Gbx2 Is Required for the Migration and Survival of a Subpopulation of Trigeminal Cranial Neural Crest Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice and Genotyping
2.2. In Situ Hybridization
2.3. Electrophoretic Mobility Shift Assay
- NRP1-F: 5′aacattccaaaaattatcaaccatttcaggaatacatttcataaaagctagattgagttctgcttgttttttatt 3′
- NRP1-R: 5′aataaaaaacaagcagaactcaatctagcttttatgaaatgtattcctgaaatggttgataatttttggaatgtt 3′
- NRP1-F: mutated 5′aacattccaaaaattatcaagattgagttctgcttgttttttatt 3′
- NRP1-R: 5′aataaaaaacaagcagaactcaatcttgataatttttggaatgtt 3′
2.4. Immunohistochemistry
2.5. Statistical Analysis
2.6. Animal Ethics Statement
3. Results
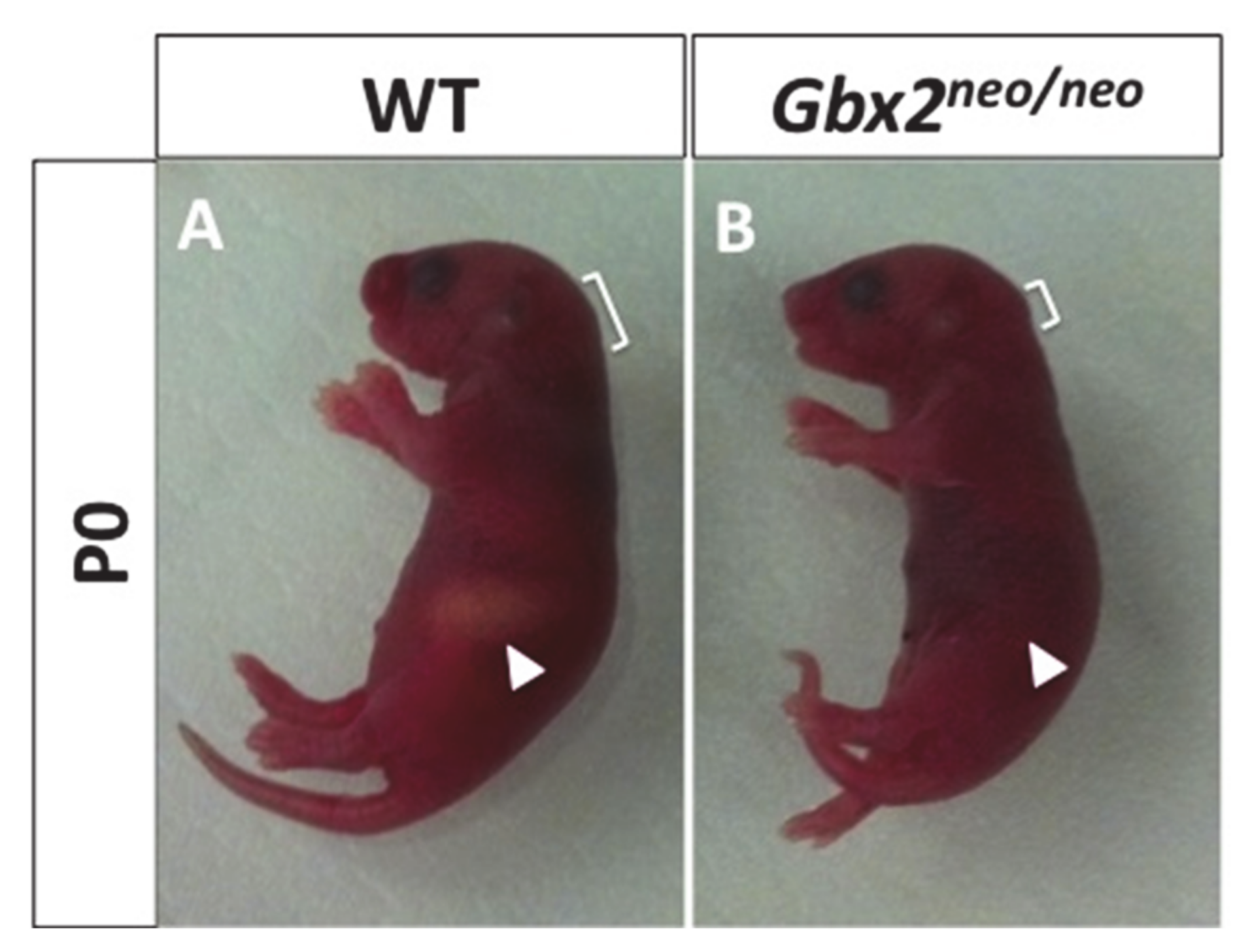
3.1. Loss of Gbx2 Results in an Inability to Suckle and Trigeminal Motor Neuron Defects
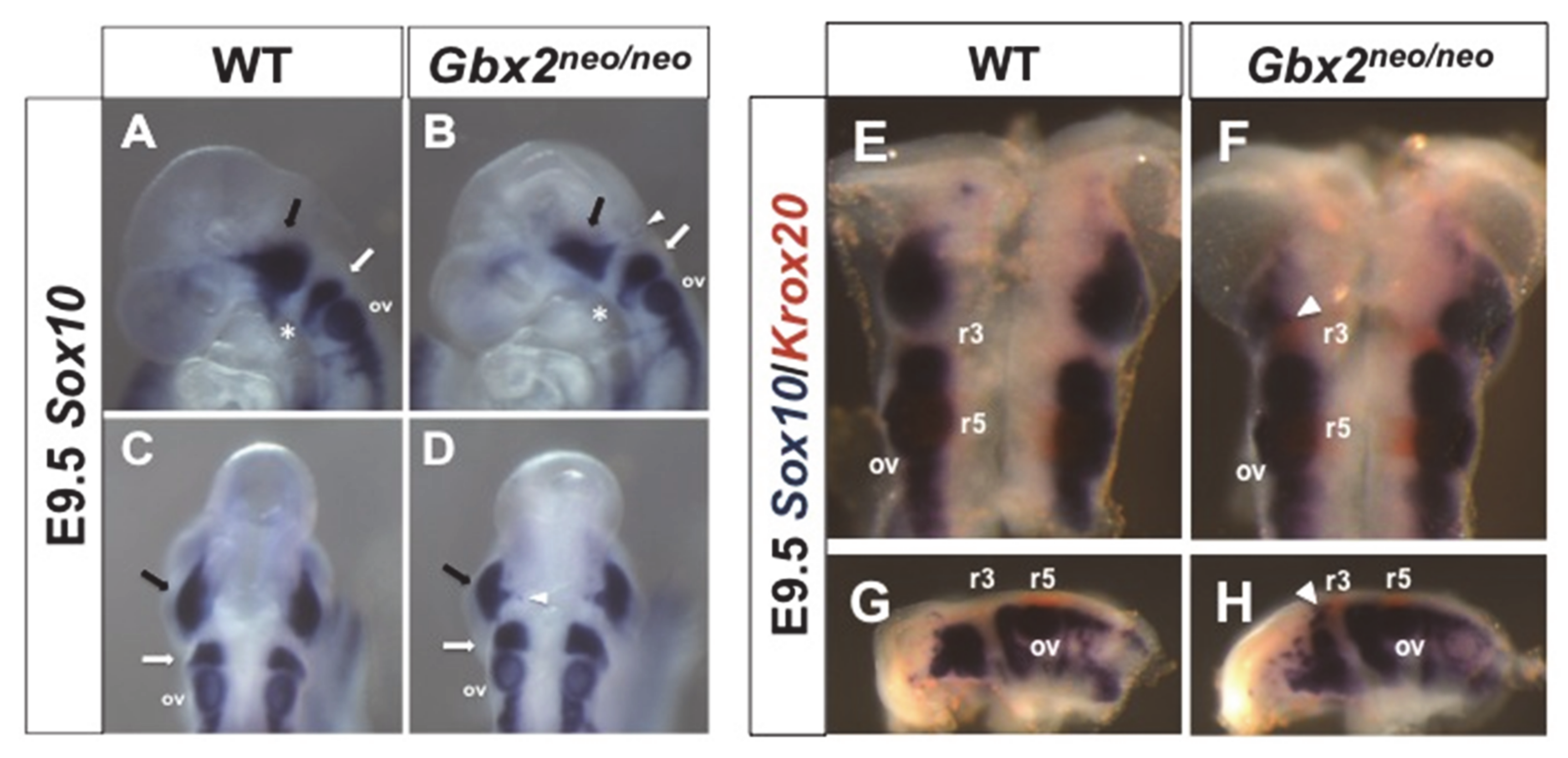
3.2. Reduction in Gbx2 Results in Trigeminal Cranial NC Cell Defects
3.3. Loss of GBX2 Target Gene, Nrp1, Expression in a Subpopulation of Trigeminal Cranial NC Cells in Gbx2neoneo Embryos
3.4. Loss of Gbx2 Results in an Increase in Apoptosis in Migrating Trigeminal NCC
4. Discussion
4.1. Gbx2 Is a Critical Factor for nV Gangliogenesis
4.2. Loss of Gbx2 and Anterior Hindbrain Tissue Disrupts r2 Motor Neuron Development
4.3. Loss of Gbx2 Alters the Temporal Expression of Krox20 in r3
4.4. Misregulation of GBX2 Target Gene, Nrp1, in Gbx2neo/neo Mutants
4.5. Gbx2 Is Required for the Migration and Survival of Trigeminal Cranial Neural Crest Cells
Author Contributions
Funding
Conflicts of Interest
References
- Li, B.; Kuriyama, S.; Moreno, M.; Mayor, R. The posteriorizing gene Gbx2 is a direct target of Wnt signalling and the earliest factor in neural crest induction. Development 2009, 136, 3267–3278. [Google Scholar] [CrossRef] [PubMed]
- Wassarman, K.M.; Lewandoski, M.; Campbell, K.; Joyner, A.L.; Rubenstein, J.L.; Martinez, S.; Martin, G.R. Specification of the anterior hindbrain and establishment of a normal mid/hindbrain organizer is dependent on Gbx2 gene function. Development 1997, 124, 2923–2934. [Google Scholar] [PubMed]
- Byrd, N.A.; Meyers, E.N. Loss of Gbx2 results in neural crest cell patterning and pharyngeal arch artery defects in the mouse embryo. Dev. Biol. 2005, 284, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Burroughs-Garcia, J.; Sittaramane, V.; Chandrasekhar, A.; Waters, S.T. Evolutionarily conserved function of Gbx2 in anterior hindbrain development. Dev. Dyn. 2011, 240, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Roeseler, D.A.; Sachdev, S.; Buckley, D.M.; Joshi, T.; Wu, D.K.; Xu, D.; Hannink, M.; Waters, S.T. Elongation factor 1 alpha1 and genes associated with Usher syndromes are downstream targets of GBX2. PLoS ONE 2012, 7, e47366. [Google Scholar] [CrossRef] [PubMed]
- Calmont, A.; Ivins, S.; Van Bueren, K.L.; Papangeli, I.; Kyriakopoulou, V.; Andrews, W.D.; Martin, J.F.; Moon, A.M.; Illingworth, E.A.; Basson, M.A.; et al. Tbx1 controls cardiac neural crest cell migration during arch artery development by regulating Gbx2 expression in the pharyngeal ectoderm. Development 2009, 136, 3173–3183. [Google Scholar] [CrossRef]
- Chai, Y.; Jiang, X.; Ito, Y.; Bringas, P., Jr.; Han, J.; Rowitch, D.H.; Soriano, P.; McMahon, A.P.; Sucov, H.M. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 2000, 127, 1671–1679. [Google Scholar]
- Kontges, G.; Lumsden, A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development 1996, 122, 3229–3242. [Google Scholar]
- Serbedzija, G.; Bronner-Fraser, M.; Fraser, S. Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development 1992, 116, 297–307. [Google Scholar]
- Uehara, M.; Yashiro, K.; Mamiya, S.; Nishino, J.; Chambon, P.; Dolle, P.; Sakai, Y. CYP26A1 and CYP26C1 cooperatively regulate anterior-posterior patterning of the developing brain and the production of migratory cranial neural crest cells in the mouse. Dev. Biol. 2007, 302, 399–411. [Google Scholar] [CrossRef]
- Davenne, M.; Maconochie, M.K.; Neun, R.; Pattyn, A.; Chambon, P.; Krumlauf, R.; Rijli, F.M. Hoxa2 and Hoxb2 control dorsoventral patterns of neuronal development in the rostral hindbrain. Neuron 1999, 22, 677–691. [Google Scholar] [CrossRef]
- De, S.; Turman, J.J. Krox-20 gene expression: Influencing hindbrain-craniofacial developmental interactions. Arch. Histol. Cytol. 2005, 68, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Irving, C.; Nieto, M.A.; DasGupta, R.; Charnay, P.; Wilkinson, D.G. Progressive spatial restriction of Sek-1 and Krox-20 gene expression during hindbrain segmentation. Dev. Biol. 1996, 173, 26–38. [Google Scholar] [CrossRef]
- Voiculescu, O.; Taillebourg, E.; Pujades, C.; Kress, C.; Buart, S.; Charnay, P.; Schneider-Maunoury, S. Hindbrain patterning: Krox20 couples segmentation and specification of regional identity. Development 2001, 128, 4967–4978. [Google Scholar] [PubMed]
- Maro, G.S.; Vermeren, M.; Voiculescu, O.; Melton, L.; Cohen, J.; Charnay, P.; Topilko, P. Neural crest boundary cap cells constitute a source of neuronal and glial cells of the PNS. Nat. Neurosci. 2004, 77, 930–938. [Google Scholar] [CrossRef]
- Nonchev, S.; Vesque, C.; Maconochie, M.; Seitanidou, T.; Ariza-McNaughton, L.; Frain, M.; Marshall, H.; Sham, M.H.; Krumlauf, R.; Charnay, P. Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20. Development 1996, 122, 543–554. [Google Scholar]
- Theil, T.; Frain, M.; Gilardi-Hebenstreit, P.; Flenniken, A.; Charnay, P.; Wilkinson, D.G. Segmental expression of the EphA4 (Sek-1) receptor tyrosine kinase in the hindbrain is under direct transcriptional control of Krox-20. Development 1998, 125, 443–452. [Google Scholar]
- Levi, G.; Topilko, P.; Schneider-Maunoury, S.; Lasagna, M.; Mantero, S.; Cancedda, R.; Charnay, P. Defective bone formation in Krox-20 mutant mice. Development 1996, 122, 113–120. [Google Scholar]
- Li, J.Y.; Joyner, A.L. Otx2 and Gbx2 are required for refinement and not induction of mid-hindbrain gene expression. Development 2001, 128, 4979–4991. [Google Scholar]
- Li, J.Y.; Lao, Z.; Joyner, A.L. Changing requirements for Gbx2 in development of the cerebellum and maintenance of the mid/hindbrain organizer. Neuron 2002, 36, 31–43. [Google Scholar] [CrossRef]
- Li, J.Y.; Lao, Z.; Joyner, A.L. New regulatory interactions and cellular responses in the isthmic organizer region revealed by altering Gbx2 expression. Development 2005, 132, 1971–1981. [Google Scholar] [CrossRef] [PubMed]
- Millet, S.; Campbell, K.B.; Epstein, D.J.; Losos, K.; Harris, E.; Joyner, A.L. A role for Gbx2 in repression of Otx2 and positioning the mid/hindbrain organizer. Nature 1999, 401, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Bouillet, P.; Chazaud, C.; Oulad-Abdelghani, M.; Dollé, P.; Chambon, P. Sequence and expression pattern of the Stra7 (Gbx-2) homeobox-containing gene induced by retinoic acid in P19 embryonal carcinoma cells. Dev. Dyn. 1995, 204, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.T.; Lewandoski, M. A threshold requirement for Gbx2 levels in hindbrain development. Development 2006, 133, 1991–2000. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.T.; Wilson, C.P.; Lewandoski, M. Cloning and embryonic expression analysis of the mouse Gbx1 gene. Gene Expr. Patterns 2003, 33, 313–317. [Google Scholar] [CrossRef]
- Chandrasekhar, A. Turning heads: Development of vertebrate branchiomotor neurons. Dev. Dyn. 2004, 229, 143–161. [Google Scholar] [CrossRef] [PubMed]
- Gavalas, A.; Davenne, M.; Lumsden, A.; Chambon, P.; Rijli, F.M. Role of Hoxa-2 in axon pathfinding and rostral hindbrain patterning. Development 1997, 124, 3693–3702. [Google Scholar]
- Kraus, F.; Haenig, B.; Kispert, A. Cloning and expression analysis of the mouse T-box gene tbx20. Mech. Dev. 2001, 100, 87–91. [Google Scholar] [CrossRef]
- Theveneau, E.; Mayor, R. Neural crest delamination and migration: From epithelium-to-mesenchyme transition to collective cell migration. Dev. Biol. 2012, 366, 34–54. [Google Scholar] [CrossRef]
- Dixon, J.; Jones, N.C.; Sandell, L.L.; Jayasinghe, S.M.; Crane, J.; Rey, J.P.; Dixon, M.J.; Trainor, P.A. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl. Acad. Sci. USA 2006, 103, 13403–13408. [Google Scholar] [CrossRef]
- Giudicelli, F.; Taillebourg, E.; Charnay, P.; Gilardi-Hebenstreit, P. Krox-20 patterns the hindbrain through both cell-autonomous and non-cell-autonomous mechanisms. Genes Dev. 2001, 15, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, Q.; Vieira, J.M.; Howard, B.; Eickholt, B.J.; Ruhrberg, C. Neuropilin 1 and 2 control cranial gangliogenesis and axon guidance through neural crest cells. Development 2008, 135, 1605–1613. [Google Scholar] [CrossRef] [PubMed]
- Fantin, A.; Schwarz, Q.; Davidson, K.; Normando, E.M.; Denti, L.; Ruhrberg, C. The cytoplasmic domain of neuropilin 1 is dispensable for angiogenesis, but promotes the spatial separation of retinal arteries and veins. Development 2011, 138, 4185–4191. [Google Scholar] [CrossRef] [PubMed]
- Lumb, R.; Wiszniak, S.; Kabbara, S.; Scherer, M.; Harvey, N.L.; Schwarz, Q. Neuropilins define distinct populations of neural crest cells. Neural Dev. 2014, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Meyers, E.N.; Martin, G.R. Differences in left-right axis pathways in mouse and chick: Functions of FGF8 and SHH. Science 1999, 285, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Abu-Issa, R.; Smyth, G.; Smoak, I.; Yamamura, K.-I.; Meyers, E.N. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development 2002, 129, 4613–4625. [Google Scholar]
- Inoue, F.; Kurokawa, D.; Takahashi, M.; Aizawa, S. Gbx2 directly restricts Otx2 expression to forebrain and midbrain, competing with class III POU factors. Mol. Cell. Biol. 2012, 32, 2618–2627. [Google Scholar] [CrossRef]
- Sunmonu, N.A.; Li, K.; Guo, Q.; Li, J.Y. Gbx2 and Fgf8 are sequentially required for formation of the midbrain-hindbrain compartment boundary. Development 2011, 138, 725–734. [Google Scholar] [CrossRef]
- Wassef, M.A.; Chomette, D.; Pouilhe, M.; Stedman, A.; Havis, E.; Desmarquet-Trin Dinh, C.; Schneider-Maunoury, S.; Gilardi-Hebenstreit, P.; Charnay, P.; Ghislain, J. Rostral hindbrain patterning involves the direct activation of a Krox20 transcriptional enhancer by Hox/Pbx and Meis factors. Development 2008, 135, 3369–3378. [Google Scholar] [CrossRef]
- Kayam, G.; Kohl, A.; Magen, Z.; Peretz, Y.; Weisinger, K.; Bar, A.; Novikov, O.; Brodski, C.; Sela-Donenfeld, D. A novel role for Pax6 in the segmental organization of the hindbrain. Development 2013, 140, 2190–2202. [Google Scholar] [CrossRef][Green Version]
- Labalette, C.; Bouchoucha, Y.X.; Wassef, M.A.; Gongal, P.A.; Le Men, J.; Becker, T.; Gilardi-Hebenstreit, P.; Charnay, P. Hindbrain patterning requires fine-tuning of early krox20 transcription by Sprouty 4. Development 2010, 138, 317–326. [Google Scholar] [CrossRef]
- Marshall, H.; Nonchev, S.; Sham, M.H.; Muchamore, I.; Lumsden, A.; Krumlauf, R. Retinoic acid alters hindbrain Hox code and induces transformation of rhombomeres 2/3 into a 4/5 identity. Nature 1992, 360, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Vermot, J.; Schuhbaur, B.; Le Mouellic, H.; McCaffery, P.; Garnier, J.-M.; Hentsch, D.; Brûlet, P.; Niederreither, K.; Chambon, P.; Dollé, P.; et al. Retinaldehyde dehydrogenase 2 and Hoxc8 are required in the murine brachial spinal cord for the specification of Lim1+ motoneurons and the correct distribution of Islet1+ motoneurons. Development 2005, 132, 1611–1621. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Limberg, B.J.; Whitaker, G.B.; Perman, B.; Leahy, D.J.; Rosenbaum, J.S.; Ginty, D.D.; Kolodkin, A.L. Characterization of neuropilin-1 structural features that confer binding to semaphorin 3A and vascular endothelial growth factor 165. J. Biol. Chem. 2002, 277, 18069–18076. [Google Scholar] [CrossRef]
- Yaron, A.; Huang, P.-H.; Cheng, H.-J.; Tessier-Lavigne, M. Differential requirement for Plexin-A3 and -A4 in mediating responses of sensory and sympathetic neurons to distinct class 3 Semaphorins. Neuron 2005, 45, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Soker, S.; Takashima, S.; Miao, H.Q.; Neufeld, G.; Klagsbrun, M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 1998, 92, 735–745. [Google Scholar] [CrossRef]
- Chauvet, S.; Cohen, S.; Yoshida, Y.; Fekrane, L.; Livet, J.; Gayet, O.; Segu, L.; Buhot, M.-C.; Jessell, T.M.; Henderson, C.E.; et al. Gating of Sema3E/PlexinD1 signaling by neuropilin-1 switches axonal repulsion to attraction during brain development. Neuron 2007, 56, 807–822. [Google Scholar] [CrossRef] [PubMed]
- Kitsukawa, T.; Shimizu, M.; Sanbo, M.; Hirata, T.; Taniguchi, M.; Bekku, Y.; Yagi, T.; Fujisawa, H. Neuropilin-semaphorin III/D-mediated chemorepulsive signals play a crucial role in peripheral nerve projection in mice. Neuron 1997, 19, 995–1005. [Google Scholar] [CrossRef]
- Schwarz, Q.; Gu, C.; Fujisawa, H.; Sabelko, K.; Gertsenstein, M.; Nagy, A.; Taniguchi, M.; Kolodkin, A.L.; Ginty, D.D.; Shima, D.T.; et al. Vascular endothelial growth factor controls neuronal migration and cooperates with Sema3A to pattern distinct compartments of the facial nerve. Genes Dev. 2004, 18, 2822–2834. [Google Scholar] [CrossRef]
- Taniguchi, M.; Yuasa, S.; Fujisawa, H.; Naruse, I.; Saga, S.; Mishina, M.; Yagi, T. Disruption of semaphorin III/D gene causes severe abnormality in peripheral nerve projection. Neuron 1997, 19, 519–530. [Google Scholar] [CrossRef]
- Solowska, J.M.; Mazurek, A.M.; Weinberger, L.; Baird, D.H. Pontocerebellar axon guidance: Neuropilin-1- and semaphorin 3A-sensitivity gradients across basilar pontine nuclei and semaphorin 3A variation across cerebellum. Mol. Cell. Neurosci. 2002, 21, 266–284. [Google Scholar] [CrossRef] [PubMed]
- Cioni, J.-M.; Telley, L.; Saywell, V.; Cadilhac, C.; Jourdan, C.; Huber, A.B.; Huang, J.Z.; Jahannault-Talignani, C.; Ango, F.; Huang, J. SEMA3A signaling controls layer-specific interneuron branching in the cerebellum. Curr. Biol. 2013, 23, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Kitsukawa, T.; Shimono, A.; Kawakami, A.; Kondoh, H.; Fujisawa, H. Overexpression of a membrane protein, neuropilin, in chimeric mice causes anomalies in the cardiovascular system, nervous system and limbs. Development 1995, 121, 4309–4318. [Google Scholar]
- Kawasaki, T.; Kitsukawa, T.; Bekku, Y.; Matsuda, Y.; Sanbo, M.; Yagi, T.; Fujisawa, H. A requirement for neuropilin-1 in embryonic vessel formation. Development 1999, 126, 4895–4902. [Google Scholar] [PubMed]
- Giger, R.; Cloutier, J.-F.; Sahay, A.; Prinjha, R.K.; Levengood, D.V.; E Moore, S.; Pickering, S.; Simmons, D.; Rastan, S.; Walsh, F.S.; et al. Neuropilin-2 is required in vivo for selective axon guidance responses to secreted semaphorins. Neuron 2000, 25, 29–41. [Google Scholar] [CrossRef]
- Toyofuku, T.; Yoshida, J.; Sugimoto, T.; Yamamoto, M.; Makino, N.; Takamatsu, H.; Takegahara, N.; Suto, F.; Hori, M.; Fujisawa, H.; et al. Repulsive and attractive semaphorins cooperate to direct the navigation of cardiac neural crest cells. Dev. Biol. 2008, 321, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.B.; Feiner, L.; Lu, M.M.; Li, J.; Ma, X.; Webber, A.L.; Jia, L.; Raper, J.A.; Epstein, J.A. PlexinA2 and semaphorin signaling during cardiac neural crest development. Development 2001, 128, 3071–3080. [Google Scholar]
- McLennan, R.; Kulesa, P.M. Neuropilin-1 interacts with the second branchial arch microenvironment to mediate chick neural crest cell dynamics. Dev. Dyn. 2010, 239, 1664–1673. [Google Scholar] [CrossRef][Green Version]
- Xu, Q.; Alldus, G.; Holder, N.; Wilkinson, D.G. Expression of truncated Sek-1 receptor tyrosine kinase disrupts the segmental restriction of gene expression in the Xenopus and zebrafish hindbrain. Development 1995, 121, 4005–4016. [Google Scholar]
- Heimbucher, T.; Murko, C.; Bajoghli, B.; Aghaallaei, N.; Huber, A.; Stebegg, R.; Eberhard, D.; Fink, M.; Simeone, A.; Czerny, T. Gbx2 and Otx2 interact with the WD40 domain of Groucho/Tle corepressors. Mol. Cell. Biol. 2007, 27, 340–351. [Google Scholar] [CrossRef]
- Schanke, J.T.; Van Ness, B.G. Organization of the transcription factor binding sites in the kappa Ig intron enhancer. Effects of position, orientation, and spacing. J. Immunol. 1994, 153, 4565–4572. [Google Scholar] [PubMed]
- Schanke, J.T.; Marcuzzi, A.; Podzorski, R.P.; Van Ness, B. An AP1 binding site upstream of the kappa immunoglobulin intron enhancer binds inducible factors and contributes to expression. Nucleic Acids Res. 1994, 22, 5425–5432. [Google Scholar] [CrossRef] [PubMed][Green Version]
- LaBonne, C.; Bronner-Frase, M. Neural crest induction in Xenopus: Evidence for a two-signal model. Development 1998, 125, 2403–2414. [Google Scholar] [PubMed]
- Garcia-Castro, M.I.; Marcelle, C.; Bronner-Fraser, M. Ectodermal Wnt function as a neural crest inducer. Science 2002, 297, 848–851. [Google Scholar] [PubMed]
- Bachelder, R.E.; Crago, A.; Chung, J.; Wendt, M.A.; Shaw, L.M.; Robinson, G.; Mercurio, A.M. Vascular endothelial growth factor is an autocrine survival factor for neuropilin-expressing breast carcinoma cells. Cancer Res. 2001, 61, 5736–5740. [Google Scholar]
- Brusselmans, K.; Bono, F.; Collen, D.; Herbert, J.-M.; Carmeliet, P.; Dewerchin, M. A novel role for vascular endothelial growth factor as an autocrine survival factor for embryonic stem cells during hypoxia. J. Biol. Chem. 2005, 280, 3493–3499. [Google Scholar] [CrossRef]
- Cariboni, A.; Davidson, K.; Dozio, E.; Memi, F.; Schwarz, Q.; Stossi, F.; Parnavelas, J.G.; Ruhrberg, C. VEGF signalling controls GnRH neuron survival via NRP1 independently of KDR and blood vessels. Development 2011, 138, 3723–3733. [Google Scholar] [CrossRef]
- Ishii, M.; Han, J.; Yen, H.-Y.; Sucov, H.M.; Chai, Y.; Maxson, R.E. Combined deficiencies of Msx1 and Msx2 cause impaired patterning and survival of the cranial neural crest. Development 2005, 132, 4937–4950. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roeseler, D.A.; Strader, L.; Anderson, M.J.; Waters, S.T. Gbx2 Is Required for the Migration and Survival of a Subpopulation of Trigeminal Cranial Neural Crest Cells. J. Dev. Biol. 2020, 8, 33. https://doi.org/10.3390/jdb8040033
Roeseler DA, Strader L, Anderson MJ, Waters ST. Gbx2 Is Required for the Migration and Survival of a Subpopulation of Trigeminal Cranial Neural Crest Cells. Journal of Developmental Biology. 2020; 8(4):33. https://doi.org/10.3390/jdb8040033
Chicago/Turabian StyleRoeseler, David A., Lona Strader, Matthew J. Anderson, and Samuel T. Waters. 2020. "Gbx2 Is Required for the Migration and Survival of a Subpopulation of Trigeminal Cranial Neural Crest Cells" Journal of Developmental Biology 8, no. 4: 33. https://doi.org/10.3390/jdb8040033
APA StyleRoeseler, D. A., Strader, L., Anderson, M. J., & Waters, S. T. (2020). Gbx2 Is Required for the Migration and Survival of a Subpopulation of Trigeminal Cranial Neural Crest Cells. Journal of Developmental Biology, 8(4), 33. https://doi.org/10.3390/jdb8040033




