A Comparative Study of 5S rDNA Non-Transcribed Spacers in Elaeagnaceae Species
Abstract
1. Introduction
2. Results
2.1. Polymerase Chain Reaction (PCR) Amplification of Elaeagnus and Shepherdia 5S rDNA Non-Transcribed Spacers (NTSs)
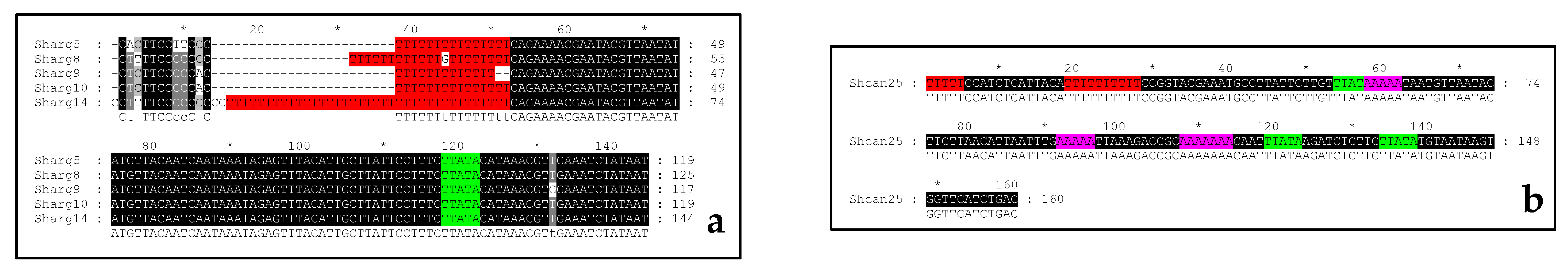
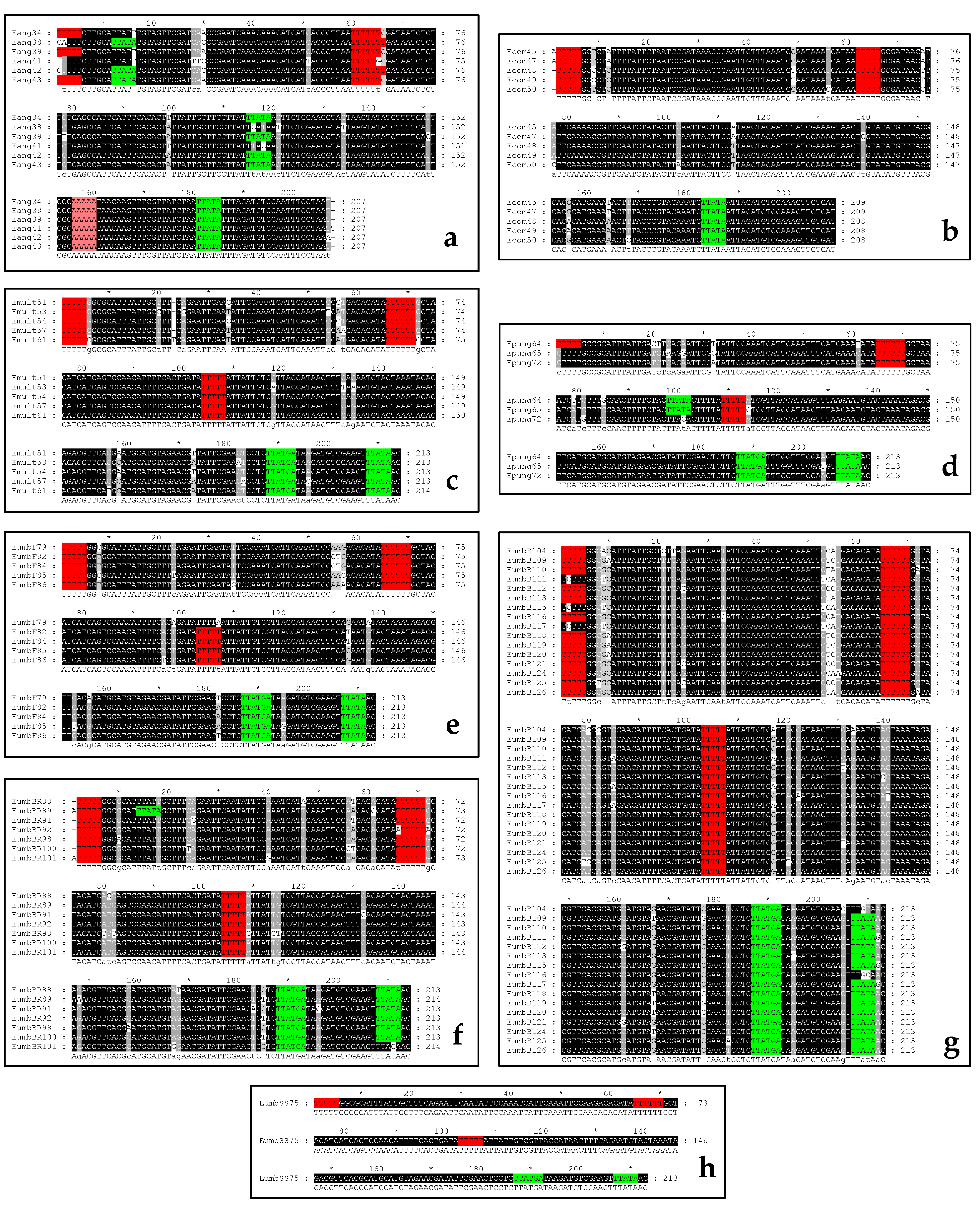
2.2. Sequencing of the PCR Products and Analysis of the Intraspecifc NTS Alignments
2.3. Interspecific, Intervarietal, and Intrageneric Level of NTS Identity in the Studied Elaeagnaceae Plants
3. Discussion
4. Materials and Methods
4.1. Plant Material and DNA Isolation
4.2. The PCR Experiments, Electrophoresis and Sequencing
4.3. Analysis of the Sequences and Statistical Tests
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Szymanski, M.; Barciszewska, M.Z.; Erdmann, V.A.; Barciszewski, J.; Barciszewska, M.Z. 5 S rRNA: Structure and interactions. Biochem. J. 2003, 371, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Szymański, M.; Karlowski, W.M. Assessing the 5S ribosomal RNA heterogeneity in Arabidopsis thaliana using short RNA next generation sequencing data. Acta Biochim. Pol. 2006, 63, 841–844. [Google Scholar] [CrossRef] [PubMed]
- Pendás, Á.M.; Moran, P.; Martínez, P.M.; Vázquez, E.G. Applications of 5S rDNA in Atlantic salmon, brown trout, and in Atlantic salmon brown trout hybrid identification. Mol. Ecol. 1995, 4, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-L.; Zhang, D.; Wang, X.; Ma, X.-F.; Wang, X.-R. Intragenomic and interspecific 5S rDNA sequence variation in five Asian pines. Am. J. Bot. 2003, 90, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Avadhani, M.N.M.; Selvaraj, C.I.; Tharachand, C.; Rajasekharan, P.E. Molecular characterization of medicinal and aromatic plants by 5S rRNA NTS and PCR RFLP—A mini review. Res. Biotechnol. 2012, 3, 41–48. [Google Scholar]
- Alexandrov, O.S.; Karlov, G.I. Development of 5S rDNA-based molecular markers for the identification of Populus deltoides Bartr. ex Marshall, Populus nigra L., and their Hybrids. Forests 2018, 9, 604. [Google Scholar] [CrossRef]
- Alexandrov, O.S.; Karlov, G.I. The development of Populus alba L. and Populus tremula L. species specific molecular markers based on 5S rDNA non-transcribed spacer polymorphism. Forests 2019, 10, 1092. [Google Scholar] [CrossRef]
- Bartish, I.V.; Swenson, U. Elaeagnaceae. In The Families and Genera of Vascular Plants; Kubitzki, K., Ed.; Springer: Berlin/Heidelgerg, Germany, 2004; pp. 131–134. [Google Scholar]
- Lian, Y.S.; Chen, X.; Lian, H. Systematic classification of the genus Hippophae L. Seabuckthorn Res. 1998, 1, 13–23. [Google Scholar]
- Rousi, A. The genus Hippophae L.: A taxomic study. Ann. Bot. Fenn. 1971, 8, 177–227. [Google Scholar]
- Veldkamp, J.F. Elaeagnaceae. Fl. Males. Ser. I 1986, 10, 151–156. [Google Scholar]
- Djamali, M.; De Beaulieu, J.-L.; Shah-Hosseini, M.; Andrieu-Ponel, V.; Ponel, P.; Amini, A.; Akhani, H.; Leroy, S.A.; Stevens, L.; Lahijani, H.; et al. A late Pleistocene long pollen record from Lake Urmia, Nw Iran. Quat. Res. 2008, 69, 413–420. [Google Scholar] [CrossRef]
- Raunkiaer, C. The Life Forms of Plants and Statistical Geography; The Claderon Press: Oxford, UK, 1934; p. 623. [Google Scholar]
- Gardner, I.C. Nitrigen fixation in Elaeagnus root nodules. Nature 1958, 181, 717–718. [Google Scholar] [CrossRef]
- Gatner, E.M.S.; Gardner, I.C. Observations on the fine structure of the root nodule endophyte of Hippophae rhamnoides L. Arch. Mikrobiol. 1970, 70, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Stangeland, R.J. The Symbiotic Nitrogen-Fixing Endophyte of Shepherdia argentea. Master’s Thesis, South Dakota State University, Brookings, SD, USA, 1970. [Google Scholar]
- Patel, S. Plant genus Elaeagnus: Underutilized lycopene and linoleic acid reserve with permaculture potential. Fruits 2015, 70, 191–199. [Google Scholar] [CrossRef]
- Xu, M. The medical research and exploitation of sea buckthorn. Hippophae 1994, 7, 32–34. [Google Scholar]
- Bernath, J.; Foldesi, D. Sea buckthorn (Hippophae rhamnoides L.): A promising new medicinal and food crop. J. Herbs Spices Med. Plants 1992, 1, 27–35. [Google Scholar] [CrossRef]
- Knowles, D.; Wilk, I. Vitamin C (ascorbic acid) content of the buffalo berry. Science 1943, 97, 43. [Google Scholar] [CrossRef]
- Wyman, D. Woody plants with ornamental fruits. Bull. Pop. Inf. Arnold Arbor. Harv. Univ. Ser. 4 1936, 4, 71–82. [Google Scholar]
- Puterova, J.; Razumova, O.; Martinek, T.; Alexandrov, O.; Divashuk, M.; Kubat, Z.; Hobza, R.; Karlov, G.; Kejnovsky, E. Satellite DNA and Transposable Elements in Seabuckthorn (Hippophae rhamnoides ), a Dioecious Plant with Small Y and Large X Chromosomes. Genome Biol. Evol. 2017, 9, 197–212. [Google Scholar] [CrossRef]
- Bone, K.D.; Razumova, O.V.; Karlov, G.I. Silver buffalo berry is a new perspective berry crop with an unknown sex determination system. In Proceedings of the International Scientific Conference of Young Scientists and Specialists Dedicated to the 160th anniversary of V.A. Michelson, Moscow, Russia, 9–11 June 2020; Publishing House of the RSAU-Moscow Agricultural Academy: Moscow, Russia, 2020; pp. 87–88. (In Russian). [Google Scholar]
- Bone, K.D.; Razumova, O.V.; Karlov, G.I.; Kirov, I.V. Analysis tandem repeats and retrotransposons of Shepherdia argentea (Pursh) Nutt. In Proceedings of the 12th International Multiconference Bioinformatics of Genome Regulation and Structure/Systems Biology (BGRS/SB-2020), Novosibirsk, Russia, 6–10 July 2020; Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, Novosibirsk State University: Novosibirsk, Russia, 2020; p. 18. [Google Scholar]
- Alexandrov, O.S.; Evtukhov, A.V.; Kiselev, I.I.; Karlov, G.I. Molecular genetic features of 5S rDNA non-transcribed spacer in Hippophae rhamnoides L. Moscow Univ. Biol. Sci. Bull. 2016, 4, 57–60. [Google Scholar]
- Islam, A.; Sinha, P.; Sharma, S.S.; Negi, M.S.; Tripathi, S.B. Isolation and Characterization of Novel Polymorphic Microsatellite Loci in Hippophae rhamnoides. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2015, 87, 727–732. [Google Scholar] [CrossRef]
- Gaskin, J.F.; Hufbauer, R.; Bogdanowicz, S.M. Microsatellite Markers for Russian Olive (Elaeagnus angustifolia; Elaeagnaceae). Appl. Plant Sci. 2013, 1, 1300013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhu, X.-T. Microsatellite DNA loci from the drought desert plant Calligonum mongolicum Turcz. (Polygonaceae). Conserv. Genet. 2009, 10, 1891–1893. [Google Scholar] [CrossRef]
- Falistocco, E.; Passeri, V.; Marconi, G. Investigations of 5S rDNA of Vitis vinifera L.: Sequence analysis and physical mapping. Genome 2007, 50, 927–938. [Google Scholar] [CrossRef]
- Sastri, D.C.; Hilu, K.; Apples, R.; Lagudah, E.S.; Playford, J.; Baum, B.R. An overview of evolution in plant 5S DNA. Plant Syst. Evol. 1992, 183, 169–181. [Google Scholar] [CrossRef]
- Feodorova, T.A.; Aleksandrov, O.S. The molecular-phylogenetic study of Petrosimonia species of Chenopodiaceae Juss. family. Izv. TAA 2015, 5, 54–60. [Google Scholar]
- Alexaandrov, O.S.; Karlov, G.I.; Sorokin, A.N.; Evtukhov, A.V. Comparative analysis of 5S rDNA non-transcribed spacers in some poplar species of Tacamahaca Spach. section. Int. J. Appl. Fund. Res. 2015, 12, 1084–1086. (In Russian) [Google Scholar]
- Alexandrov, O.S.; Evtukhov, A.V. Study of 5S rDNA non-transcribed spacers of Tacamahaca Far Eastern poplar species. In Conservation of the Variety of the Plant World of Tuva and the Adjacent Regions of Central Asia: History, Modernity, Prospects, Proceedings of the 1st International Scientific and Practical Conference, Kyzyl, Russia 5–7 June 2016; Sambuu, A.D., Ed.; Tuva Institute for the Integrated Development of Natural Resources of the Siberian Branch of the Russian Academy of Sciences: Kyzyl, Russia, 2016; pp. 55–56. (In Russian) [Google Scholar]
- Negi, M.S.; Rajagopal, J.; Chauhan, N.; Cronn, R.; Lakshmikumaran, M. Length and sequence heterogeneity in 5S rDNA of Populus deltoides. Genome 2002, 45, 1181–1188. [Google Scholar] [CrossRef]
- Alexandrov, O.S.; Karlov, G.I.; Sorokin, A.N.; Potapenko, N.C. Development of the molecular marker system for species identification of poplars and analysis of hybrids. In Proceedings of the III (V) All-Russia Youth Conference with Participation of Prospects of Development and Problems of Modern Botany, Novosibirsk, Russia, 10–14 November 2014; Asbaganov, S.V., Ed.; Akademizdat: Novosibirsk, Russia, 2014; pp. 123–124. [Google Scholar]
- Wilson, N. Genome Analysis of Populus Species: Assessment of Genetic Diversity of P. deltoides, Characterization of Wide Hybrids and Phylogenetic Analysis Using Molecular Markers. Master’s Thesis, Teri University, New Delhi, India, 2013. [Google Scholar]
- Volkov, R.A.; Panchuk, I.I.; Borisjuk, N.V.; Hosiawa-Baranska, M.; Maluszynska, J.; Hemleben, V. Evolutional dynamics of 45S and 5S ribosomal DNA in ancient allohexaploid Atropa belladonna. BMC Plant Biol. 2017, 17, 21. [Google Scholar] [CrossRef]
- Baum, B.; Bailey, L. The molecular diversity of the 5S rRNA gene in Kengyilia alatavica (Drobov) J.L. Yang, Yen and Baum (Poaceae: Triticeae): Potential genomic assignment of different rDNA units. Genome 1997, 40, 215–228. [Google Scholar] [CrossRef]
- Diao, Y.; Lin, X.M.; Liao, C.L.; Tang, C.Z.; Chen, Z.J.; Hu, Z.L. Authentication of Panax ginseng from its adulterants by PCR-RFLP and ARMS. Planta Med. 2009, 75, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Mahelka, V.; Kopecky, D.; Baum, B.R. Contrasting patterns of evolution of 45S and 5S rDNA families uncover new aspects in the genome constitution of the agronomically important grass Thinopyrum intermedium (Triticeae). Mol. Biol. Evol. 2013, 30, 2065–2086. [Google Scholar] [CrossRef] [PubMed]
- Maughan, P.J.; Kolano, A.B.; Maluszynska, J.; Coles, N.D.; Bonifacio, A.; Rojas, J.; Coleman, E.C.; Stevens, M.R.; Fairbanks, D.J.; Parkinson, E.S.; et al. Molecular and cytological characterization of ribosomal RNA genes in Chenopodium quinoa and Chenopodium berlandieri. Genome 2006, 49, 825–839. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.S.; Rivera, M.; Lourenço, L.B. Molecular organization and chromosomal localization of 5S rDNA in Amazonian Engystomops (Anura, Leiuperidae). BMC Genet. 2012, 20, 17. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Pérez, J.; Nantón, A.; Méndez, J. Sequence characterization of the 5S ribosomal DNA and the internal transcribed spacer (ITS) region in four European Donax species (Bivalvia: Donacidae). BMC Genet. 2018, 19, 97. [Google Scholar] [CrossRef]
- He, W.; Qin, Q.; Liu, S.; Li, T.; Wang, J.; Xiao, J.; Xie, L.; Zhang, C.; Liu, Y. Organization and variation analysis of 5S rDNA in different ploidy-level hybrids of red crucian carp × topmouth culter. PLoS ONE 2012, 7, e38976. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Stumph, W.E. RNA polymerase II/III transcription specificity determined by TATA box orientation. Proc. Natl. Acad. Sci. USA 1995, 92, 8606–8610. [Google Scholar] [CrossRef]
- Duchêne, S.; Ho, S.Y.; Holmes, E.C. Declining transition/transversion ratios through time reveal limitations to the accuracy of nucleotide substitution models. BMC Evol. Biol. 2005, 15, 36. [Google Scholar] [CrossRef]
- Alexandrov, O.S. Study of the upstream ricin gene sequences in different castor (Ricinus communis) varieties as a preliminary step in CRISPR/Cas9 editing. Res. Crops 2020, 21, 344–348. [Google Scholar]
- Tynkevich, Y.O.; Volkov, R.A. Novel structural class of 5S rDNA of Rosa wichurana Crep. Dopov. Nac. Akad. Nauk Ukr. 2014, 5, 143–148. [Google Scholar] [CrossRef]
- Clarke, L.A.; Rebelo, C.S.; Gonçalves, J.; Boavida, M.G.; Jordan, P. PCR amplification introduces errors into mononucleotide and dinucleotide repeat sequences. Mol. Pathol. 2001, 54, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Kieleczawa, J. Fundamentals of sequencing of difficult templates—An overview. J. Biomol. Tech. 2006, 17, 207–217. [Google Scholar] [PubMed]
- Handell-Mazzetti, H.R.E. Elaeagnus umbellata var. siphonantha (Nakai) Hand.-Mazz. Symb. Sin. 1933, 7, 540. [Google Scholar]
- Fulnecek, J.; Lim, K.Y.; Leitch, A.R.; Kovarík, A.; Matyásek, R. Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity 2002, 88, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.Q.; Duan, J.A.; Zhu, D.Y.; Dong, T.T.X.; Tsim, K.W.K. Species identification of Radix Astragali (Huangqi) by DNA sequence of its 5S-rRNA spacer domain. Phytochemistry 2000, 54, 363–368. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Razumova, O.V.; Alexandrov, O.S.; Divashuk, M.G.; Sukhorada, T.I.; Karlov, G.I. Molecular cytogenetic analysis of monoecious hemp (Cannabis sativa L.) cultivars reveals its karyotype variations and sex chromosomes constitution. Protoplasma 2016, 253, 895–901. [Google Scholar] [CrossRef]
- GeneDoc: Analysis and Visualization of Genetic Variation. Available online: http://www.nrbsc.org/gfx/genedoc/ebinet.htm (accessed on 26 November 2020).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexandrov, O.S.; Razumova, O.V.; Karlov, G.I. A Comparative Study of 5S rDNA Non-Transcribed Spacers in Elaeagnaceae Species. Plants 2021, 10, 4. https://doi.org/10.3390/plants10010004
Alexandrov OS, Razumova OV, Karlov GI. A Comparative Study of 5S rDNA Non-Transcribed Spacers in Elaeagnaceae Species. Plants. 2021; 10(1):4. https://doi.org/10.3390/plants10010004
Chicago/Turabian StyleAlexandrov, Oleg S., Olga V. Razumova, and Gennady I. Karlov. 2021. "A Comparative Study of 5S rDNA Non-Transcribed Spacers in Elaeagnaceae Species" Plants 10, no. 1: 4. https://doi.org/10.3390/plants10010004
APA StyleAlexandrov, O. S., Razumova, O. V., & Karlov, G. I. (2021). A Comparative Study of 5S rDNA Non-Transcribed Spacers in Elaeagnaceae Species. Plants, 10(1), 4. https://doi.org/10.3390/plants10010004





