Quantitative Craniofacial Analysis and Generation of Human Induced Pluripotent Stem Cells for Muenke Syndrome: A Case Report
Abstract
1. Introduction
2. Materials and Methods
2.1. Genetic Diagnosis
2.2. Consents, Questionnaire, Medical History, Medication, and Dental History
2.3. Clinical Evaluation
2.4. Photographs, X-rays, and Scans
2.4.1. Photographs
2.4.2. Dental Cone Beam Computed Tomography Scans
2.4.3. Craniofacial Cephalometric Analysis
2.4.4. Geometric Morphometric Analysis
2.5. Derivation and Analysis of Human Induced Pluripotent Stem Cells
3. Results
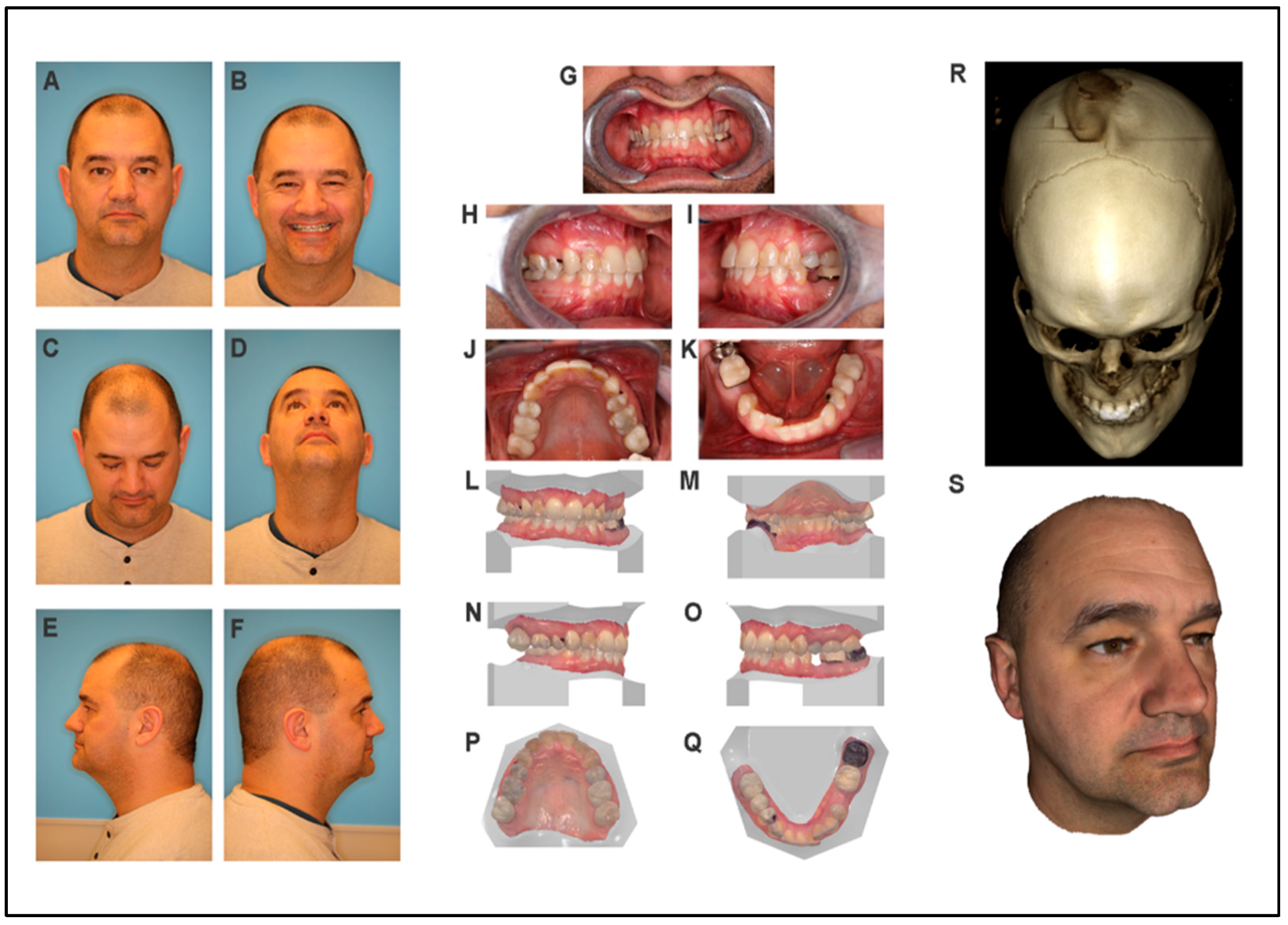
3.1. Participant One (Proband)
3.1.1. Craniofacial Exam
3.1.2. Craniofacial Measurements
3.1.3. Intraoral Examination
3.1.4. CBCT Scan and 3D Photos
3.1.5. Cephalometric Analysis
3.2. Participant Two (Mother)
3.2.1. Craniofacial Exam
3.2.2. Craniofacial Measurements
3.2.3. Intraoral Exam
3.2.4. CBCT Scan and 3D Photos
3.2.5. Cephalometric Analysis
3.3. Participant Three (Father)
3.3.1. Craniofacial Exam
3.3.2. Craniofacial Measurements
3.3.3. Intraoral Exam
3.3.4. CBCT Scan and 3D Photos
3.3.5. Cephalometric Analysis
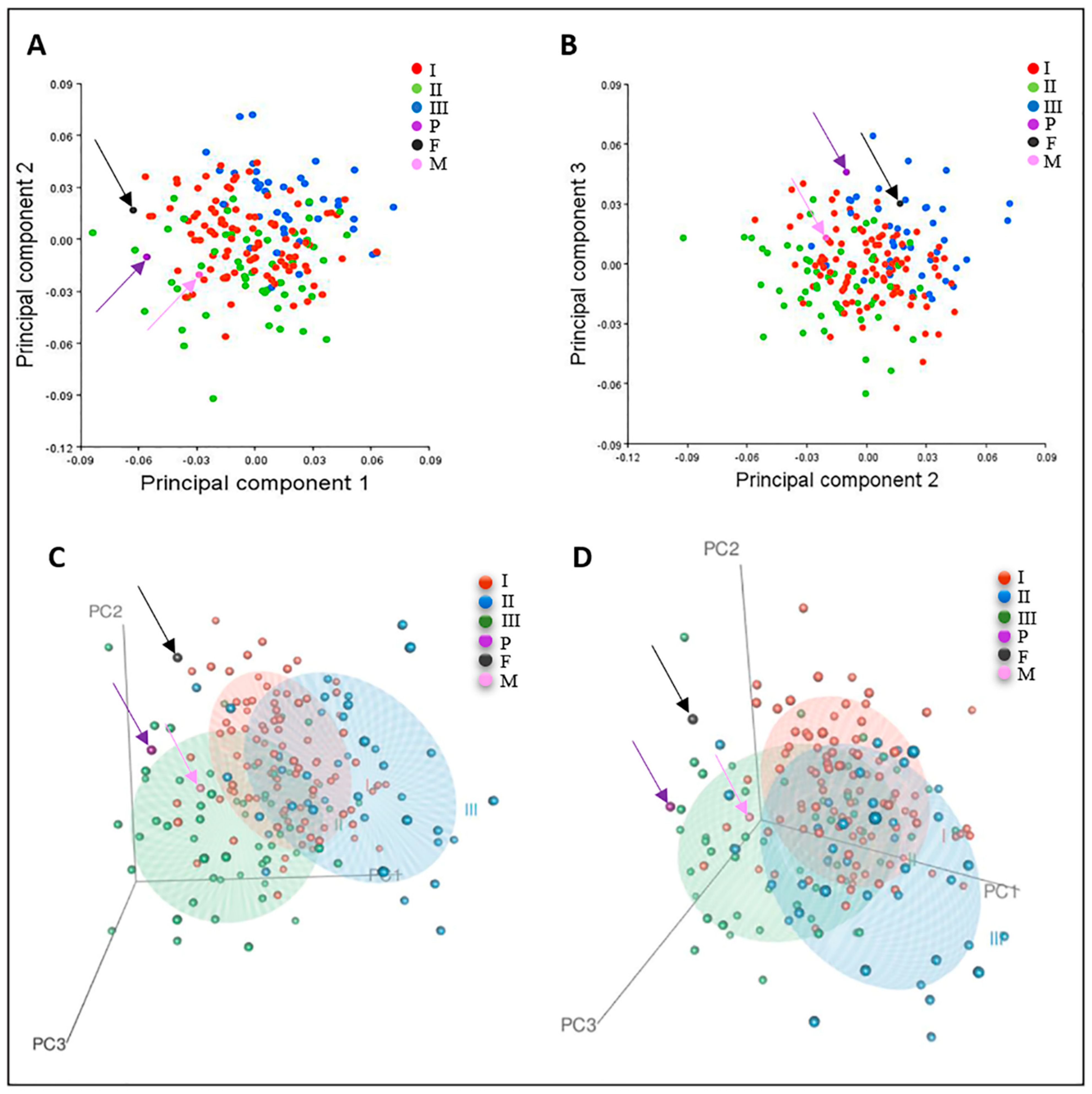
3.4. Geometric Morphometric Analysis
3.4.1. Participant One (Proband)
3.4.2. Participant Two (Mother)
3.4.3. Participant Three (Father)
3.5. Human Induced Pluripotent Stem Cell Derived from Proband and Healthy Family Members
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cunningham, M.L.; Seto, M.L.; Ratisoontorn, C.; Heike, C.L.; Hing, A.V. Syndromic craniosynostosis: From history to hydrogen bonds. Orthod. Craniofac. Res. 2007, 10, 67–81. [Google Scholar] [CrossRef]
- Moloney, D.M.; Wall, S.A.; Ashworth, G.J.; Oldridge, M.; Glass, I.A.; Francomano, C.A.; Muenke, M.; Wilkie, A.O. Prevalence of Pro250Arg mutation of fibroblast growth factor receptor 3 in coronal craniosynostosis. Lancet 1997, 349, 1059–1062. [Google Scholar] [CrossRef]
- González-Del Angel, A.; Estandía-Ortega, B.; Alcántara-Ortigoza, M.A.; Martínez-Cruz, V.; Gutiérrez-Tinajero, D.J.; Rasmussen, A.; Gómez-González, C.S. Expansion of the variable expression of Muenke syndrome: Hydrocephalus without craniosynostosis. Am. J. Med. Genet. A 2016, 170, 3189–3196. [Google Scholar] [CrossRef] [PubMed]
- Agochukwu, N.B.; Solomon, B.D.; Gropman, A.L.; Muenke, M. Epilepsy in Muenke syndrome: FGFR3-related craniosynostosis. Pediatr. Neurol. 2012, 47, 355–361. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Anderson, P.J.; Snell, B.; Moore, M.H. Muencke syndrome with cleft lip and palate. J. Craniofac. Surg. 2013, 24, 1484–1485. [Google Scholar] [CrossRef] [PubMed]
- Aravidis, C.; Konialis, C.P.; Pangalos, C.G.; Kosmaidou, Z. A familial case of Muenke syndrome. Diverse expressivity of the FGFR3 Pro252Arg mutation--case report and review of the literature. J. Matern. Fetal Neonatal Med. 2014, 27, 1502–1506. [Google Scholar] [CrossRef] [PubMed]
- den Ottelander, B.K.; de Goederen, R.; van Veelen, M.C.; van de Beeten, S.D.C.; Lequin, M.H.; Dremmen, M.H.G.; Loudon, S.E.; Telleman, M.A.J.; de Gier, H.H.W.; Wolvius, E.B.; et al. Muenke syndrome: Long-term outcome of a syndrome-specific treatment protocol. J. Neurosurg. Pediatr. 2019, 24, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Doherty, E.S.; Lacbawan, F.; Hadley, D.W.; Brewer, C.; Zalewski, C.; Kim, H.J.; Solomon, B.; Rosenbaum, K.; Domingo, D.L.; Hart, T.C.; et al. Muenke syndrome (FGFR3-related craniosynostosis): Expansion of the phenotype and review of the literature. Am. J. Med. Genet. A 2007, 143a, 3204–3215. [Google Scholar] [CrossRef]
- Kruszka, P.; Addissie, Y.A.; Yarnell, C.M.; Hadley, D.W.; Guillen Sacoto, M.J.; Platte, P.; Paelecke, Y.; Collmann, H.; Snow, N.; Schweitzer, T.; et al. Muenke syndrome: An international multicenter natural history study. Am. J. Med. Genet. A 2016, 170a, 918–929. [Google Scholar] [CrossRef]
- Nah, H.D.; Koyama, E.; Agochukwu, N.B.; Bartlett, S.P.; Muenke, M. Phenotype profile of a genetic mouse model for Muenke syndrome. Childs Nerv. Syst. 2012, 28, 1483–1493. [Google Scholar] [CrossRef]
- Öwall, L.; Kreiborg, S.; Dunø, M.; Hermann, N.V.; Darvann, T.A.; Hove, H. Phenotypic variability in Muenke syndrome-observations from five Danish families. Clin. Dysmorphol. 2020, 29, 1–9. [Google Scholar] [CrossRef]
- Roscioli, T.; Elakis, G.; Cox, T.C.; Moon, D.J.; Venselaar, H.; Turner, A.M.; Le, T.; Hackett, E.; Haan, E.; Colley, A.; et al. Genotype and clinical care correlations in craniosynostosis: Findings from a cohort of 630 Australian and New Zealand patients. Am. J. Med. Genet. C Semin. Med. Genet. 2013, 163c, 259–270. [Google Scholar] [CrossRef]
- Keller, M.K.; Hermann, N.V.; Darvann, T.A.; Larsen, P.; Hove, H.D.; Christensen, L.; Schwartz, M.; Marsh, J.L.; Kreiborg, S. Craniofacial morphology in Muenke syndrome. J. Craniofac. Surg. 2007, 18, 374–386. [Google Scholar] [CrossRef]
- Samra, F.; Bauder, A.R.; Swanson, J.W.; Whitaker, L.A.; Bartlett, S.P.; Taylor, J.A. Assessing the midface in Muenke syndrome: A cephalometric analysis and review of the literature. J. Plast. Reconstr. Aesthet. Surg. 2016, 69, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Liberton, D.K.; Verma, P.; Almpani, K.; Fung, P.W.; Mishra, R.; Oberoi, S.; Şenel, F.; Mah, J.K.; Huang, J.; Padwa, B.L.; et al. Craniofacial Analysis May Indicate Co-Occurrence of Skeletal Malocclusions and Associated Risks in Development of Cleft Lip and Palate. J. Dev. Biol. 2020, 8, 2. [Google Scholar] [CrossRef]
- Jani, P.; Nguyen, Q.C.; Almpani, K.; Keyvanfar, C.; Mishra, R.; Liberton, D.; Orzechowski, P.; Frischmeyer-Guerrerio, P.A.; Duverger, O.; Lee, J.S. Severity of oro-dental anomalies in Loeys-Dietz syndrome segregates by gene mutation. J. Med. Genet. 2020, 57, 699–707. [Google Scholar] [CrossRef]
- Anderson, R.H.; Francis, K.R. Modeling rare diseases with induced pluripotent stem cell technology. Mol. Cell. Probes 2018, 40, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Twigg, S.R.; Healy, C.; Babbs, C.; Sharpe, J.A.; Wood, W.G.; Sharpe, P.T.; Morriss-Kay, G.M.; Wilkie, A.O. Skeletal analysis of the Fgfr3(P244R) mouse, a genetic model for the Muenke craniosynostosis syndrome. Dev. Dyn. 2009, 238, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Liberton, D.K.; Verma, P.; Contratto, A.; Lee, J.S. Development and Validation of Novel Three-Dimensional Craniofacial Landmarks on Cone-Beam Computed Tomography Scans. J. Craniofac. Surg. 2019, 30, e611–e615. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, C.P. MorphoJ: An integrated software package for geometric morphometrics. Mol. Ecol. Resour. 2011, 11, 353–357. [Google Scholar] [CrossRef]
- Mui, B.W.H.; Arora, D.; Mallon, B.S.; Martinez, A.F.; Lee, J.S.; Muenke, M.; Kruszka, P.; Kidwai, F.K.; Robey, P.G. Generation of human induced pluripotent stem cell line (NIDCRi001-A) from a Muenke syndrome patient with an FGFR3 p.Pro250Arg mutation. Stem Cell Res. 2020, 46, 101823. [Google Scholar] [CrossRef]
- Wakui, T.; Matsumoto, T.; Matsubara, K.; Kawasaki, T.; Yamaguchi, H.; Akutsu, H. Method for evaluation of human induced pluripotent stem cell quality using image analysis based on the biological morphology of cells. J. Med. Imaging 2017, 4, 044003. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ibrahimi, O.A.; Zhang, F.; Eliseenkova, A.V.; Linhardt, R.J.; Mohammadi, M. Proline to arginine mutations in FGF receptors 1 and 3 result in Pfeiffer and Muenke craniosynostosis syndromes through enhancement of FGF binding affinity. Hum. Mol. Genet. 2004, 13, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Plotnikov, A.N.; Hubbard, S.R.; Schlessinger, J.; Mohammadi, M. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell 2000, 101, 413–424. [Google Scholar] [CrossRef]
- MacArthur, M.W.; Thornton, J.M. Influence of proline residues on protein conformation. J. Mol. Biol. 1991, 218, 397–412. [Google Scholar] [CrossRef]
- Perálvarez-Marín, A.; Lórenz-Fonfría, V.A.; Simón-Vázquez, R.; Gomariz, M.; Meseguer, I.; Querol, E.; Padrós, E. Influence of proline on the thermostability of the active site and membrane arrangement of transmembrane proteins. Biophys. J. 2008, 95, 4384–4395. [Google Scholar] [CrossRef]
- Berezin, M.Y.; Achilefu, S. Fluorescence lifetime measurements and biological imaging. Chem. Rev. 2010, 110, 2641–2684. [Google Scholar] [CrossRef]
- Becker, W. Fluorescence lifetime imaging--techniques and applications. J. Microsc. 2012, 247, 119–136. [Google Scholar] [CrossRef]
- Hallgrimsson, B.; Percival, C.J.; Green, R.; Young, N.M.; Mio, W.; Marcucio, R. Morphometrics, 3D Imaging, and Craniofacial Development. Curr. Top. Dev. Biol. 2015, 115, 561–597. [Google Scholar] [CrossRef]
- Ridgway, E.B.; Wu, J.K.; Sullivan, S.R.; Vasudavan, S.; Padwa, B.L.; Rogers, G.F.; Mulliken, J.B. Craniofacial growth in patients with FGFR3Pro250Arg mutation after fronto-orbital advancement in infancy. J. Craniofac. Surg. 2011, 22, 455–461. [Google Scholar] [CrossRef]
- Rice, D.P.; Rice, R.; Thesleff, I. Fgfr mRNA isoforms in craniofacial bone development. Bone 2003, 33, 14–27. [Google Scholar] [CrossRef]
- Billington, C.J., Jr.; Schmidt, B.; Marcucio, R.S.; Hallgrimsson, B.; Gopalakrishnan, R.; Petryk, A. Impact of retinoic acid exposure on midfacial shape variation and manifestation of holoprosencephaly in Twsg1 mutant mice. Dis. Models Mech. 2015, 8, 139–146. [Google Scholar] [CrossRef]
- Phillips, M.D.; Kuznetsov, S.A.; Cherman, N.; Park, K.; Chen, K.G.; McClendon, B.N.; Hamilton, R.S.; McKay, R.D.; Chenoweth, J.G.; Mallon, B.S.; et al. Directed differentiation of human induced pluripotent stem cells toward bone and cartilage: In vitro versus in vivo assays. Stem Cells Transl. Med. 2014, 3, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Tremoleda, J.L.; Forsyth, N.R.; Khan, N.S.; Wojtacha, D.; Christodoulou, I.; Tye, B.J.; Racey, S.N.; Collishaw, S.; Sottile, V.; Thomson, A.J.; et al. Bone tissue formation from human embryonic stem cells in vivo. Cloning Stem Cells 2008, 10, 119–132. [Google Scholar] [CrossRef]
- Zou, L.; Kidwai, F.K.; Kopher, R.A.; Motl, J.; Kellum, C.A.; Westendorf, J.J.; Kaufman, D.S. Use of RUNX2 expression to identify osteogenic progenitor cells derived from human embryonic stem cells. Stem Cell Rep. 2015, 4, 190–198. [Google Scholar] [CrossRef]
- Kidwai, F.; Mui, B.W.H.; Arora, D.; Iqbal, K.; Hockaday, M.; de Castro Diaz, L.F.; Cherman, N.; Martin, D.; Myneni, V.D.; Ahmad, M.; et al. Lineage-specific differentiation of osteogenic progenitors from pluripotent stem cells reveals the FGF1-RUNX2 association in neural crest-derived osteoprogenitors. Stem Cells 2020, 38, 1107–1123. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kidwai, F.K.; Mui, B.W.H.; Almpani, K.; Jani, P.; Keyvanfar, C.; Iqbal, K.; Paravastu, S.S.; Arora, D.; Orzechowski, P.; Merling, R.K.; et al. Quantitative Craniofacial Analysis and Generation of Human Induced Pluripotent Stem Cells for Muenke Syndrome: A Case Report. J. Dev. Biol. 2021, 9, 39. https://doi.org/10.3390/jdb9040039
Kidwai FK, Mui BWH, Almpani K, Jani P, Keyvanfar C, Iqbal K, Paravastu SS, Arora D, Orzechowski P, Merling RK, et al. Quantitative Craniofacial Analysis and Generation of Human Induced Pluripotent Stem Cells for Muenke Syndrome: A Case Report. Journal of Developmental Biology. 2021; 9(4):39. https://doi.org/10.3390/jdb9040039
Chicago/Turabian StyleKidwai, Fahad K., Byron W. H. Mui, Konstantinia Almpani, Priyam Jani, Cyrus Keyvanfar, Kulsum Iqbal, Sriram S. Paravastu, Deepika Arora, Pamela Orzechowski, Randall K. Merling, and et al. 2021. "Quantitative Craniofacial Analysis and Generation of Human Induced Pluripotent Stem Cells for Muenke Syndrome: A Case Report" Journal of Developmental Biology 9, no. 4: 39. https://doi.org/10.3390/jdb9040039
APA StyleKidwai, F. K., Mui, B. W. H., Almpani, K., Jani, P., Keyvanfar, C., Iqbal, K., Paravastu, S. S., Arora, D., Orzechowski, P., Merling, R. K., Mallon, B., Myneni, V. D., Ahmad, M., Kruszka, P., Muenke, M., Woodcock, J., Gilman, J. W., Robey, P. G., & Lee, J. S. (2021). Quantitative Craniofacial Analysis and Generation of Human Induced Pluripotent Stem Cells for Muenke Syndrome: A Case Report. Journal of Developmental Biology, 9(4), 39. https://doi.org/10.3390/jdb9040039





