunez, J. A highly sensitive immunocapture polymerase chain reaction method for plum pox potyvirus detection. J. Virol. Methods 1992, 39, 27–37. [Google Scholar] [CrossRef]
- Untiveros, M.; Perez-Egusquiza, Z.; Clover, G. PCR assays for the detection of members of the genus Ilarvirus and family Bromoviridae. J. Virol. Methods 2010, 165, 97–104. [Google Scholar] [CrossRef] [PubMed]
- James, D.; Sanderson, D.; Varga, A.; Sheveleva, A.; Chirkov, S. Genome sequence analysis of new isolates of the Winona strain of Plum pox virus and the first definitive evidence of intra-strain recombination events. Phytopathology 2016, 106, 407–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheveleva, A.A.; Mitrofanova, I.V.; Gorina, V.M.; Chirkov, S.N. Molecular analysis of new Crimean isolates of the plum pox virus. Mosc. Univ. Biol. Sci. Bull. 2020, 75, 77–82. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Cultivar/Hybrid | Number of Trees Infected/Tested with PPV | Fruit yield, kg/Tree a | Yield Loss, % | p-Value b | |
|---|---|---|---|---|---|
| Uninfected Trees | PPV-Infected Trees | ||||
| Obilnaya | 0/17 | 7.7 ± 0.7 | - | - | |
| Zonalnaya | 0/13 | 6.8 ± 1.5 | - | - | |
| Hybrid 102-8 | 0/13 | 8.1 ± 1.1 | - | - | |
| Sevastyanovskaya | 3/12 | 7.2 ± 1.1 | 4.2 ± 1.2 | 42 | <0.05 |
| Shakirovskaya | 17/23 | 5.5 ± 0.7 | 3.4 ± 1.3 | 38 | <0.01 |
| Nizhnekamskaya | 10/24 | 5.9 ± 1.4 | 5.8 ± 1.2 | 2 | |
| Hybrid 88-2 | 5/12 | 4.7 ± 1.1 | 2.6 ± 1.4 | 45 | <0.05 |
| Hybrid 80-8 | 8/21 | 6.9 ± 1.4 | 4.0 ± 1.2 | 42 | <0.01 |
| Truzhenitsa Tatarii | 17/17 | - | 5.9 ± 1.4 | - | |
 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheveleva, A.; Osipov, G.; Gasanova, T.; Ivanov, P.; Chirkov, S. Plum Pox Virus Strain C Isolates Can Reduce Sour Cherry Productivity. Plants 2021, 10, 2327. https://doi.org/10.3390/plants10112327
Sheveleva A, Osipov G, Gasanova T, Ivanov P, Chirkov S. Plum Pox Virus Strain C Isolates Can Reduce Sour Cherry Productivity. Plants. 2021; 10(11):2327. https://doi.org/10.3390/plants10112327
Chicago/Turabian StyleSheveleva, Anna, Gennady Osipov, Tatiana Gasanova, Peter Ivanov, and Sergei Chirkov. 2021. "Plum Pox Virus Strain C Isolates Can Reduce Sour Cherry Productivity" Plants 10, no. 11: 2327. https://doi.org/10.3390/plants10112327
APA StyleSheveleva, A., Osipov, G., Gasanova, T., Ivanov, P., & Chirkov, S. (2021). Plum Pox Virus Strain C Isolates Can Reduce Sour Cherry Productivity. Plants, 10(11), 2327. https://doi.org/10.3390/plants10112327





