Cerebellar Morphology and Behavioral Profiles in Mice Lacking Heparan Sulfate Ndst Gene Function
Abstract
1. Introduction
2. Materials and Methods
3. Results
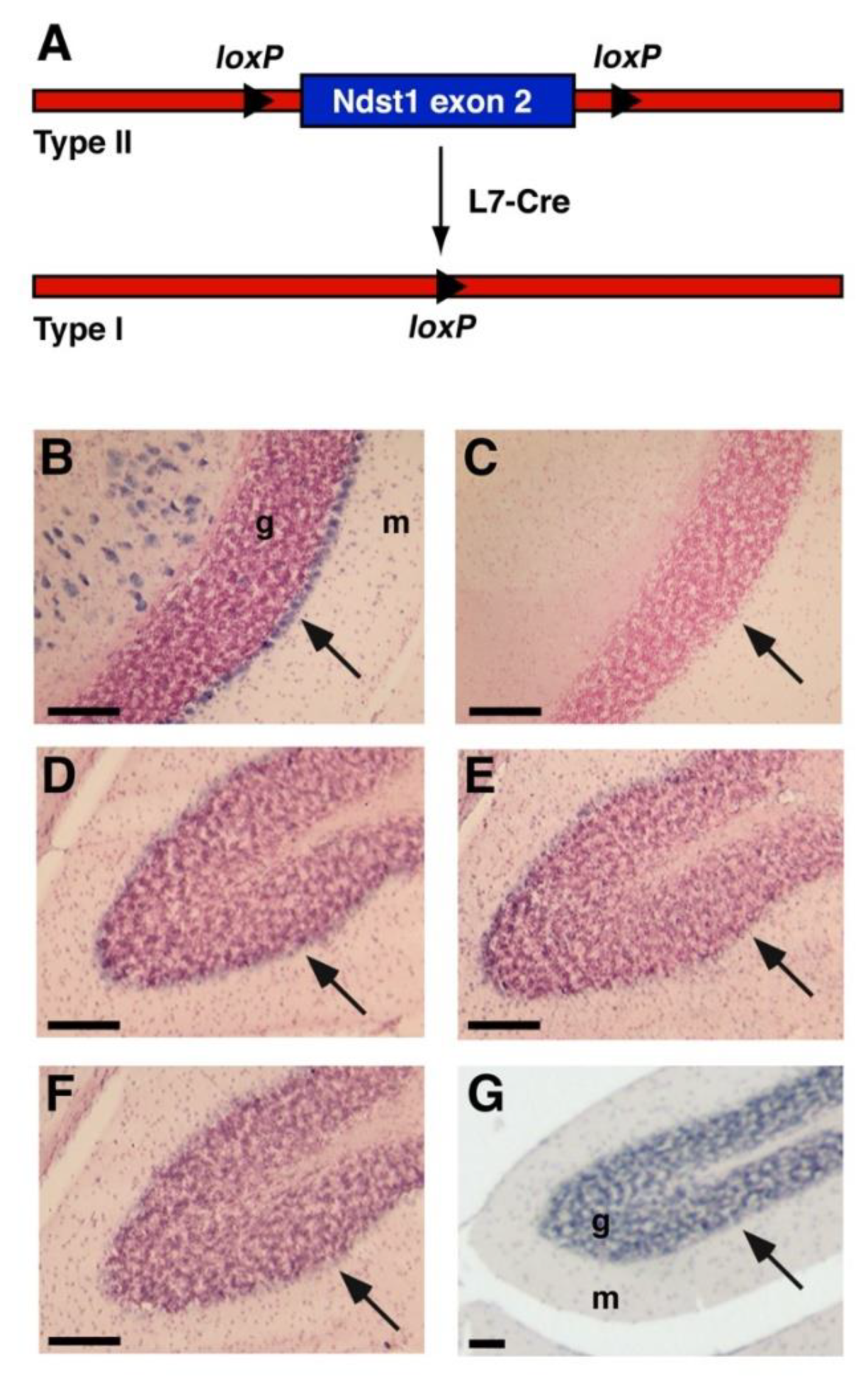
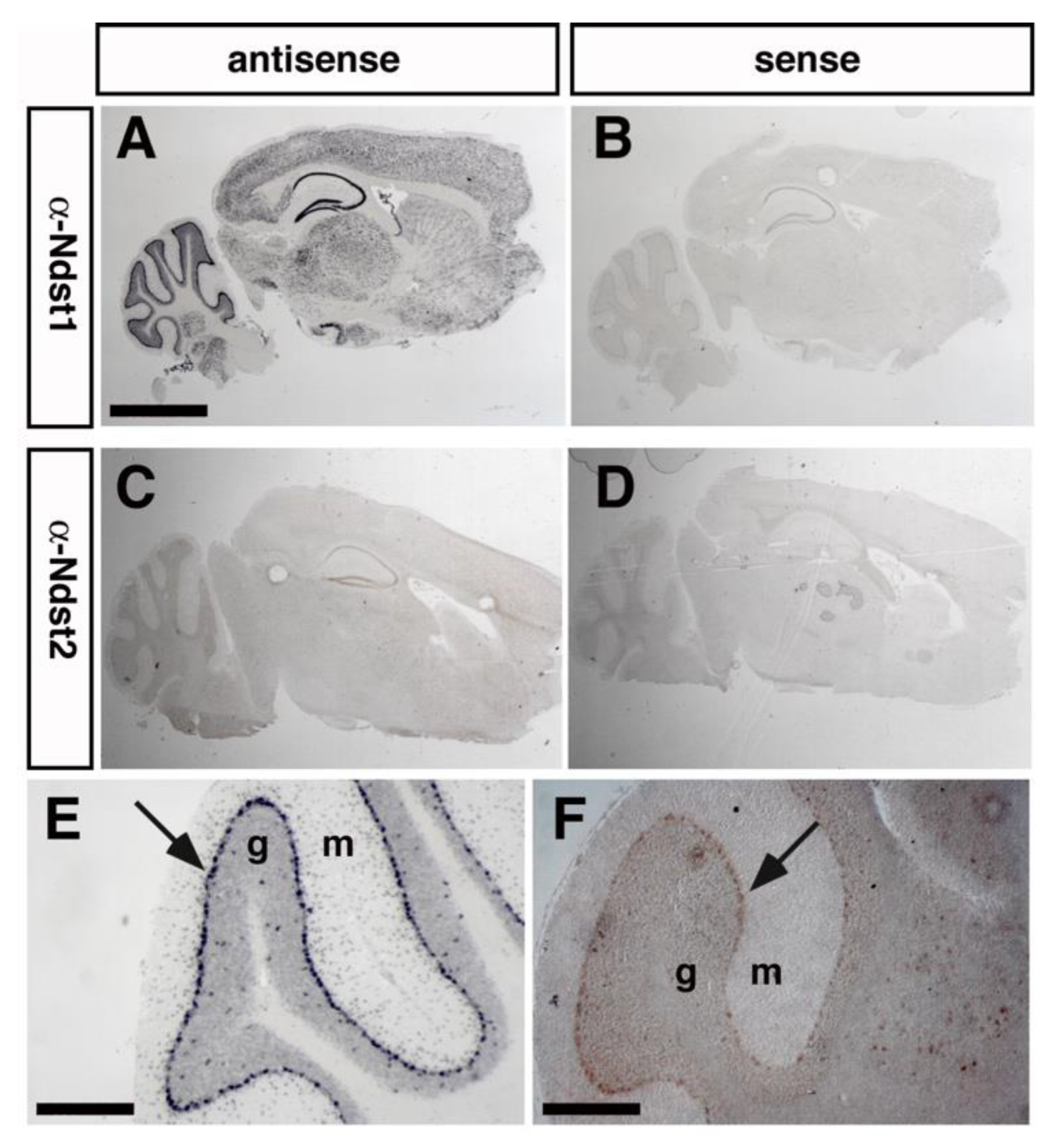
3.1. Ndst1 Expression in the Mouse Cerebellum
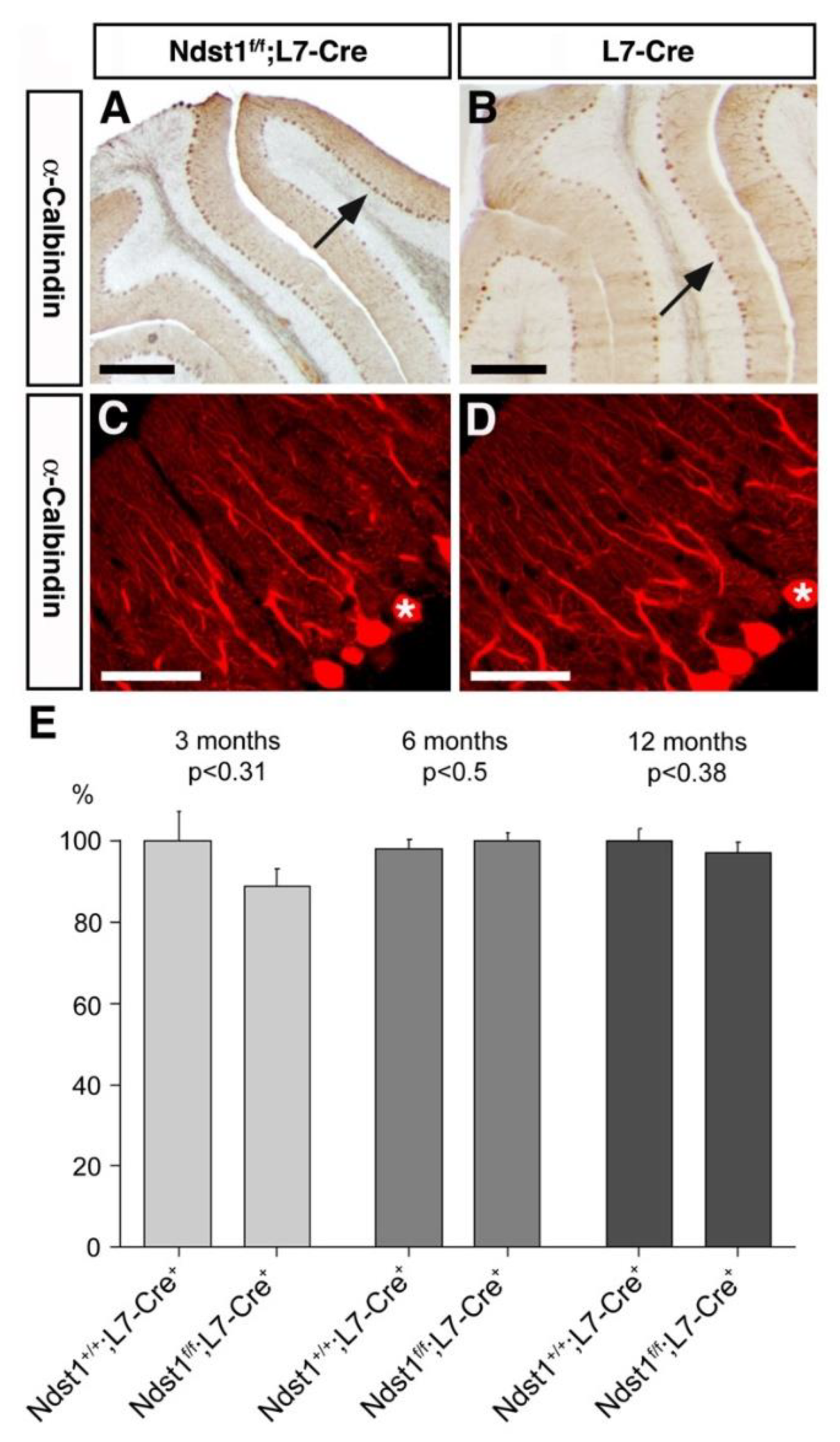
3.2. Cerebellar Architecture after L7 Mediated Ndst1 Deletion
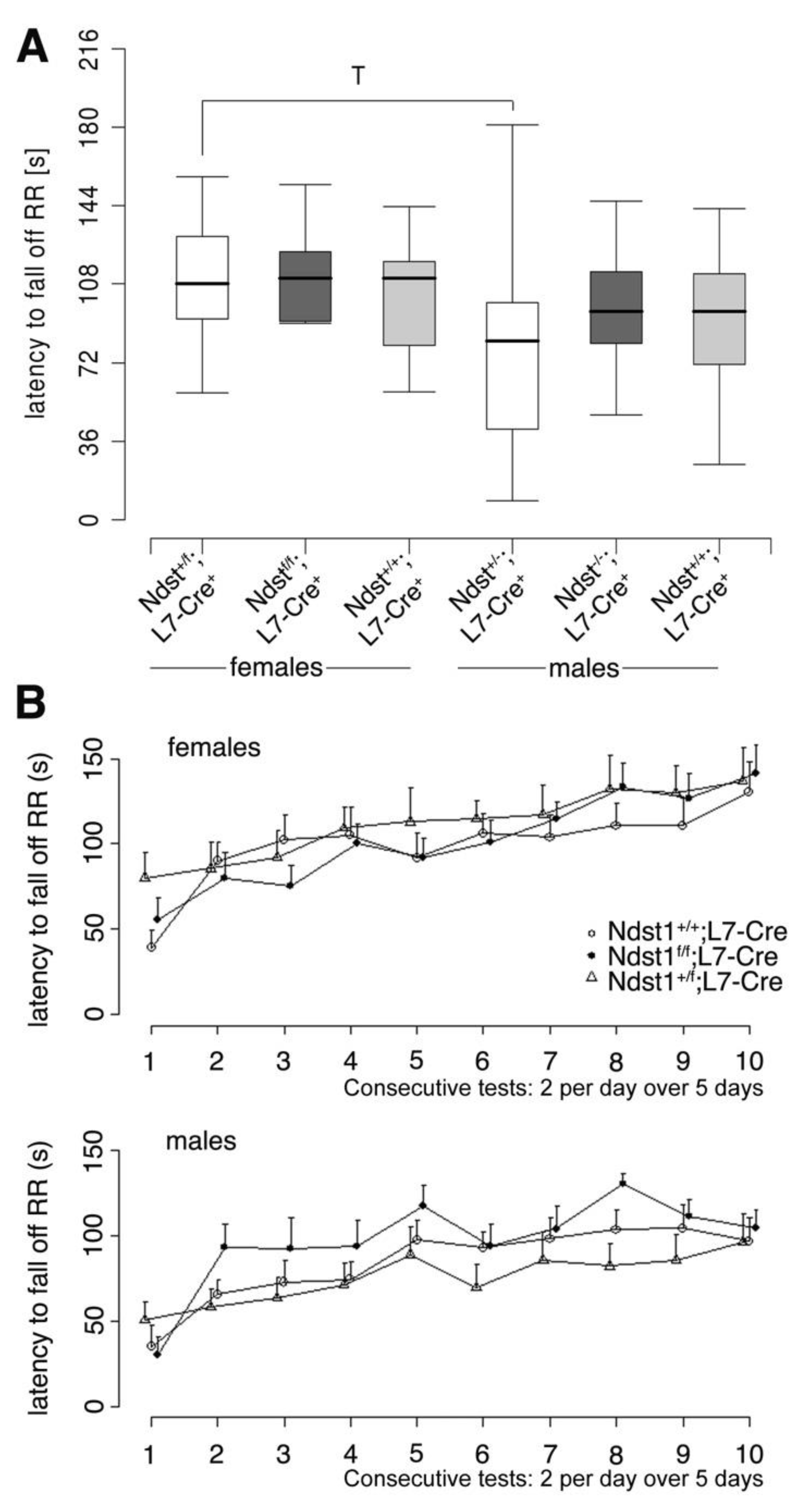
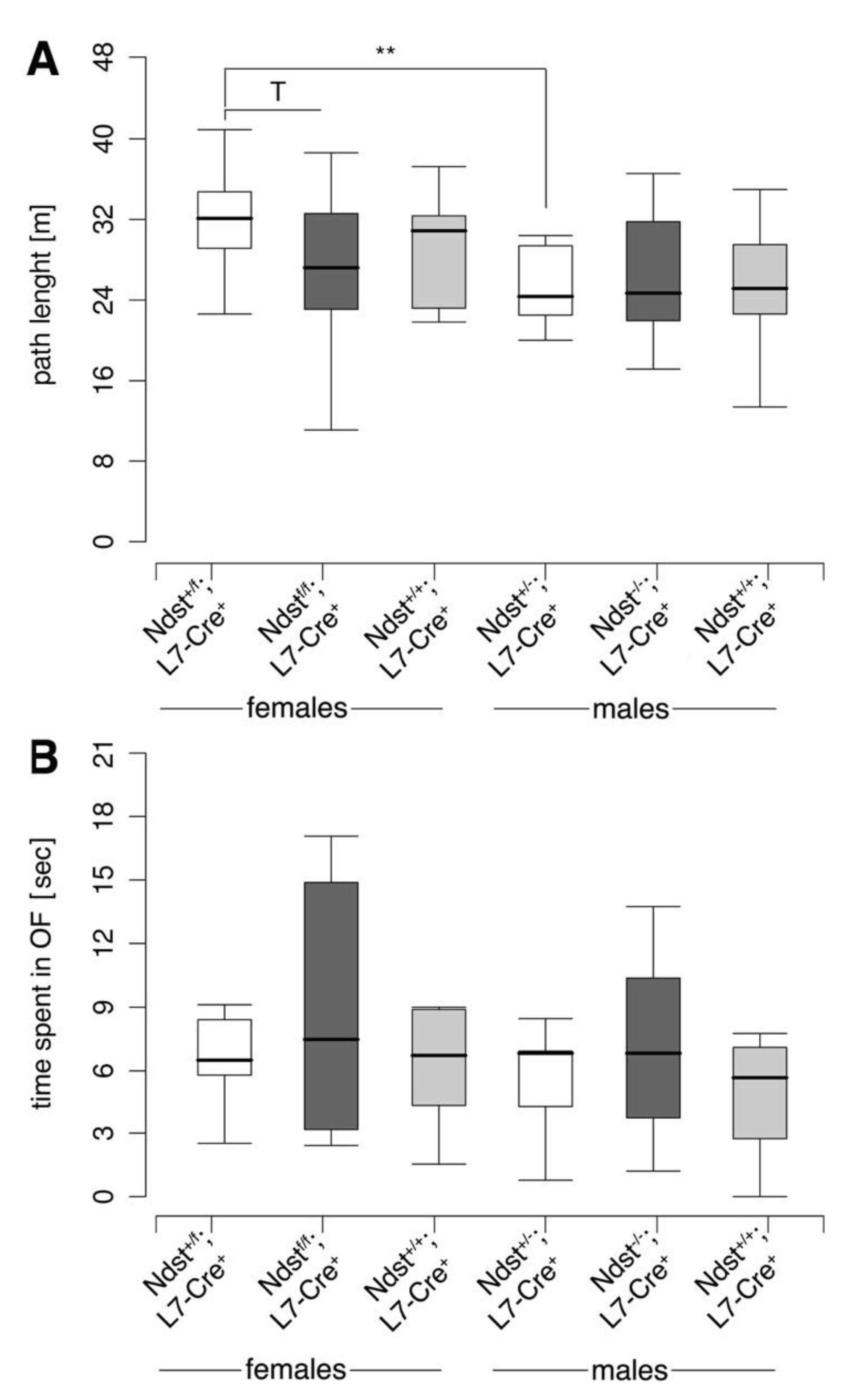
3.3. Normal Motor Function in Ndst1f/f;L7-Cre+ Mutant Mice
3.4. Analysis of Ndst1 Expression and Function in the Adult Brain
3.5. Reproductive Behavior in Compound Mutant Mice
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Esko, J.D.; Lindahl, U. Molecular diversity of heparan sulfate. J. Clin. Investig. 2001, 108, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, U.; Kusche-Gullberg, M.; Kjellén, L. Regulated diversity of heparan sulfate. J. Biol. Chem. 1998, 273, 24979–24982. [Google Scholar] [CrossRef]
- Aikawa, J.; Grobe, K.; Tsujimoto, M.; Esko, J.D. Multiple isozymes of heparan sulfate/heparin glcnac n-deacetylase/n-sulfotransferase: Structure and activity of the fourth member, ndst4. J. Biol. Chem. 2001, 276, 5876–5882. [Google Scholar] [CrossRef] [PubMed]
- Perrimon, N.; Bernfield, M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature 2000, 404, 725–728. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.H.; Perrimon, N. Dally cooperates with drosophila frizzled 2 to transduce wingless signalling. Nature 1999, 400, 281–284. [Google Scholar] [CrossRef]
- Lin, X.H.; Buff, E.M.; Perrimon, N.; Michelson, A.M. Heparan sulfate proteoglycans are essential for fgf receptor signaling during drosophila embryonic development. Development 1999, 126, 3715–3723. [Google Scholar]
- The, I.; Bellaiche, Y.; Perrimon, N. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol. Cell 1999, 4, 633–639. [Google Scholar] [CrossRef]
- Bullock, S.L.; Fletcher, J.M.; Beddington, R.S.P.; Wilson, V.A. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev. 1998, 12, 1894–1906. [Google Scholar] [CrossRef]
- Grobe, K.; Inatani, M.; Pallerla, S.R.; Castagnola, J.; Yamaguchi, Y.; Esko, J.D. Cerebral hypoplasia and craniofacial defects in mice lacking heparan sulfate ndst1 gene function. Development 2005, 132, 3777–3786. [Google Scholar] [CrossRef]
- Inatani, M.; Irie, F.; Plump, A.S.; Tessier-Lavigne, M.; Yamaguchi, Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science 2003, 302, 1044–1046. [Google Scholar] [CrossRef]
- Li, J.P.; Gong, F.; Hagner-McWhirter, A.; Forsberg, E.; Abrink, M.; Kisilevsky, R.; Zhang, X.; Lindahl, U. Targeted disruption of a murine glucuronyl c5-epimerase gene results in heparan sulfate lacking l-iduronic acid and in neonatal lethality. J. Biol. Chem. 2003, 278, 28363–28366. [Google Scholar] [CrossRef]
- McLaughlin, D.; Karlsson, F.; Tian, N.; Pratt, T.; Bullock, S.L.; Wilson, V.A.; Price, D.J.; Mason, J.O. Specific modification of heparan sulphate is required for normal cerebral cortical development. Mech. Dev. 2003, 120, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Pallerla, S.R.; Pan, Y.; Zhang, X.; Esko, J.D.; Grobe, K. Heparan sulfate ndst1 gene function variably regulates multiple signaling pathways during mouse development. Dev. Dyn. 2007, 236, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Irie, F.; Badie-Mahdavi, H.; Yamaguchi, Y. Autism-like socio-communicative deficits and stereotypies in mice lacking heparan sulfate. Proc. Natl. Acad. Sci. USA 2012, 109, 5052–5056. [Google Scholar] [CrossRef]
- Blanchard, D.C.; Defensor, E.B.; Meyza, K.Z.; Pobbe, R.L.; Pearson, B.L.; Bolivar, V.J.; Blanchard, R.J. Btbr t+tf/j mice: Autism-relevant behaviors and reduced fractone-associated heparan sulfate. Neurosci. Biobehav. Rev. 2012, 36, 285–296. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kalus, I.; Salmen, B.; Viebahn, C.; von Figura, K.; Schmitz, D.; D’Hooge, R.; Dierks, T. Differential involvement of the extracellular 6-o-endosulfatases sulf1 and sulf2 in brain development and neuronal and behavioural plasticity. J. Cell. Mol. Med. 2009, 13, 4505–4521. [Google Scholar] [CrossRef]
- Zcharia, E.; Metzger, S.; Chajek-Shaul, T.; Aingorn, H.; Elkin, M.; Friedmann, Y.; Weinstein, T.; Li, J.P.; Lindahl, U.; Vlodavsky, I. Transgenic expression of mammalian heparanase uncovers physiological functions of heparan sulfate in tissue morphogenesis, vascularization, and feeding behavior. FASEB J. 2004, 18, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Reizes, O.; Lincecum, J.; Wang, Z.; Goldberger, O.; Huang, L.; Kaksonen, M.; Ahima, R.; Hinkes, M.T.; Barsh, G.S.; Rauvala, H.; et al. Transgenic expression of syndecan-1 uncovers a physiological control of feeding behavior by syndecan-3. Cell 2001, 106, 105–116. [Google Scholar] [CrossRef]
- Strader, A.D.; Reizes, O.; Woods, S.C.; Benoit, S.C.; Seeley, R.J. Mice lacking the syndecan-3 gene are resistant to diet-induced obesity. J. Clin. Investig. 2004, 114, 1354–1360. [Google Scholar] [CrossRef] [PubMed]
- Reizes, O.; Benoit, S.C.; Clegg, D.J. The role of syndecans in the regulation of body weight and synaptic plasticity. Int. J. Biochem. Cell Biol. 2008, 40, 28–45. [Google Scholar] [CrossRef]
- Pallerla, S.R.; Lawrence, R.; Lewejohann, L.; Pan, Y.; Fischer, T.; Schlomann, U.; Zhang, X.; Esko, J.D.; Grobe, K. Altered heparan sulfate structure in mice with deleted ndst3 gene function. J. Biol. Chem. 2008, 283, 16885–16894. [Google Scholar] [CrossRef] [PubMed]
- Lencz, T.; Guha, S.; Liu, C.; Rosenfeld, J.; Mukherjee, S.; DeRosse, P.; John, M.; Cheng, L.; Zhang, C.; Badner, J.A.; et al. Genome-wide association study implicates ndst3 in schizophrenia and bipolar disorder. Nat. Commun. 2013, 4, 2739. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, S.; Teramoto, T.; Whalen, M.J.; Irizarry, M.C.; Takagi, Y.; Qiu, J.; Harada, J.; Waeber, C.; Breakefield, X.O.; Moskowitz, M.A. Fgf-2 regulates neurogenesis and degeneration in the dentate gyrus after traumatic brain injury in mice. J. Clin. Investig. 2003, 112, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Charytoniuk, D.; Porcel, B.; Rodriguez Gomez, J.; Faure, H.; Ruat, M.; Traiffort, E. Sonic hedgehog signalling in the developing and adult brain. J. Physiol. Paris 2002, 96, 9–16. [Google Scholar] [CrossRef]
- Ernfors, P.; Lonnerberg, P.; Ayer-LeLievre, C.; Persson, H. Developmental and regional expression of basic fibroblast growth factor mrna in the rat central nervous system. J. Neurosci. Res. 1990, 27, 10–15. [Google Scholar] [CrossRef]
- Traiffort, E.; Charytoniuk, D.; Watroba, L.; Faure, H.; Sales, N.; Ruat, M. Discrete localizations of hedgehog signalling components in the developing and adult rat nervous system. Eur. J. Neurosci. 1999, 11, 3199–3214. [Google Scholar] [CrossRef]
- Traiffort, E.; Charytoniuk, D.A.; Faure, H.; Ruat, M. Regional distribution of sonic hedgehog, patched, and smoothened mrna in the adult rat brain. J. Neurochem. 1998, 70, 1327–1330. [Google Scholar] [CrossRef]
- Traiffort, E.; Moya, K.L.; Faure, H.; Hassig, R.; Ruat, M. High expression and anterograde axonal transport of aminoterminal sonic hedgehog in the adult hamster brain. Eur. J. Neurosci. 2001, 14, 839–850. [Google Scholar] [CrossRef]
- Mark, R.J.; Fuson, K.S.; Keane-Lazar, K.; May, P.C. Fibroblast growth factor-8 protects cultured rat hippocampal neurons from oxidative insult. Brain Res. 1999, 830, 88–93. [Google Scholar] [CrossRef]
- Fontaine, V.; Kinkl, N.; Sahel, J.; Dreyfus, H.; Hicks, D. Survival of purified rat photoreceptors in vitro is stimulated directly by fibroblast growth factor-2. J. Neurosci. 1998, 18, 9662–9672. [Google Scholar] [CrossRef]
- Reilly, J.O.; Karavanova, I.D.; Williams, K.P.; Mahanthappa, N.K.; Allendoerfer, K.L. Cooperative effects of sonic hedgehog and ngf on basal forebrain cholinergic neurons. Mol. Cell. Neurosci. 2002, 19, 88–96. [Google Scholar] [CrossRef]
- Rafuse, V.F.; Soundararajan, P.; Leopold, C.; Robertson, H.A. Neuroprotective properties of cultured neural progenitor cells are associated with the production of sonic hedgehog. Neuroscience 2005, 131, 899–916. [Google Scholar] [CrossRef] [PubMed]
- Arenas, E. Stem cells in the treatment of parkinson’s disease. Brain Res. Bull. 2002, 57, 795–808. [Google Scholar] [CrossRef]
- Sibilia, M.; Steinbach, J.P.; Stingl, L.; Aguzzi, A.; Wagner, E.F. A strain-independent postnatal neurodegeneration in mice lacking the egf receptor. EMBO J. 1998, 17, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Smeyne, R.J.; Klein, R.; Schnapp, A.; Long, L.K.; Bryant, S.; Lewin, A.; Lira, S.A.; Barbacid, M. Severe sensory and sympathetic neuropathies in mice carrying a disrupted trk/ngf receptor gene. Nature 1994, 368, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Klein, R. Role of neurotrophins in mouse neuronal development. FASEB J. 1994, 8, 738–744. [Google Scholar] [CrossRef]
- Kornblum, H.I.; Hussain, R.; Wiesen, J.; Miettinen, P.; Zurcher, S.D.; Chow, K.; Derynck, R.; Werb, Z. Abnormal astrocyte development and neuronal death in mice lacking the epidermal growth factor receptor. J. Neurosci. Res. 1998, 53, 697–717. [Google Scholar] [CrossRef]
- Cohlberg, J.A.; Li, J.; Uversky, V.N.; Fink, A.L. Heparin and other glycosaminoglycans stimulate the formation of amyloid fibrils from alpha-synuclein in vitro. Biochemistry 2002, 41, 1502–1511. [Google Scholar] [CrossRef]
- Leteux, C.; Chai, W.; Nagai, K.; Herbert, C.G.; Lawson, A.M.; Feizi, T. 10e4 antigen of scrapie lesions contains an unusual nonsulfated heparan motif. J. Biol. Chem. 2001, 276, 12539–12545. [Google Scholar] [CrossRef]
- McBride, P.A.; Wilson, M.I.; Eikelenboom, P.; Tunstall, A.; Bruce, M.E. Heparan sulfate proteoglycan is associated with amyloid plaques and neuroanatomically targeted prp pathology throughout the incubation period of scrapie-infected mice. Exp. Neurol. 1998, 149, 447–454. [Google Scholar] [CrossRef]
- Van Horssen, J.; Kleinnijenhuis, J.; Maass, C.N.; Rensink, A.A.; Otte-Holler, I.; David, G.; van den Heuvel, L.P.; Wesseling, P.; de Waal, R.M.; Verbeek, M.M. Accumulation of heparan sulfate proteoglycans in cerebellar senile plaques. Neurobiol. Aging 2002, 23, 537–545. [Google Scholar] [CrossRef]
- Verbeek, M.M.; Otte-Höller, I.; Van den Born, J.; Van den Heuvel, L.P.W.J.; David, G.; Wesseling, P.; De Waal, R.M.W. Agrin is a major heparan sulfate proteoglycan accumulating in alzheimer’s disease brain. Am. J. Pathol. 1999, 155, 2115–2125. [Google Scholar] [CrossRef]
- Barski, J.J.; Dethleffsen, K.; Meyer, M. Cre recombinase expression in cerebellar purkinje cells. Genesis 2000, 28, 93–98. [Google Scholar] [CrossRef]
- Rogers, D.C.; Peters, J.; Martin, J.E.; Ball, S.; Nicholson, S.J.; Witherden, A.S.; Hafezparast, M.; Latcham, J.; Robinson, T.L.; Quilter, C.A.; et al. Shirpa, a protocol for behavioral assessment: Validation for longitudinal study of neurological dysfunction in mice. Neurosci. Lett. 2001, 306, 89–92. [Google Scholar] [CrossRef]
- Pellow, S.; Chopin, P.; File, S.E.; Briley, M. Validation of open:Closed arm entries in an elevated plus-maze as a measure of anxiety in the rat. J. Neurosci. Methods 1985, 14, 149–167. [Google Scholar] [CrossRef]
- Wang, L.; Fuster, M.; Sriramarao, P.; Esko, J.D. Endothelial heparan sulfate deficiency impairs l-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat. Immunol. 2005, 6, 902–910. [Google Scholar] [CrossRef]
- Garner, O.B.; Yamaguchi, Y.; Esko, J.D.; Videm, V. Small changes in lymphocyte development and activation in mice through tissue-specific alteration of heparan sulphate. Immunology 2008, 125, 420–429. [Google Scholar] [CrossRef]
- Pan, Y.; Woodbury, A.; Esko, J.D.; Grobe, K.; Zhang, X. Heparan sulfate biosynthetic gene ndst1 is required for fgf signaling in early lens development. Development 2006, 133, 4933–4944. [Google Scholar] [CrossRef]
- Zhang, X.M.; Ng, A.H.; Tanner, J.A.; Wu, W.T.; Copeland, N.G.; Jenkins, N.A.; Huang, J.D. Highly restricted expression of cre recombinase in cerebellar purkinje cells. Genesis 2004, 40, 45–51. [Google Scholar] [CrossRef]
- Grobe, K.; Ledin, J.; Ringvall, M.; Holborn, K.; Forsberg, E.; Esko, J.D.; Kjellen, L. Heparan sulfate and development: Differential roles of the n-acetylglucosamine n-deacetylase/n-sulfotransferase (ndst) isozymes. Biochim. Biophys. Acta 2002, 1573, 209–215. [Google Scholar] [CrossRef]
- Pan, Y.; Carbe, C.; Powers, A.; Zhang, E.E.; Esko, J.D.; Grobe, K.; Feng, G.S.; Zhang, X. Bud specific n-sulfation of heparan sulfate regulates shp2-dependent fgf signaling during lacrimal gland induction. Development 2008, 135, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.M.; Gritli-Linde, A.; Smeyne, R.; Kottmann, A.; McMahon, A.P. Sonic hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Dev. Biol. 2004, 270, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Wechsler-Reya, R.J.; Scott, M.P. Control of neuronal precursor proliferation in the cerebellum by sonic hedgehog. Neuron 1999, 22, 103–114. [Google Scholar] [CrossRef]
- Wallace, V.A. Purkinje-cell-derived sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr. Biol. 1999, 9, 445–448. [Google Scholar] [CrossRef]
- Dahmane, N.; Ruiz-i-Altaba, A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 1999, 126, 3089–3100. [Google Scholar] [PubMed]
- Rubin, J.B.; Choi, Y.J.; Segal, R.A. Cerebellar proteoglycans regulate sonic hedgehog responses during development. Development 2002, 129, 2223–2232. [Google Scholar]
- Kaksonen, M.; Pavlov, I.; Voikar, V.; Lauri, S.E.; Hienola, A.; Riekki, R.; Lakso, M.; Taira, T.; Rauvala, H. Syndecan-3-deficient mice exhibit enhanced ltp and impaired hippocampus-dependent memory. Mol. Cell. Neurosci. 2002, 21, 158–172. [Google Scholar] [CrossRef]
- Pankonin, M.S.; Gallagher, J.T.; Loeb, J.A. Specific structural features of heparan sulfate proteoglycans potentiate neuregulin-1 signaling. J. Biol. Chem. 2005, 280, 383–388. [Google Scholar] [CrossRef]
- Holmborn, K.; Ledin, J.; Smeds, E.; Eriksson, I.; Kusche-Gullberg, M.; Kjellen, L. Heparan sulfate synthesized by mouse embryonic stem cells deficient in ndst1 and ndst2 is 6-o-sulfated but contains no n-sulfate groups. J. Biol. Chem. 2004, 279, 42355–42358. [Google Scholar] [CrossRef]
- Esko, J.D.; Selleck, S.B. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 2002, 71, 435–471. [Google Scholar] [CrossRef]
- Ringvall, M.; Ledin, J.; Holmborn, K.; Van Kuppevelt, T.; Ellin, F.; Eriksson, I.; Olofsson, A.M.; Kjellén, L.; Forsberg, E. Defective heparan sulfate biosynthesis and neonatal lethality in mice lacking n-deacetylase/n-sulfotransferase-1. J. Biol. Chem. 2000, 275, 25926–25930. [Google Scholar] [CrossRef] [PubMed]
- Handel, M.A.; Dawson, M. Effects on spermiogenesis in the mouse of a male sterile neurological mutation, purkinje cell degeneration. Gamete Res. 1981, 4, 185–192. [Google Scholar] [CrossRef]
- Garg, H.G.; Linhardt, R.J.; Hales, C.A. Chemistry and Biology of Heparin and Heparan Sulfate; Elsevier Ltd.: Oxford, UK, 2005. [Google Scholar]
- Najmabadi, H.; Hu, H.; Garshasbi, M.; Zemojtel, T.; Abedini, S.S.; Chen, W.; Hosseini, M.; Behjati, F.; Haas, S.; Jamali, P.; et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 2011, 478, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Reuter, M.S.; Musante, L.; Hu, H.; Diederich, S.; Sticht, H.; Ekici, A.B.; Uebe, S.; Wienker, T.F.; Bartsch, O.; Zechner, U.; et al. Ndst1 missense mutations in autosomal recessive intellectual disability. Am. J. Med. Genet. A 2014, 164A, 2753–2763. [Google Scholar] [CrossRef]
- Hope, K.A.; Flatten, D.; Cavitch, P.; May, B.; Sutcliffe, J.S.; O’Donnell, J.; Reiter, L.T. The drosophila gene sulfateless modulates autism-like behaviors. Front. Genet. 2019, 10, 574. [Google Scholar] [CrossRef] [PubMed]
- Dowling, C.; Allen, N.J. Mice lacking glypican 4 display juvenile hyperactivity and adult social interaction deficits. Brain Plast. 2018, 4, 197–209. [Google Scholar] [CrossRef]
- Paganini, L.; Hadi, L.A.; Chetta, M.; Rovina, D.; Fontana, L.; Colapietro, P.; Bonaparte, E.; Pezzani, L.; Marchisio, P.; Tabano, S.M.; et al. A hs6st2 gene variant associated with x-linked intellectual disability and severe myopia in two male twins. Clin. Genet. 2019, 95, 368–374. [Google Scholar] [CrossRef]
- Forsberg, M.; Holmborn, K.; Kundu, S.; Dagalv, A.; Kjellen, L.; Forsberg-Nilsson, K. Under-sulfation of heparan sulfate restricts the differentiation potential of mouse embryonic stem cells. J. Biol. Chem. 2012, 287, 10853–10862. [Google Scholar] [CrossRef]
- Jakobsson, L.; Kreuger, J.; Holmborn, K.; Lundin, L.; Eriksson, I.; Kjellen, L.; Claesson-Welsh, L. Heparan sulfate in trans potentiates vegfr-mediated angiogenesis. Dev. Cell 2006, 10, 625–634. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lewejohann, L.; Pallerla, S.R.; Schreiber, R.S.; Gerula, J.; Grobe, K. Cerebellar Morphology and Behavioral Profiles in Mice Lacking Heparan Sulfate Ndst Gene Function. J. Dev. Biol. 2020, 8, 13. https://doi.org/10.3390/jdb8030013
Lewejohann L, Pallerla SR, Schreiber RS, Gerula J, Grobe K. Cerebellar Morphology and Behavioral Profiles in Mice Lacking Heparan Sulfate Ndst Gene Function. Journal of Developmental Biology. 2020; 8(3):13. https://doi.org/10.3390/jdb8030013
Chicago/Turabian StyleLewejohann, Lars, Srinivas R. Pallerla, Rebecca S. Schreiber, Joanna Gerula, and Kay Grobe. 2020. "Cerebellar Morphology and Behavioral Profiles in Mice Lacking Heparan Sulfate Ndst Gene Function" Journal of Developmental Biology 8, no. 3: 13. https://doi.org/10.3390/jdb8030013
APA StyleLewejohann, L., Pallerla, S. R., Schreiber, R. S., Gerula, J., & Grobe, K. (2020). Cerebellar Morphology and Behavioral Profiles in Mice Lacking Heparan Sulfate Ndst Gene Function. Journal of Developmental Biology, 8(3), 13. https://doi.org/10.3390/jdb8030013





