Requirement of the Dynein-Adaptor Spindly for Mitotic and Post-Mitotic Functions in Drosophila
Abstract
1. Introduction
2. Results
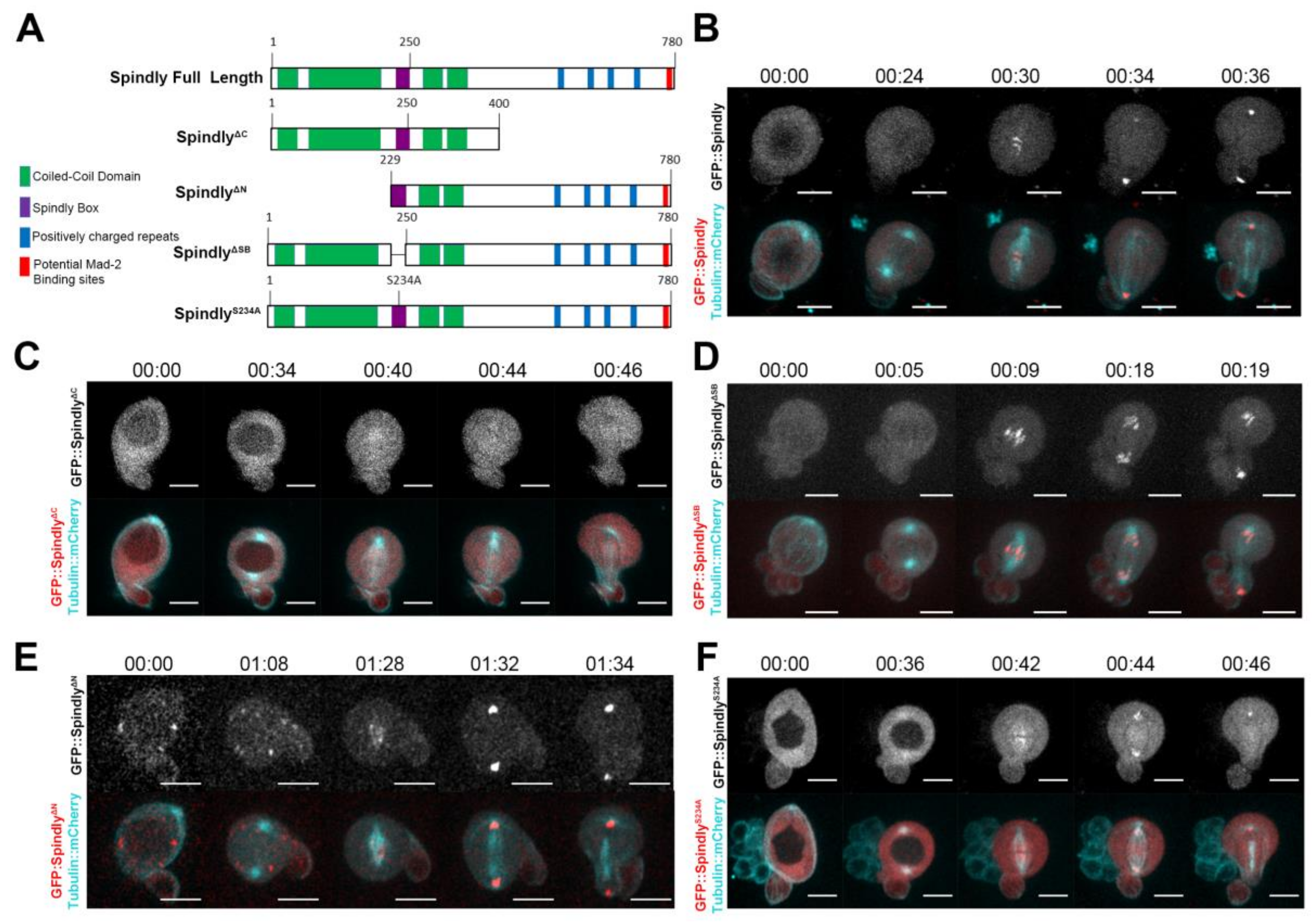
2.1. Requirements for Dynamic Subcellular Localisation of Spindly
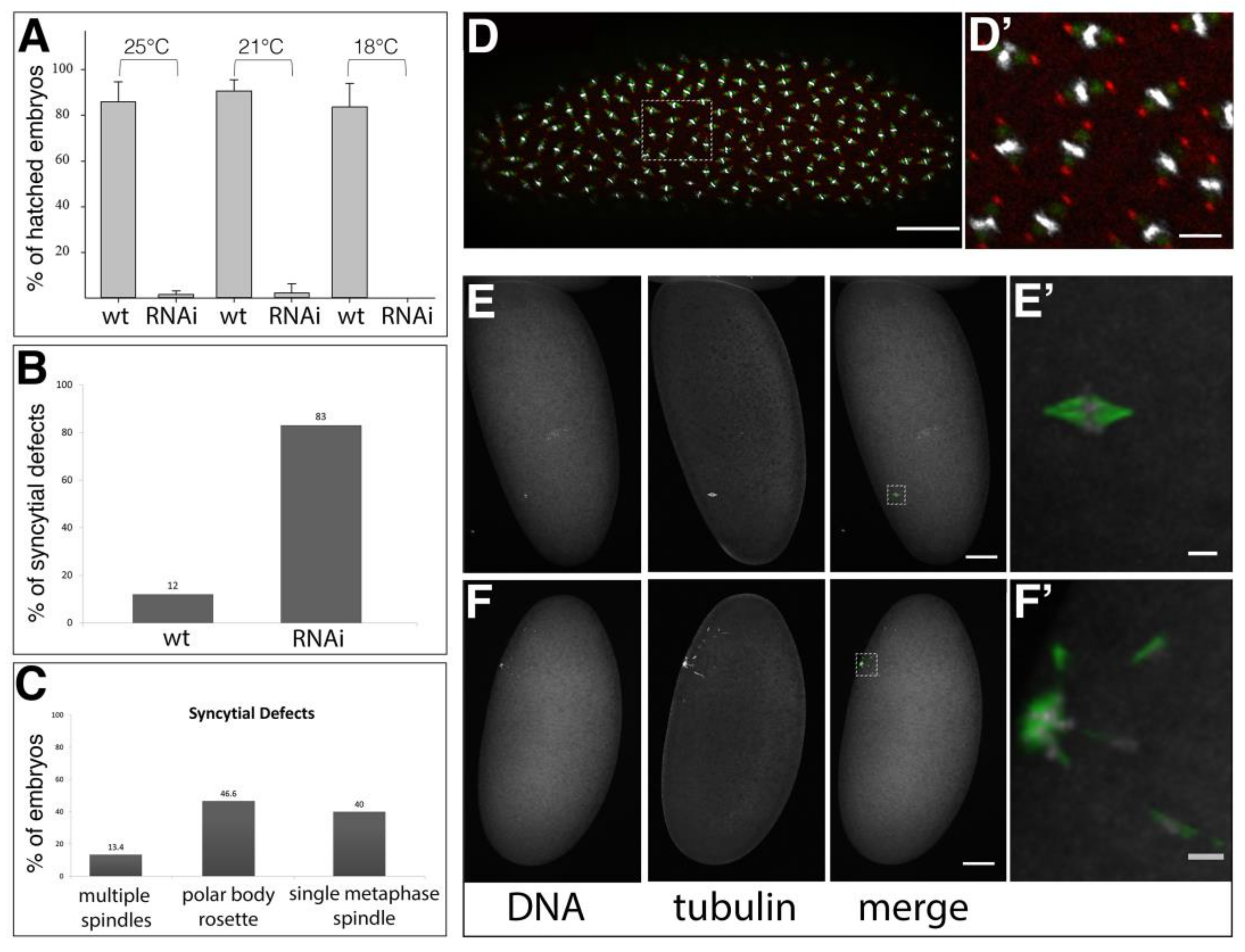
2.2. Requirement of Spindly for Initiation of Syncytial Cleavage Division
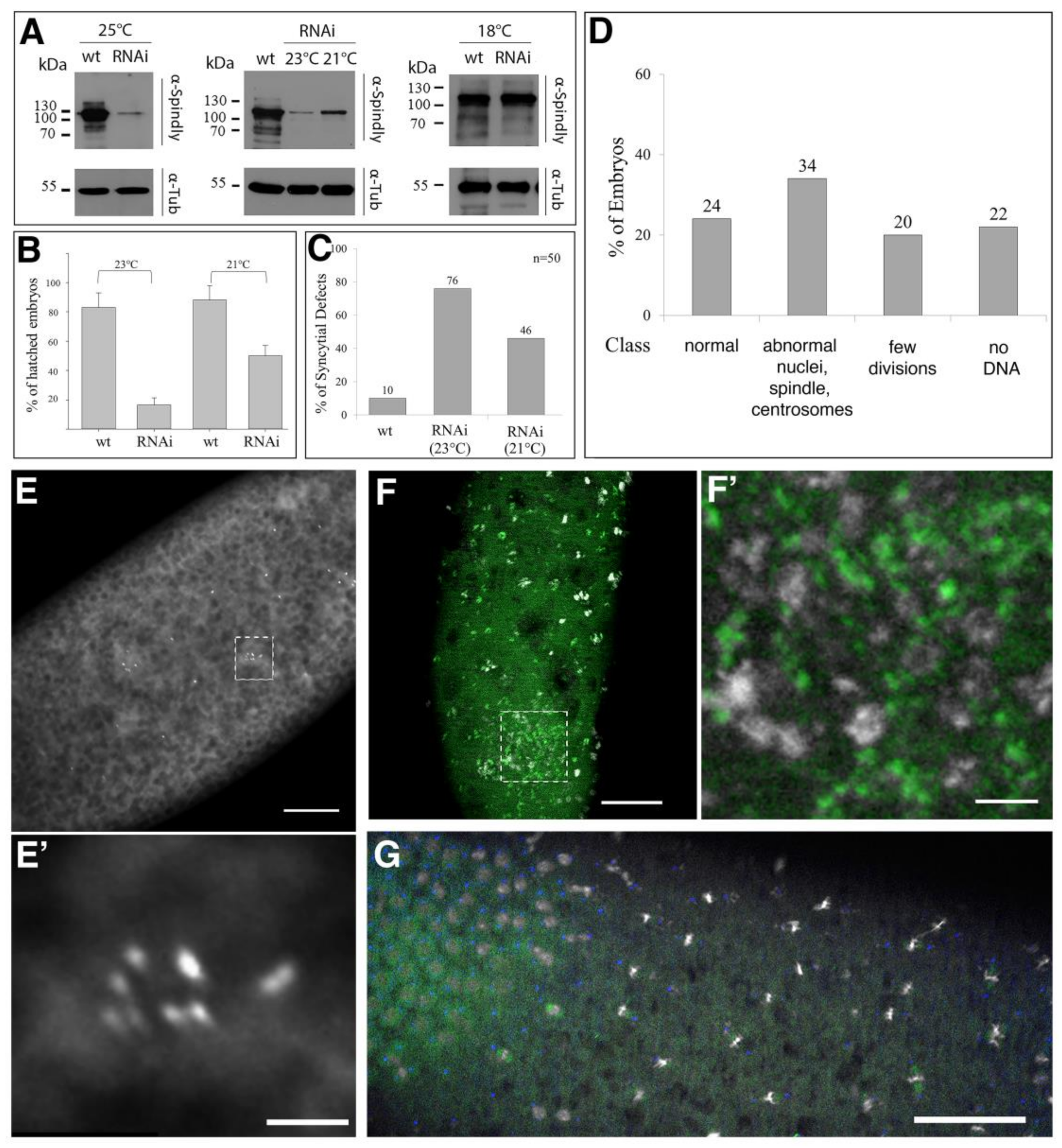
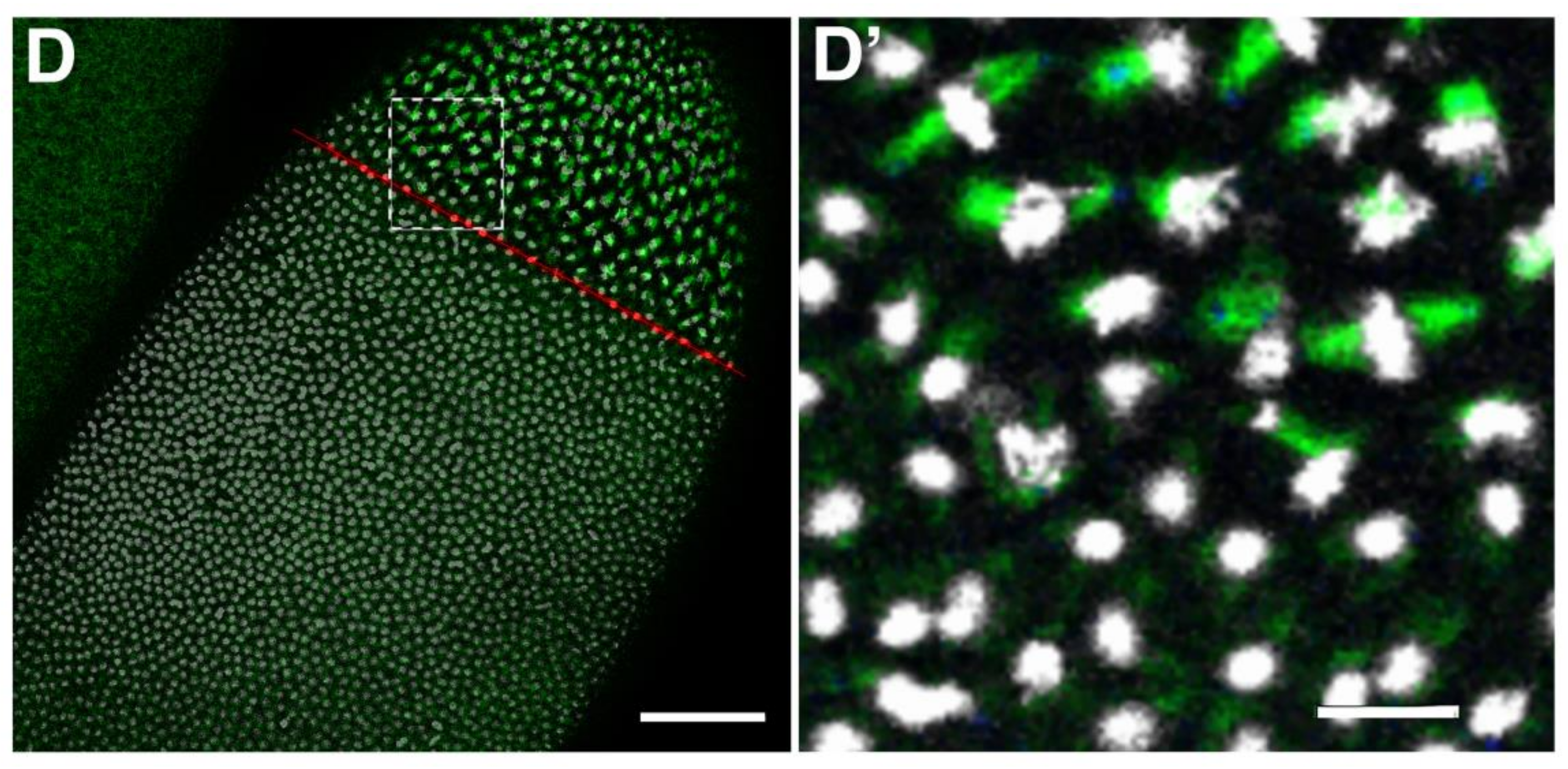
2.3. Requirement of Spindly for Syncytial Cleavage Divisions
2.4. Requirements of Spindly Protein Domains in Early Development
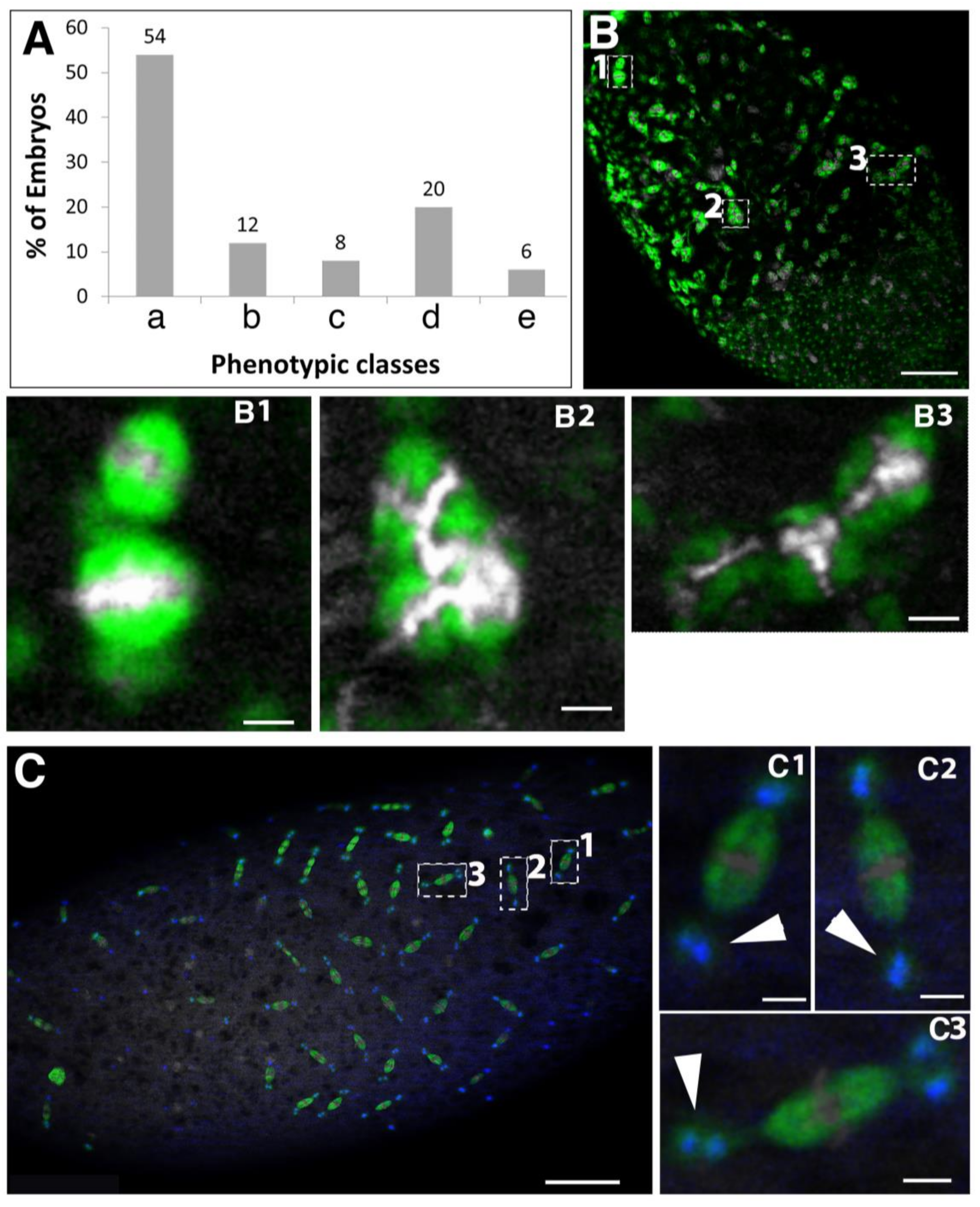
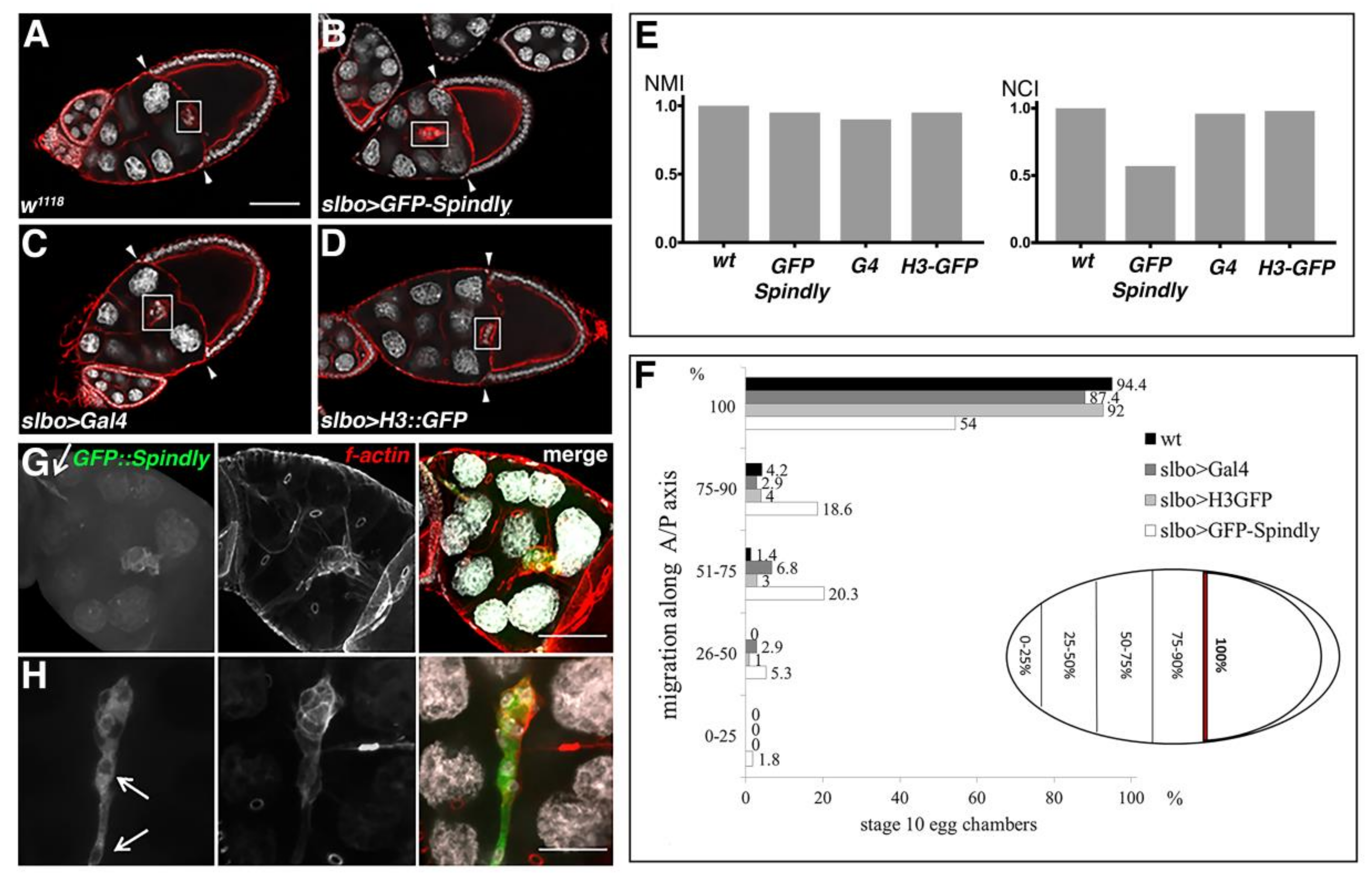
2.5. Spindly Overexpression Causes Defects in Egg Shape and Embryogenesis
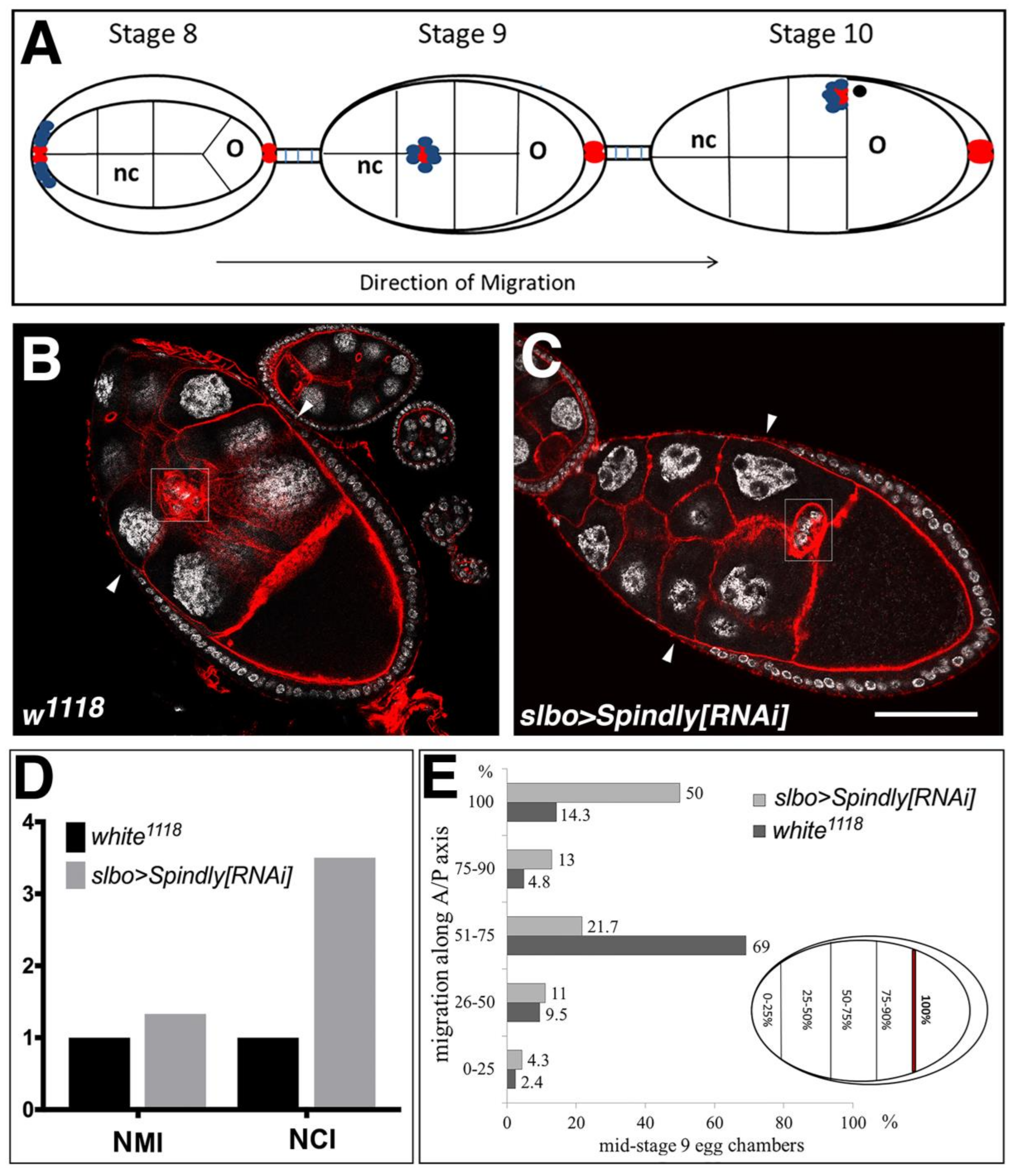
2.6. Requirement of Spindly in Ovarian Border Cell Migration
3. Discussion
3.1. Spindly Function in Early Drosophila Embryogenesis
3.2. Spindly Overexpression Affects Dynein-Dependent Processes in Egg Patterning
3.3. A Role for Spindly in Cell Migration
4. Materials and Methods
4.1. Drosophila Strains and Culture
4.2. Embryo Collection
4.3. Culture and Live Imaging of Larval Neuroblasts and Embryos
4.4. Site-Directed Mutagenesis
4.5. Protein Extraction and Western Blotting
4.6. Embryo Immunostaining
4.7. Ovary Dissection and Immunolabeling
4.8. Egg Chamber Staging
4.9. Quantification of the Migration Phenotypes
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Foe, V.E.; Odell, G.M.; Edgar, B. Mitosis and morphogenesis in the Drosophila embryo: Point and counterpoint. In The Development of Drosophila Melanogaster; Bate, M., Martinez-Arias, A., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1993; pp. 149–300. [Google Scholar]
- Lee, L.A.; Orr-Weaver, T.L. Regulation of cell cycles in Drosophila development: Intrinsic and extrinsic cues. Annu. Rev. Genet. 2003, 37, 545–578. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Raff, J.W. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 1999, 18, 2184–2195. [Google Scholar] [CrossRef] [PubMed]
- Su, T.T.; Sprenger, F.; DiGregorio, P.J.; Campbell, S.D.; O’Farrell, P.H. Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes Dev. 1998, 12, 1495–1503. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A. Spindle assembly checkpoint: The third decade. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 3595–3604. [Google Scholar] [CrossRef] [PubMed]
- Lara-Gonzalez, P.; Westhorpe, F.G.; Taylor, S.S. The spindle assembly checkpoint. Curr. Biol. 2012, 22, R966–R980. [Google Scholar] [CrossRef]
- Jia, L.; Kim, S.; Yu, H. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem. Sci. 2013, 38, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Howell, B.J.; McEwen, B.F.; Canman, J.C.; Hoffman, D.B.; Farrar, E.M.; Rieder, C.L.; Salmon, E.D. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001, 155, 1159–1172. [Google Scholar] [CrossRef] [PubMed]
- Griffis, E.R.; Stuurman, N.; Vale, R.D. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 2007, 177, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Mische, S.; He, Y.; Ma, L.; Li, M.; Serr, M.; Hays, T.S. Dynein light intermediate chain: An essential subunit that contributes to spindle checkpoint inactivation. Mol. Biol. Cell 2008, 19, 4918–4929. [Google Scholar] [CrossRef] [PubMed]
- Basto, R.; Scaerou, F.; Mische, S.; Wojcik, E.; Lefebvre, C.; Gomes, R.; Hays, T.; Karess, R. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr. Biol. 2004, 14, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Buffin, E.; Lefebvre, C.; Huang, J.; Gagou, M.E.; Karess, R.E. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 2005, 15, 856–861. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, E.; Basto, R.; Serr, M.; Scaerou, F.; Karess, R.; Hays, T. Kinetochore dynein: Its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 2001, 3, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Buffin, E.; Emre, D.; Karess, R.E. Flies without a spindle checkpoint. Nat. Cell Biol. 2007, 9, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, Z.; Gagou, M.E.; Lefebvre, C.; Emre, D.; Karess, R.E. Separating the spindle, checkpoint, and timer functions of BubR1. J. Cell Biol. 2009, 187, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Basu, J.; Bousbaa, H.; Logarinho, E.; Li, Z.; Williams, B.C.; Lopes, C.; Sunkel, C.E.; Goldberg, M.L. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol. 1999, 146, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.C.; Karr, T.L.; Montgomery, J.M.; Goldberg, M.L. The Drosophila l(1)zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J. Cell Biol. 1992, 118, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.S.; Sampaio, P.; Williams, B.; Goldberg, M.; Sunkel, C.E. The Drosophila Bub3 protein is required for the mitotic checkpoint and for normal accumulation of cyclins during G2 and early stages of mitosis. J. Cell Sci. 2005, 118, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Karess, R.E.; Glover, D.M. rough deal: A gene required for proper mitotic segregation in Drosophila. J. Cell Biol. 1989, 109, 2951–2961. [Google Scholar] [CrossRef] [PubMed]
- Althoff, F.; Karess, R.E.; Lehner, C.F. Spindle checkpoint-independent inhibition of mitotic chromosome segregation by Drosophila Mps1. Mol. Biol. Cell 2012, 23, 2275–2291. [Google Scholar] [CrossRef] [PubMed]
- Karess, R. Rod-Zw10-Zwilch: A key player in the spindle checkpoint. Trends Cell. Biol. 2005, 15, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Kops, G.J.; Kim, Y.; Weaver, B.A.; Mao, Y.; McLeod, I.; Yates, J.R., 3rd; Tagaya, M.; Cleveland, D.W. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 2005, 169, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Starr, D.A.; Williams, B.C.; Hays, T.S.; Goldberg, M.L. ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 1998, 142, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.C.; Li, Z.; Liu, S.; Williams, E.V.; Leung, G.; Yen, T.J.; Goldberg, M.L. Zwilch, a new component of the ZW10/ROD complex required for kinetochore functions. Mol. Biol. Cell 2003, 14, 1379–1391. [Google Scholar] [CrossRef] [PubMed]
- Bader, J.R.; Kasuboski, J.M.; Winding, M.; Vaughan, P.S.; Hinchcliffe, E.H.; Vaughan, K.T. Polo-like kinase1 is required for recruitment of dynein to kinetochores during mitosis. J. Biol. Chem. 2011, 286, 20769–20777. [Google Scholar] [CrossRef] [PubMed]
- Whyte, J.; Bader, J.R.; Tauhata, S.B.; Raycroft, M.; Hornick, J.; Pfister, K.K.; Lane, W.S.; Chan, G.K.; Hinchcliffe, E.H.; Vaughan, P.S.; et al. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J. Cell Biol. 2008, 183, 819–834. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, R.; Essex, A.; Hu, J.S.; Maddox, P.S.; Motegi, F.; Sugimoto, A.; O’Rourke, S.M.; Bowerman, B.; McLeod, I.; Yates, J.R., 3rd; et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 2008, 22, 2385–2399. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.W.; Fava, L.L.; Uldschmid, A.; Schmitz, M.H.; Gerlich, D.W.; Nigg, E.A.; Santamaria, A. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J. Cell Biol. 2009, 185, 859–874. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, R.; Holland, A.J.; Varma, D.; Wan, X.; Civril, F.; Cleveland, D.W.; Oegema, K.; Salmon, E.D.; Desai, A. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010, 24, 957–971. [Google Scholar] [CrossRef] [PubMed]
- Barisic, M.; Sohm, B.; Mikolcevic, P.; Wandke, C.; Rauch, V.; Ringer, T.; Hess, M.; Bonn, G.; Geley, S. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol. Biol. Cell 2010, 21, 1968–1981. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.J.; Reis, R.M.; Niessen, S.; Pereira, C.; Andres, D.A.; Spielmann, H.P.; Cleveland, D.W.; Desai, A.; Gassmann, R. Preventing farnesylation of the dynein adaptor Spindly contributes to the mitotic defects caused by farnesyltransferase inhibitors. Mol. Biol. Cell 2015, 26, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- Moudgil, D.K.; Westcott, N.; Famulski, J.K.; Patel, K.; Macdonald, D.; Hang, H.; Chan, G.K. A novel role of farnesylation in targeting a mitotic checkpoint protein, human Spindly, to kinetochores. J. Cell Biol. 2015, 208, 881–896. [Google Scholar] [CrossRef] [PubMed]
- Mosalaganti, S.; Keller, J.; Altenfeld, A.; Winzker, M.; Rombaut, P.; Saur, M.; Petrovic, A.; Wehenkel, A.; Wohlgemuth, S.; Muller, F.; et al. Structure of the RZZ complex and molecular basis of its interaction with Spindly. J. Cell Biol. 2017, 216, 961–981. [Google Scholar] [CrossRef] [PubMed]
- Varma, D.; Wan, X.; Cheerambathur, D.; Gassmann, R.; Suzuki, A.; Lawrimore, J.; Desai, A.; Salmon, E.D. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J. Cell Biol. 2013, 202, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Schlager, M.A.; Hoang, H.T.; Urnavicius, L.; Bullock, S.L.; Carter, A.P. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014, 33, 1855–1868. [Google Scholar] [CrossRef] [PubMed]
- Hoogenraad, C.C.; Akhmanova, A. Bicaudal D Family of Motor Adaptors: Linking Dynein Motility to Cargo Binding. Trends Cell Biol. 2016, 26, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [PubMed]
- Pampalona, J.; Januschke, J.; Sampaio, P.; Gonzalez, C. Time-lapse recording of centrosomes and other organelles in Drosophila neuroblasts. Methods Cell Biol. 2015, 129, 301–315. [Google Scholar] [PubMed]
- Ni, J.Q.; Liu, L.P.; Binari, R.; Hardy, R.; Shim, H.S.; Cavallaro, A.; Booker, M.; Pfeiffer, B.D.; Markstein, M.; Wang, H.; et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 2009, 182, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Page, A.W.; Orr-Weaver, T.L. Activation of the meiotic divisions in Drosophila oocytes. Dev. Biol. 1997, 183, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Tastan, O.Y.; Maines, J.Z.; Li, Y.; McKearin, D.M.; Buszczak, M. Drosophila ataxin 2-binding protein 1 marks an intermediate step in the molecular differentiation of female germline cysts. Development 2010, 137, 3167–3176. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dickinson, L.K.; Lehmann, R. Genetics of nanos localization in Drosophila. Dev. Dyn. 1994, 199, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Perez-Mongiovi, D.; Malmanche, N.; Bousbaa, H.; Sunkel, C. Maternal expression of the checkpoint protein BubR1 is required for synchrony of syncytial nuclear divisions and polar body arrest in Drosophila melanogaster. Development 2005, 132, 4509–4520. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Montell, D.J. Border-cell migration: The race is on. Nat. Rev. Mol. Cell Biol. 2003, 4, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Rorth, P. Specification of C/EBP function during Drosophila development by the bZIP basic region. Science 1994, 266, 1878–1881. [Google Scholar] [CrossRef] [PubMed]
- Assaker, G.; Ramel, D.; Wculek, S.K.; Gonzalez-Gaitan, M.; Emery, G. Spatial restriction of receptor tyrosine kinase activity through a polarized endocytic cycle controls border cell migration. Proc. Natl. Acad. Sci. USA 2010, 107, 22558–22563. [Google Scholar] [CrossRef] [PubMed]
- Sharp, D.J.; Rogers, G.C.; Scholey, J.M. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2000, 2, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Sharp, D.J.; Brown, H.M.; Kwon, M.; Rogers, G.C.; Holland, G.; Scholey, J.M. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell 2000, 11, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Brust-Mascher, I.; Scholey, J.M. Sliding filaments and mitotic spindle organization. Nat. Cell Biol. 2014, 16, 737–739. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Wojcik, E.J.; Sanders, M.A.; McGrail, M.; Hays, T.S. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 1999, 146, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Morales-Mulia, S.; Scholey, J.M. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell 2005, 16, 3176–3186. [Google Scholar] [CrossRef] [PubMed]
- Gama, J.B.; Pereira, C.; Simoes, P.A.; Celestino, R.; Reis, R.M.; Barbosa, D.J.; Pires, H.R.; Carvalho, C.; Amorim, J.; Carvalho, A.X.; et al. Molecular mechanism of dynein recruitment to kinetochores by the Rod-Zw10-Zwilch complex and Spindly. J. Cell Biol. 2017, 216, 943–960. [Google Scholar] [CrossRef] [PubMed]
- Defachelles, L.; Hainline, S.G.; Menant, A.; Lee, L.A.; Karess, R.E. A maternal effect rough deal mutation suggests that multiple pathways regulate Drosophila RZZ kinetochore recruitment. J. Cell Sci. 2015, 128, 1204–1216. [Google Scholar] [CrossRef] [PubMed]
- Horne-Badovinac, S. The Drosophila egg chamber-a new spin on how tissues elongate. Integr. Comp. Biol. 2014, 54, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Haigo, S.L.; Bilder, D. Global tissue revolutions in a morphogenetic movement controlling elongation. Science 2011, 331, 1071–1074. [Google Scholar] [CrossRef] [PubMed]
- Crest, J.; Diz-Munoz, A.; Chen, D.Y.; Fletcher, D.A.; Bilder, D. Organ sculpting by patterned extracellular matrix stiffness. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Luschnig, S.; Moussian, B.; Krauss, J.; Desjeux, I.; Perkovic, J.; Nusslein-Volhard, C. An F1 genetic screen for maternal-effect mutations affecting embryonic pattern formation in Drosophila melanogaster. Genetics 2004, 167, 325–342. [Google Scholar] [CrossRef] [PubMed]
- Hudson, A.M.; Cooley, L. Understanding the function of actin-binding proteins through genetic analysis of Drosophila oogenesis. Annu. Rev. Genet. 2002, 36, 455–488. [Google Scholar] [CrossRef] [PubMed]
- Januschke, J.; Gervais, L.; Dass, S.; Kaltschmidt, J.A.; Lopez-Schier, H.; St Johnston, D.; Brand, A.H.; Roth, S.; Guichet, A. Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr. Biol. 2002, 12, 1971–1981. [Google Scholar] [CrossRef]
- Forlani, S.; Ferrandon, D.; Saget, O.; Mohier, E. A regulatory function for K10 in the establishment of dorsoventral polarity in the Drosophila egg and embryo. Mech. Dev. 1993, 41, 109–120. [Google Scholar] [CrossRef]
- Neuman-Silberberg, F.S.; Schupbach, T. The Drosophila TGF-alpha-like protein Gurken: Expression and cellular localization during Drosophila oogenesis. Mech. Dev. 1996, 59, 105–113. [Google Scholar] [CrossRef]
- MacDougall, N.; Clark, A.; MacDougall, E.; Davis, I. Drosophila gurken (TGFalpha) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev. Cell 2003, 4, 307–319. [Google Scholar] [CrossRef]
- Rom, I.; Faicevici, A.; Almog, O.; Neuman-Silberberg, F.S. Drosophila Dynein light chain (DDLC1) binds to gurken mRNA and is required for its localization. Biochim. Biophys. Acta 2007, 1773, 1526–1533. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Swan, A.; Nguyen, T.; Suter, B. Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat. Cell Biol. 1999, 1, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Warrior, R. The Drosophila Lissencephaly1 (DLis1) gene is required for nuclear migration. Dev. Biol. 2000, 226, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Duncan, J.E.; Warrior, R. The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr. Biol. 2002, 12, 1982–1991. [Google Scholar] [CrossRef]
- Sitaram, P.; Merkle, J.A.; Lee, E.; Lee, L.A. Asunder is required for dynein localization and dorsal fate determination during Drosophila oogenesis. Dev. Biol. 2014, 386, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Swan, A.; Suter, B. Role of Bicaudal-D in patterning the Drosophila egg chamber in mid-oogenesis. Development 1996, 122, 3577–3586. [Google Scholar] [PubMed]
- Yang, N.; Inaki, M.; Cliffe, A.; Rorth, P. Microtubules and Lis-1/NudE/dynein regulate invasive cell-on-cell migration in Drosophila. PLoS ONE 2012, 7, e40632. [Google Scholar] [CrossRef] [PubMed]
- Horne-Badovinac, S.; Bilder, D. Dynein regulates epithelial polarity and the apical localization of stardust A mRNA. PLoS Genet. 2008, 4, e8. [Google Scholar] [CrossRef] [PubMed]
- Quintyne, N.J.; Gill, S.R.; Eckley, D.M.; Crego, C.L.; Compton, D.A.; Schroer, T.A. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 1999, 147, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Schroer, T.A. Motors, clutches and brakes for membrane traffic: A commemorative review in honor of Thomas Kreis. Traffic 2000, 1, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Albertson, R.; Chabu, C.; Sheehan, A.; Doe, C.Q. Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. J. Cell Sci. 2004, 117, 6061–6070. [Google Scholar] [CrossRef] [PubMed]
- Rusan, N.M.; Peifer, M. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 2007, 177, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Karr, T.L.; Alberts, B.M. Organization of the cytoskeleton in early Drosophila embryos. J. Cell Biol. 1986, 102, 1494–1509. [Google Scholar] [CrossRef] [PubMed]
- Abramoff, M. Image processing with Image J. Biophotonic Int. 2004, 11, 36–42. [Google Scholar]



| Primer | 5′➔3′ |
|---|---|
| Forward | caagatgccgtggacattaagacggagttggaagctccagaattaattcc |
| Reverse | cttaatgtccacggcatcttgctgttccaagtttaagtcgatttctcg |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clemente, G.D.; Hannaford, M.R.; Beati, H.; Kapp, K.; Januschke, J.; Griffis, E.R.; Müller, H.-A.J. Requirement of the Dynein-Adaptor Spindly for Mitotic and Post-Mitotic Functions in Drosophila. J. Dev. Biol. 2018, 6, 9. https://doi.org/10.3390/jdb6020009
Clemente GD, Hannaford MR, Beati H, Kapp K, Januschke J, Griffis ER, Müller H-AJ. Requirement of the Dynein-Adaptor Spindly for Mitotic and Post-Mitotic Functions in Drosophila. Journal of Developmental Biology. 2018; 6(2):9. https://doi.org/10.3390/jdb6020009
Chicago/Turabian StyleClemente, Giuliana D., Matthew R. Hannaford, Hamze Beati, Katja Kapp, Jens Januschke, Eric R. Griffis, and Hans-Arno J. Müller. 2018. "Requirement of the Dynein-Adaptor Spindly for Mitotic and Post-Mitotic Functions in Drosophila" Journal of Developmental Biology 6, no. 2: 9. https://doi.org/10.3390/jdb6020009
APA StyleClemente, G. D., Hannaford, M. R., Beati, H., Kapp, K., Januschke, J., Griffis, E. R., & Müller, H.-A. J. (2018). Requirement of the Dynein-Adaptor Spindly for Mitotic and Post-Mitotic Functions in Drosophila. Journal of Developmental Biology, 6(2), 9. https://doi.org/10.3390/jdb6020009






