Regulation of Silk Genes by Hox and Homeodomain Proteins in the Terminal Differentiated Silk Gland of the Silkworm Bombyx mori
Abstract
:1. Introduction
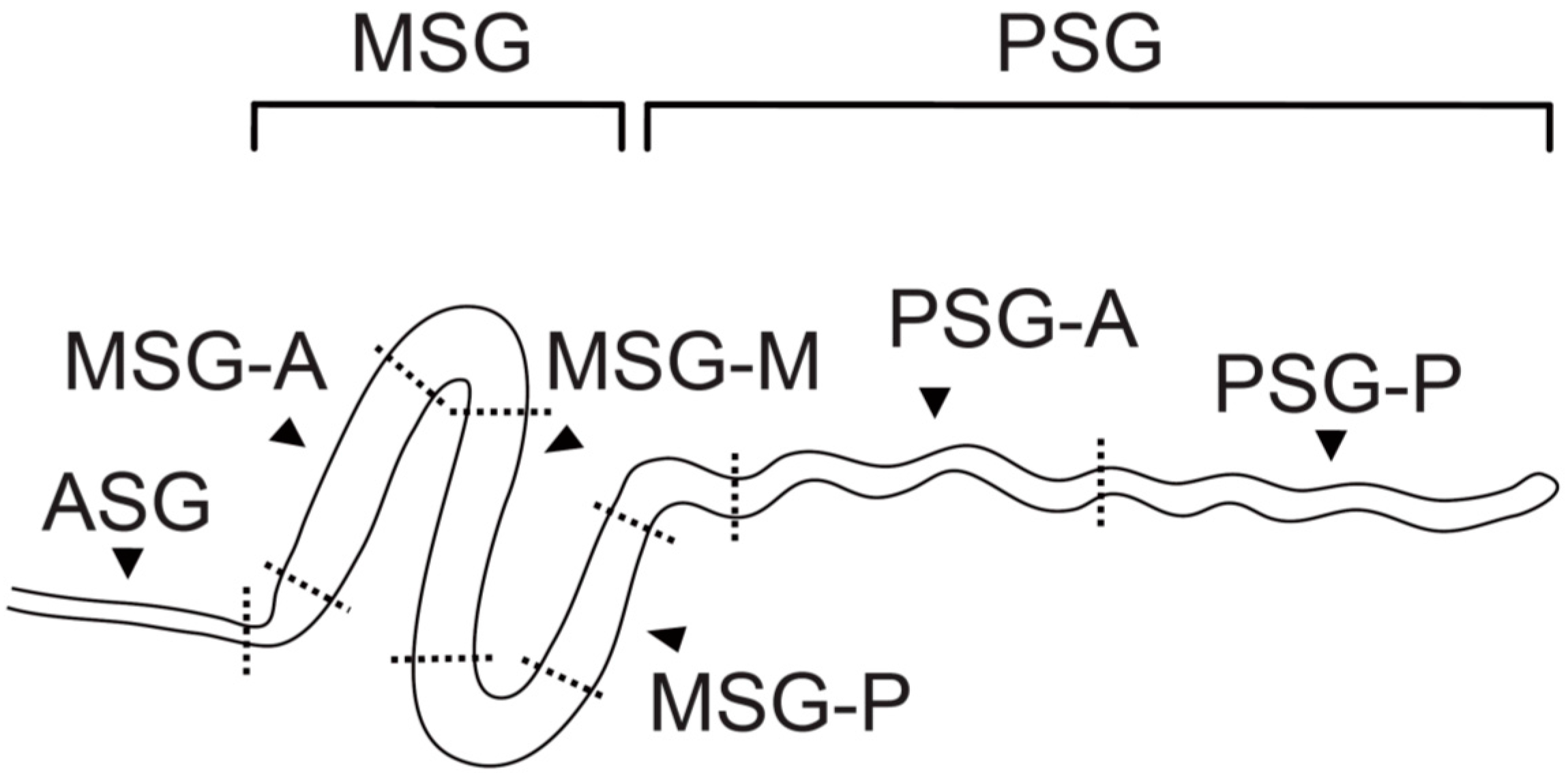
2. Region-Specific Expression of Silk Genes in the Silk Gland
3. Region-Specific Expression of Hox and Several Homeobox Genes in the Fully Differentiated Silk Gland
4. Antp Regulates the MSG-Specific Expression of ser1
5. Antp Activates Other MSG-Specific Silk Genes
6. The LIM-Homeodomain Protein, Arrowhead, Regulates the PSG-Specific Expression of Silk Fiber Protein Genes
7. Perspective
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tazima, Y. The Genetics of the Silkworm; Logos Press: London, UK; Academic Press: London, UK, 1964. [Google Scholar]
- Suzuki, Y. Genes that are involved in Bombyx body plan and silk gene regulation. Int. J. Dev. Biol. 1994, 38, 231–235. [Google Scholar] [PubMed]
- Ueno, K.; Nagata, T.; Suzuki, Y. Roles of homeotic genes in the Bombyx body plan. In Molecular Model Systems in the Lepidoptera; Goldsmith, M.R., Wilkins, A.S., Eds.; Cambridge University Press: New York, NY, USA, 1995; pp. 165–180. [Google Scholar]
- Chai, C.-L.; Zhang, Z.; Huang, F.-F.; Wang, X.-Y.; Yu, Q.-Y.; Liu, B.-B.; Tian, T.; Xia, Q.-Y.; Lu, C.; Xiang, Z.-H. A genomewide survey of homeobox genes and identification of novel structure of the Hox cluster in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Masumoto, M.; Yaginuma, T.; Niimi, T. Functional analysis of Ultrabithorax in the silkworm, Bombyx mori using RNAi. Dev. Genes Evol. 2009, 219, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Nagata, T.; Suzuki, Y.; Ueno, K.; Kokubo, H.; Xu, X.; Hui, C.-C.; Hara, W.; Fukuta, M. Developmental expression of the Bombyx Antennapedia homologue and homeotic changes in the Nc mutant. Genes Cells 1996, 1, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.-H.; Wang, X.-Y.; Chai, C.-L.; Zhang, C.-D.; Lu, C.; Xiang, Z.-H. Identification and function of abdominal-A in the silkworm, Bombyx mori. Insect Mol. Biol. 2009, 18, 155–160. [Google Scholar] [CrossRef] [PubMed]
- The International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1036–1045. [Google Scholar]
- Tomita, S.; Kikuchi, A. Abd-B suppresses lepidopteran proleg development in posterior abdomen. Dev. Biol. 2009, 328, 403–409. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ueno, K.; Hui, C.-C.; Fukuta, M.; Suzuki, Y. Molecular analysis of the deletion mutants in the E homeotic complex of the silkworm Bombyx mori. Development 1992, 114, 555–563. [Google Scholar] [PubMed]
- Yasukochi, Y.; Ashakumary, L.A.; Wu, C.; Yoshido, A.; Nohata, J.; Mita, K.; Sahara, K. Organization of the Hox gene cluster of the silkworm, Bombyx mori: A split of the Hox cluster in a non-Drosophila insect. Dev. Genes Evol. 2004, 214, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.S.; Carroll, S.B. Moleculae mechanisms of selector gene function and evolution. Curr. Opin. Genet. Dev. 2002, 12, 592–600. [Google Scholar] [CrossRef]
- Mann, R.S.; Morata, G. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu. Rev. Cell Dev. Biol. 2000, 16, 243–271. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, W.; Krumlauf, R. Homeobox genes and axial patterning. Cell 1992, 68, 283–302. [Google Scholar] [CrossRef]
- Berger, M.F.; Badis, G.; Gehrke, A.R.; Talukder, S.; Philippakis, A.A.; Penta-Castillo, L.; Alleyene, T.M.; Mnaimneh, S.; Botvinnik, O.B.; Chan, E.T.; et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 2008, 133, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Ekker, S.C.; Jackson, D.G.; Kessler, D.P.; Sun, B.I.; Young, K.E.; Beachy, P.A. The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. EMBO J. 1994, 13, 3551–3560. [Google Scholar] [PubMed]
- Gehring, W.J.; Qian, Y.Q.; Billeter, M.; Furukubo-Tokunaga, K.; Schier, A.F.; Resendez-Perez, D.; Affolter, M.; Otting, G.; Wuthrich, K. Homeodomain-DNA recognition. Cell 1994, 78, 211–223. [Google Scholar] [CrossRef]
- Mann, R.S. The specificity of homeotic gene function. BioEssays 1995, 17, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Noyes, M.B.; Christensen, R.G.; Wakabayashi, A.; Stormo, G.D.; Brodsky, M.H.; Wolfe, S.A. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 2008, 133, 1277–1289. [Google Scholar] [CrossRef] [PubMed]
- Ebner, A.; Cabernard, C.; Affolter, M.; Merabet, S. Recognition of distinct target sites by a unique Labial/Extradenticle/Homothorax complex. Development 2005, 132, 1591–1600. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; Scott, M.P. What determines the specificity of action of Drosophila homeodomain proteins? Cell 1990, 63, 883–894. [Google Scholar] [CrossRef]
- Huber, S.D.; Lohmann, I. Shaping segments: Hox gene function in the genomic age. BioEssays 2008, 30, 965–979. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-P.; Brocchieri, L.; Shen, W.-F.; Largman, C.; Cleary, M.L. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol. Cell Biol. 1996, 16, 1734–1745. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.-K.; Jaffe, L.; Capovilla, M.; Botas, J.; Mann, R.S. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 1994, 78, 603–615. [Google Scholar] [CrossRef]
- Chan, S.-K.; Mann, R.S. A structural model for a homeotic protein-extradenticle-DNA complex accounts for the choice of HOX protein in the heterodimer. Proc. Natl. Acad. Sci. USA 1996, 93, 5223–5228. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Passner, J.M.; Rohs, R.; Jain, R.; Sosinsky, A.; Crickmore, M.A.; Jacob, V.; Aggarwal, A.K.; Honig, B.; Mann, R. Functional specificity of a Hox protein Mediated by the recognition of minor groove structure. Cell 2007, 131, 530–543. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.S.; Lelli, K.M.; Joshi, R. Hox specificity: Unique roles for cofactors and collaborators. Curr. Top. Dev. Biol. 2009, 88, 63–101. [Google Scholar] [PubMed]
- Moens, C.B.; Selleri, L. Hox cofactors in vertebrate development. Dev. Biol. 2006, 291, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.; Riley, T.; Liu, P.; Abe, N.; Gomez-Alcala, P.; Dror, I.; Zhou, T.; Rohs, R.; Honig, B.; Bussemaker, H.J.; et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 2011, 147, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Uhl, J.D.; Cook, T.A.; Gebelein, B. Comparing anterior and posterior Hox complex formation reveals guidelines for predicting cis-regulatory elements. Dev. Biol. 2010, 343, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Habib, F.; Yelagandula, R.; Shashidhara, L.S. Genome-level identification of targets of Hox protein Ultrabitholax in Drosophila: Novel mechanisms for target selection. Sci. Rep. 2011. [Google Scholar] [CrossRef] [PubMed]
- Hersh, B.M.; Nelson, C.E.; Stoll, S.J.; Norton, J.E.; Albert, T.J.; Caroll, S.B. The UBX-regulated network in the haltere imaginal disc of D. melanogaster. Dev. Biol. 2007, 302, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Hueber, S.D.; Bezdan, D.; Henz, S.R.; Blank, M.; Wu, H.; Lohmann, I. Comparative analysis of Hox downstream genes in Drosophila. Development 2007, 134, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.; Ma, L.; Negre, N.; White, K.P.; Mann, R.S. Genome-wide tissue-specific occupancy of the Hox protein Ultrabithorax and Hox cofactor Homothorax in Drosophila. PLoS ONE 2011. [Google Scholar] [CrossRef] [PubMed]
- Galant, R.; Caroll, S.B. Evolution of a transcriptional repression domain in an insect Hox protein. Nature 2002, 415, 910–913. [Google Scholar] [CrossRef] [PubMed]
- Galant, R.; Walsh, C.M.; Caroll, S.B. Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites. Development 2002, 129, 3115–3126. [Google Scholar] [PubMed]
- Krasnow, M.A.; Saffman, E.E.; Kornfeld, K.; Hogness, D.S. Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosohila cells. Cell 1989, 57, 1031–1043. [Google Scholar] [CrossRef]
- Li, X.; Murre, C.; McGinnis, W. Activity regulation of a Hox protein and a role for the homeodomain in inhibiting transcriptional activation. EMBO J. 1999, 18, 198–211. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Merabet, S.; Kambris, Z.; Capovilla, M.; Berenger, H.; Pradel, J.; Graba, Y. The hexapeptide and linker regions of the AbdA Hox protein regulate its activating and repressive functions. Dev. Cell 2003, 4, 761–768. [Google Scholar] [CrossRef]
- Papadopoulos, D.K.; Resendez-Perez, D.; Cardenas-Chavez, D.L.; Villanueva-Segura, K.; Canales-del-Castillo, R.; Felix, D.A.; Funfschilling, R.; Ghering, W.J. Functional synthetic Antennapedia genes and the dual roles of YPWM motif and linker size in transcriptional activation and repression. Proc. Natl. Acad. Sci. USA 2011, 108, 11959–11964. [Google Scholar] [CrossRef] [PubMed]
- Prince, F.; Katuyama, T.; Oshima, Y.; Plaza, S.; Resendez-Perez, D.; Berry, M.; Kurata, S.; Ghering, W.J. The YPWM motif links Antennapedia to the basal transcriptional machinery. Development 2008, 135, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
- Ronshaugen, M.; McGinnis, N.; McGinnis, W. Hox protein mutation and macroevolution of the insect body plan. Nature 2002, 415, 914–917. [Google Scholar] [CrossRef] [PubMed]
- Saadaoi, M.; Merabet, S.; Litim-Mecheri, I.; Arbeille, E.; Sambrani, N.; Damen, W.; Brena, C.; Pradel, J.; Graba, Y. Selection of distinct Hox-Extradenticle interaction modes fine-tunes Hox protein activity. Proc. Natl. Acad. Sci. USA 2011, 108, 2276–2281. [Google Scholar] [CrossRef] [PubMed]
- Winslow, G.M.; Hayashi, S.; Krasnow, M.; Hogness, D.S.; Scott, M.P. Transcriptional activation by the Antennapedia and fushi tarazu proteins in cultured Drosophila cells. Cell 1989, 57, 1017–1030. [Google Scholar] [CrossRef]
- Kimoto, M.; Kitagawa, T.; Kobayashi, I.; Nakata, T.; Kuroiwa, A.; Takiya, S. Inhibition of the binding of MSG-intermolt-specific complex, MIC, to the sericin-1 gene promoter and sericin-1 gene expression by POU-M1/SGF3. Dev. Genes Evol. 2012, 222, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Tsubota, T.; Uchino, K.; Sezutsu, H.; Takiya, S. Hox transcription factor Antp regulates sericin-1 gene expression in the terminal differentiated silk gland of Bombyx mori. Dev. Biol. 2014, 386, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Tsubota, T.; Uchino, K.; Sezutsu, H.; Takiya, S. LIM-homeodomain transcription factor Awh is a key component activating all three fibroin genes, fibH, fibL and fhx, in the silk gland of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2015, 56, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Ohno, K.; Sawada, J.; Takiya, S.; Kimoto, M.; Matsumoto, A.; Tsubota, T.; Uchino, K.; Hui, C.-C.; Sezutsu, H.; Handa, H.; et al. Silk gland factor-2, involved in fibroin gene transcription, consists of LIM homeodomain, LIM-interactiong, and single-stranded DNA-binding proteins. J. Biol. Chem. 2013, 288, 31581–31591. [Google Scholar] [CrossRef] [PubMed]
- Takiya, S.; Inoue, H.; Kimoto, M. Novel enhancer and promoter elements indispensable for the tissue-specific expression of the sericin-1 gene of the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2011, 41, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Tsubota, T.; Tomita, S.; Uchino, K.; Kimoto, M.; Takiya, S.; Kajiwara, H.; Yamazaki, T.; Sezutsu, H. A Hox gene, Antennapedia, regulates expression of multiple major silk protein genes in the silkworm Bombyx mori. J. Biol. Chem. 2016, 291, 7087–7096. [Google Scholar] [CrossRef] [PubMed]
- Kokubo, H.; Takiya, S.; Mach, V.; Suzuki, Y. Spatial and temporal expression pattern of Bombyx fork head/SGF-1 gene in embryogenesis. Dev. Genes Evol. 1996, 206, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Kokubo, H.; Ueno, K.; Amanai, K.; Suzuki, Y. Involvement of the Bombyx Scr gene in development of the embryonic silk gland. Dev. Biol. 1997, 186, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Kokubo, H.; Xu, P.-X.; Xu, X.; Matsunami, K.; Suzuki, Y. Spatial and temporal expression pattern of POU-M1/SGF-3 in Bombyx mori embryogenesis. Dev. Genes Evol. 1997, 206, 494–502. [Google Scholar] [CrossRef]
- Matsunami, K.; Kokubo, H.; Ohno, K.; Suzuki, Y. Expression pattern analysis of SGF-3/POU-M1 in relation to sericin-1 gene expression in the silk gland. Dev. Growth Differ. 1998, 40, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S.; Suzuki, Y.; Hara, W.; Takiya, S.; Suzuki, T. Fibroin gene transcription in the embryonic stages of the silkworm, Bombyx mori. Dev. Growth Differ. 1988, 30, 293–299. [Google Scholar] [CrossRef]
- Couble, P.; Michaille, J.-J.; Garel, A.; Couble, M.-L.; Prudohmme, J.-C. Developmental switches of sericin mRNA splicing in individual cells of Bombyx mori silk gland. Dev. Biol. 1987, 124, 431–440. [Google Scholar] [CrossRef]
- Ishikawa, E.; Suzuki, Y. Tissue-and stage-specific expression of sericin genes in the middle silk gland of Bombyx mori. Dev. Growth Differ. 1985, 27, 73–82. [Google Scholar] [CrossRef]
- Michaille, J.-J.; Garel, A.; Prudhomme, J.-C. Cloning and characterization of the highly polymorphic Ser2 gene of Bombyx mori. Gene 1990, 86, 177–184. [Google Scholar] [CrossRef]
- Obara, T.; Suzuki, Y. Temporal and spatial control of silk gene transcription analyzed by nuclear run-on assay. Dev. Biol. 1988, 127, 384–391. [Google Scholar] [CrossRef]
- Suzuki, Y.; Takiya, S.; Suzuki, T.; Hui, C.-C.; Matsuno, K.; Fukuta, M.; Nagata, T.; Ueno, K. Developmental regulation of silk gene expression in Bombyx mori. In Molecular Insect Science; Hagedorn, H.H., Hildebrand, J.G., Kidwell, M.G., Law, J.H., Eds.; Plenum Press: New York, NY, USA, 1990. [Google Scholar]
- Takasu, Y.; Yamada, H.; Tamura, T.; Sezutsu, H.; Mita, K.; Tsubouchi, K. Identification and characterization of a novel sericin gene expressed in the anterior middle silk gland of the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2007, 37, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Takasu, Y.; Hata, T.; Uchino, K.; Zhan, Q. Identification of Ser2 proteins as major sericin components in the non-cocoon silk of Bombyx mori. Insect Biochem. Mol. Biol. 2010, 40, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Oyama, F.; Ueda, H.; Mizuno, S.; Shimura, K. Molecular cloning of the fibroin light chain complementary DNA and its use in the study of the expression of the light chain gene in the posterior silk gland of Bombyx mori. Experientia 1985, 41, 1167–1171. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, H.; Suzuki, Y. Repeated turn-off and turn-on of fibroin gene transcription during silk gland development of Bombyx mori. Dev. Biol. 1980, 78, 394–406. [Google Scholar] [CrossRef]
- Suzuki, Y.; Giza, P.E. Accentuated expression of silk fibroin genes in vivo and in vitro. J. Mol. Biol. 1976, 107, 183–206. [Google Scholar] [CrossRef]
- Suzuki, Y.; Suzuki, E. Quantitative measurements of fibroin messenger RNA synthesis in the posterior silk gland of normal and mutant Bombyx mori. J. Mol. Biol. 1974, 88, 393–407. [Google Scholar] [CrossRef]
- Bello, B.; Horard, B.; Couble, P. The selective expression of silk-protein encoding genes in Bombyx mori silk gland. Bull. Inst. Pasteur. 1994, 92, 81–100. [Google Scholar]
- Couble, P.; Moine, A.; Garel, A.; Prudhomme, J.-C. Developmental variations of a nonfibroin mRNA of Bombyx mori silkgland, encoding for a low-molecular-weight silk protein. Dev. Biol. 1983, 97, 398–407. [Google Scholar] [CrossRef]
- Couble, P.; Chevillard, M.; Moine, A.; Revel-Chapuis, P.; Prudhomme, J.-C. Structural organization of the P25 gene of Bombyx mori and comparative analysis of its 5′ flanking DNA with that of the fibroin gene. Nucleic Acids Res. 1985, 13, 1801–1814. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Tanaka, K.; Arisaka, F.; Kimura, S.; Ohtomo, K.; Mizuno, S. Silk fibroin of Bombyx mori is secreted, assembling a high molecular elementary unit consisting of H-chain, L-chain, and P25, with a 6:6:1 molar ratio. J. Biol. Chem. 2000, 275, 40517–40528. [Google Scholar] [CrossRef] [PubMed]
- Garel, A.; Deleage, G.; Prudhomme, J.-C. Structure and organization of the Bombyx mori sericin 1 gene and of the sericins1 deduced from the sequence of the ser 1B cDNA. Insect Biochem. Mol. Biol. 1997, 27, 469–477. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, P.; Dong, Z.; Wang, D.; Guo, P.; Guo, X.; Song, Q.; Zhang, W.; Xia, Q. Comparative proteome analysis of multi-layer cocoon of the silkworm, Bombyx mori. PLoS ONE 2015, 10, e0123403. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, S.; Gopinathan, K.P. Expression pattern of Cubitus interruptus from the mulberry silkworm Bombyx mori in late developmental stages. Dev. Genes Evol. 2003, 213, 166–177. [Google Scholar] [PubMed]
- Dhawan, S.; Gopinathan, K.P. Expression profiling of homeobox genes in silk gland development in the mulberry silkworm Bombyx mori. Dev. Genes Evol. 2003, 213, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.-C.; Matsuno, K.; Ueno, K.; Suzuki, Y. Molecular characterization and silkgland expression pattern of Bombyx engrailed and invected genes. Proc. Natl. Acad. Sci. USA 1992, 89, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Ogawa, S.; Hino, R.; Adachi, T.; Tomita, M.; Yoshizato, K. Structure and function of 5′-flanking regions of a novel transcription enhancing element with a homeodomain protein-binding motif. Insect Biochem. Mol. Biol. 2007, 37, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.-C.; Matsuno, K.; Suzuki, Y. Fibroin gene promoter contains a cluster of homeodomain binding sites that interact with three silk gland factors. J. Mol. Biol. 1990, 213, 651–670. [Google Scholar] [CrossRef]
- Takiya, S.; Kokubo, H.; Suzuki, Y. Transcriptional regulatory elements upstream and intron of the fibroin gene bind three specific factors POU-M1, Bm Fkh and FMBP-1. Biochem. J. 1997, 321, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Takiya, S.; Ishikawa, T.; Ohtsuka, K.; Nishita, Y.; Suzuki, Y. Fibroin-modulator-binding protein-1 (FMBP-1) contains a novel DNA-binding domain, repeats of the score and three amino acid peptode (STP), conserved from Caenorhabditis elegans to humans. Nucleic Acids Res. 2005, 33, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Horard, B.; Julien, E.; Nony, P.; Garel, A.; Couble, P. Differential binding of the Bombyx silk gland-specific factor SGFB to its target DNA sequence drives posterior-cell-restricted expression. Mol. Cell. Biol. 1997, 17, 1572–1579. [Google Scholar] [CrossRef] [PubMed]
- Durand, B.; Drevet, J.; Couble, P. P25 gene regulation in Bombyx mori silk gland: Two promoter-binding factors have distinct tissue and developmental specificities. Mol. Cell. Biol. 1992, 12, 5768–5777. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Julien, E.; Bordeaux, M.-C.; Garel, A.; Couble, P. Fork head alternative binding drives stage-specific gene expression in the silk gland of Bombyx mori. Insect Biochem. Mol. Biol. 2002, 32, 377–387. [Google Scholar] [CrossRef]
- Mack, V.; Takiya, S.; Ohno, K.; Handa, H.; Imai, T.; Suzuki, Y. Silk gland factor-1 involved in the regulation of Bombyx sericin-1 gene contain fork head motif. J. Biol. Chem. 1995, 270, 9340–9346. [Google Scholar]
- Matsuno, K.; Hui, C.-C.; Takiya, S.; Suzuki, T.; Ueno, K.; Suzuki, Y. Transcriptional signals and protein binding sites for sericin gene transcription in vitro. J. Biol. Chem. 1989, 264, 18707–18713. [Google Scholar] [PubMed]
- Matsuno, K.; Takiya, S.; Hui, C.-C.; Fukuta, M.; Ueno, K.; Suzuki, Y. Transcriptional stimulation via SC site of Bombyx sericin-1 gene through an interaction with a DNA binding protein SGF-3. Nucleic Acids Res. 1990, 18, 1853–1858. [Google Scholar] [CrossRef] [PubMed]
- Takiya, S.; Gazi, M.; Mach, V. The DNA binding of insect Fork head factor is strongly influenced by the negative cooperation of neighbouring bases. Insect Biochem. Mol. Biol. 2003, 33, 1145–1154. [Google Scholar] [CrossRef]
- Foos, N.; Maurel-Zaffran, C.; Mate, M.J.; Vincentelli, R.; Hainaut, M.; Berenger, H.; Pradel, J.; Saurin, A.J.; Ortiz-Lombardia, M.; Graba, Y. A flexible extension of the Drosophila Ultrabithorax homeodomain defines a novel Hox/PBC interaction mode. Structure 2015, 23, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Hudry, B.; Remacle, S.; Delfini, M.-C.; Rezsohazy, R.; Graba, Y.; Merabet, S. Hox proteins display a common and ancestral ability to diversify their interaction mode with PBC class cofactors. PLoS Biol. 2012, e1001351. [Google Scholar] [CrossRef] [PubMed]
- Hirose, S.; Suzuki, Y. In vitro transcription of eukaryotic genes is affected differently by the degree of DNA supercoiling. Proc. Natl. Acad. Sci. USA 1988, 85, 718–722. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Tsuda, M.; Takiya, S.; Hirose, S.; Suzuki, E.; Kameda, M.; Ninaki, O. Tissue-specific transcription enhancement of the fibroin gene characterized by cell-free systems. Proc. Natl. Acad. Sci. USA 1986, 83, 9522–9526. [Google Scholar] [CrossRef] [PubMed]
- Takiya, S.; Hui, C.-C.; Suzuki, Y. A contribution of the core-promoter and its surrounding regions to the preferential transcription of the fibroin gene in posterior silk gland extracts. EMBO J. 1990, 9, 489–496. [Google Scholar] [PubMed]
- Tsuda, M.; Suzuki, Y. Faithful transcription initiation of fibroin gene in a homologous cell-free system reveals an enhancing effect of 5′ flanking sequence far upstream. Cell 1981, 27, 175–182. [Google Scholar] [CrossRef]
- Tsuda, M.; Suzuki, Y. Transcription modulation in vitro of the fibroin gene exerted by a 200-base-pair region upstream from the “TATA” box. Proc. Natl. Acad. Sci. USA 1983, 80, 7442–7446. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, M.; Hirose, S.; Suzuki, Y. Participation of the upstream region of the fibroin gene in the formation of transcription complex in vitro. Mol. Cell. Biol. 1986, 6, 3928–3933. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, Y.; Suzuki, Y. The DNA sequence of Bombyx mori fibroin gene including the 5′ flanking, mRNA coding, entire intervening and fibroin protein coding regions. Cell 1979, 18, 591–600. [Google Scholar] [CrossRef]
- Tsujimoto, Y.; Suzuki, Y. Natural fibroin genes purified without using cloning procedures from fibroin-producing and nonproducing tissues reveal indistinguishable structure and function. Proc. Natl. Acad. Sci. USA 1984, 81, 1644–1648. [Google Scholar] [CrossRef] [PubMed]
- Fukuta, M.; Matsuno, K.; Hui, C.-C.; Nagata, T.; Takiya, S.; Xu, P.-X.; Ueno, K.; Suzuki, Y. Molecular cloning of a POU domain-containing factor involved in the regulation of the Bombyx sericin-1 gene. J. Biol. Chem. 1993, 268, 19471–19475. [Google Scholar] [PubMed]
- Bello, B.; Couble, P. Specific expression of a silk-encoding gene of Bombyx in the anterior salivary gland of Drosophila. Nature 1990, 346, 480–482. [Google Scholar] [CrossRef] [PubMed]
- Curtiss, J.; Heilig, J.S. Establishment of Drosophila imaginal precursor cells controlled by Arrowhead gene. Development 1995, 121, 3819–3828. [Google Scholar] [PubMed]
- Nony, P.; Prudhomme, J.-C.; Couble, P. Regulation of the P25 gene transcription in the silk gland of Bombyx. Biol. Cell. 1995, 84, 43–52. [Google Scholar] [CrossRef]
- Zhao, X.-M.; Liu, C.; Li, Q.-Y.; Hu, W.-B.; Zhou, M.-T.; Zhang, Y.-X.; Peng, Z.-C.; Zhao, P.; Xia, Q.-Y. Basic helix-loop-helix transcription factor Bmsage is involved in regulation of fibroin-H-chain gene via interaction with SGF1 in Bombyx mori. PLoS ONE 2014. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.-X.; Fukuta, M.; Takiya, S.; Matsuno, K.; Xu, X.; Suzuki, Y. Promoter of the POU-M1/SGF-3 gene involved in the expression of Bombyx silk genes. J. Biol. Chem. 1994, 269, 2733–2742. [Google Scholar] [PubMed]
- Tolkunova, E.N.; Fujioka, M.; Kobayashi, M.; Deka, D.; Jaynes, J.B. Two distinct types of repression domain in engrailed: one interacts with the groucho corepressor and is preferentially active on integrated target genes. Mol. Cell. Biol. 1998, 18, 2804–2814. [Google Scholar] [CrossRef] [PubMed]
- Gebelein, B.; McKay, D.J.; Mann, R.S. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature 2004, 431, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Merabet, S.; Pradel, J.; Graba, Y. Getting a molecular grasp on Hox contextual activity. Trends Genet. 2005, 21, 477–480. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.-C.; Suzuki, Y.; Kikuchi, Y.; Mizuno, S. Homeodomain binding sites in the 5′ flanking region of the Bombyx mori silk fibroin light-chain gene. J. Mol. Biol. 1990, 213, 395–398. [Google Scholar] [CrossRef]
- Hui, C.-C.; Suzuki, Y. Homeodomain binding sites in the promoter region of silk protein genes. Dev. Growth Differ. 1990, 32, 263–273. [Google Scholar] [CrossRef]
- Oh, S.-K.; Scott, M.P.; Sarnow, P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes Dev. 1992, 6, 1643–1653. [Google Scholar] [CrossRef] [PubMed]
- Kondrashov, N.; Pusic, A.; Stumpf, C.R.; Shimizu, K.; Hsieh, A.C.; Xue, S.; Ishijima, J.; Shiroishi, T.; Barna, M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 2011, 145, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Tian, S.; Fujii, K.; Kladwang, W.; Das, R.; Barna, M. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 2015, 517, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Kmita, M.; Duboule, D. Organizing axes in time and space; 25 years of colinear tinkering. Science 2003, 301, 331–333. [Google Scholar] [CrossRef] [PubMed]
- Noordermeer, D.; Leleu, M.; Splinter, E.; Rougemont, J.; de Laat, W.; Duboule, D. The dynamic architecture of Hox gene clusters. Science 2011, 334, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Zakany, J.; Kmita, M.; Duboule, D. A dual role for Hox genes in limb anterior-posterior asymmetry. Science 2004, 304, 1669–1672. [Google Scholar] [CrossRef] [PubMed]
- Horard, B.; Mange, A.; Pelisser, B.; Couble, P. Bombyx gene promoter analysis in transplanted silk gland transformed by particle delivery system. Insect Mol. Biol. 1994, 3, 261–265. [Google Scholar]
- Wang, H.-B.; Nita, M.; Iwanaga, M.; Kawasaki, H. βFTZ-F1 and Broad-complex positively regulate the transcription of the wing cuticle protein gene, BMWCP5, in wing discs of Bombyx mori. Insect Biochem. Mol. Biol. 2009, 39, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Kuwabara, N.; Uchino, K.; Kobayashi, I.; Kanda, T. An improved DNA injection method for silkworm eggs drastically increases the efficiency of producing transgenic silkworms. J. Insect Biotech. Sericol. 2007, 76, 155–159. [Google Scholar]
- Imamura, M.; Nakai, J.; Inoue, S.; Quan, G.X.; Kanda, T.; Tamura, T. Targeted gene expression using the GAL4/UAS system in the silkworm Bombyx mori. Genetics 2003, 165, 1329–1340. [Google Scholar] [PubMed]
- Tamura, T.; Thibert, C.; Royer, C.; Kanda, T.; Eppen, A.; Kamba, M.; Komoto, N.; Thomas, J.L.; Mauchamp, B.; Chavancy, G.; et al. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat. Biotechnol. 2000, 18, 81–84. [Google Scholar] [PubMed]
- Takasu, Y.; Sajwan, S.; Damion, T.; Osanai-Futanashi, M.; Uchino, K.; Sezutsu, H.; Tamura, T.; Zurovec, M. Efficient TALEN construction for Bombyx mori gene targeting. PLoS ONE 2013, 8, e73458. [Google Scholar] [CrossRef]
- Nakade, S.; Tsubota, T.; Sakano, Y.; Kume, S.; Sakamoto, N.; Obara, M.; Daimon, T.; Sezutsu, H.; Yamamoto, T.; Sakuma, T.; et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014, 5, 5560. [Google Scholar] [CrossRef] [PubMed]
- Toyama, K. Mendel’s low of heredity as applied to the silkworm crosses. Biol. Zbl. 1906, 26, 321–334. [Google Scholar]
| Silk Gene | ASG (1) | MSG | PSG | Reference | |||
|---|---|---|---|---|---|---|---|
| Anterior (1) | Middle (1) | Posterior (1) | Anterior (1),(2) | Posterior (1),(2) | |||
| Silk Fiber Protein | |||||||
| fibH | − | − | − | − | +++ (3) | +++ | [48] |
| fibL | − | − | − | − | +++ | +++ | [48] |
| fhx | − | − | − | − | +++ | +++ | [48] |
| Glue Protein | |||||||
| ser1 | − | − | +++ | +++ | − | − | [46,47,51] |
| ser2 | − | +++ | ++ | − | − | − | [51] |
| ser3 | − | +++ | +++ | − | − | − | [51] |
| fhxh4 | − | − | ++ | +++ | − | − | [51] |
| fhxh5 | − | − | ++ | +++ | − | − | [51] |
| Hox Gene | |||||||
| lab | ++ | +/− | + | + | + | + | [47] |
| Scr | ++ | +/− | − | − | − | − | [47] |
| Antp | − | ++ (4) | ++ | ++ | − | − | [47] |
| Ubx | − | − | − | +/− | +/− | +/− | [47] |
| abd-A | − | + | + | + | + | + | [47] |
| Abd-B | + | +/− | − | − | − | + | [47] |
| Homeobox Gene (5) | |||||||
| Awh | − | − | − | − | ++ | ++ | [48] |
| En | + | +/− | + | + | − | − | [47] |
| in | ++ | − | + | ++ | − | − | [47] |
| Pou-m1 | ++ | ++ | + | − | − | − | [46,47] |
| cad | + | +/− | +/− | +/− | − | − | [47] |
| exd | ++ | ++ | ++ | ++ | ++ | ++ | [47] |
| hth | + | +/− | ++ | ++ | + | + | [47] |
| Gene | Position | Strand | Factor | Sequence | Reference |
|---|---|---|---|---|---|
| Silk Fiber Protein | |||||
| fibH | −1620 | − | SGF-2 | CTTG CAATTA AGCACTTATTC | [77] |
| −200 | + | SGF-2 | GAT CAATTA AAT CATAATTA ATC (3) | [48,49,78] | |
| −110 | − | SGF-2 | GATA CAATTA CATAG AAATTA ATC (3) | [48,49,78] | |
| −270 | − | Fkh (1) | GTAA TATTTAAAGA ACTTA | ||
| −130 | − | Fkh, FMBP-1 | ATCT TTTTATTTAACAT AACAA (4) | [79,80] | |
| −70 | + | Fkh | TAGA TGTTTATTCT ATCG | [78] | |
| −60 | − | Fkh (2) | GACG TATTTACTTT CGAT | ||
| −130 | + | POU-M1 | TGTT ATGTTAAA TAAA | [78] | |
| −110 | + | POU-M1 | TTCT ATGTAATT GTATC | [78] | |
| fibL | −230 | + | SGF-2 (1) | GGAT CAATTA GATCGCTTTG | |
| −50 | − | SGF-2 | AAGA CAATTA AAA TAAATA TC (3) | [48] | |
| −50 | + | Fkh (1), FMBP-1 (1) | TTGA TATTTATTTT AATTG (4) | ||
| −30 | − | Fkh (1), FMBP-1 (1) | CCAC TATTTATATA TAAAA (5) | ||
| fhx | −30 | + | SGF-2 | GGAA CAAT ACTTG TATAATTA ATGTTG (6) | [48,81] |
| +100 | + | SGF-2 (1) | GGT CAATTA TAACTAC | ||
| −70 | + | Fkh, FMBP-1 (1) | ACGC TATTTATTTA ACGT (4) | [81,82,83] | |
| Glue Protein | |||||
| ser1 | −1350 | + | MIC | TAATGC AATTAATATC GTATC | [50] |
| −310 | + | MIC | AATTCC AATTAATTAT AGTCG | [50] | |
| −180 | + | MIC | GAAATC AATTAATAAC ATAAA | [50] | |
| −70 | + | MIC | GCGAA AATTTATTAC TCTCT | [50] | |
| −160 | − | Fkh (1) | ATTT TGTTTGCCTA TTTTA | ||
| −120 | + | Fkh (1) | AGAA CGTTTGTTGA ACAA | ||
| −90 | + | Fkh | ACAT TGTTTGCACA AATGTT | [84,85,86] | |
| −200 | + | POU-M1 | AGCC ATGAATAA ATTAG | [85,86] | |
| −140 | − | POU-M1 | CTCT ATGTAAAT GGTTT | [85,86] | |
| ser3 | −90 | + | MIC, Fkh (2), FMBP-1 (1) | AAAT AATTAATTATTTATTTT ATTG (4) | [51] |
| −100 | − | Fkh (2) | TAAT TATTTGTTTA ATACAC | ||
| −30 | − | Fkh (1) | CGGC TATTTATACT AATTT (7) | ||
| −70 | − | POU-M1 (1) | CTTT ATGAATAA ACAG | ||
| fhxh4 | −1660 | − | MIC | AAATT GATTTATGAC AGAG | [51] |
| −180 | − | Fkh (1) | TTTT TGTTTAATTA TTAT | ||
| −160 | − | Fkh (1) | TTTT TGTTTAATTT TTTT | ||
| −60 | + | Fkh (1) | TAAA TGTTTATTTT CTTAT | ||
| −40 | − | Fkh (1) | ACTG TGTTTAAATT ATGTT | ||
| −30 | + | Fkh (1) | TGCT TATTTATATG TAAG (8) | ||
| fhxh5 | −300 | − | MIC | AATGA GATTTATAAT ATTGAT | [51] |
| −200 | + | Fkh (1) | CTTG TATTTAGATT ATTTA | ||
| −130 | + | Fkh (1) | ATATAA TATTTAATGT AAACG | ||
| −60 | − | Fkh (1) | GTGTAA TATTTGCTGG ATATTA | ||
| −30 | − | Fkh (1) | AAACT AGTTTGTATA ATTCC |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takiya, S.; Tsubota, T.; Kimoto, M. Regulation of Silk Genes by Hox and Homeodomain Proteins in the Terminal Differentiated Silk Gland of the Silkworm Bombyx mori. J. Dev. Biol. 2016, 4, 19. https://doi.org/10.3390/jdb4020019
Takiya S, Tsubota T, Kimoto M. Regulation of Silk Genes by Hox and Homeodomain Proteins in the Terminal Differentiated Silk Gland of the Silkworm Bombyx mori. Journal of Developmental Biology. 2016; 4(2):19. https://doi.org/10.3390/jdb4020019
Chicago/Turabian StyleTakiya, Shigeharu, Takuya Tsubota, and Mai Kimoto. 2016. "Regulation of Silk Genes by Hox and Homeodomain Proteins in the Terminal Differentiated Silk Gland of the Silkworm Bombyx mori" Journal of Developmental Biology 4, no. 2: 19. https://doi.org/10.3390/jdb4020019
APA StyleTakiya, S., Tsubota, T., & Kimoto, M. (2016). Regulation of Silk Genes by Hox and Homeodomain Proteins in the Terminal Differentiated Silk Gland of the Silkworm Bombyx mori. Journal of Developmental Biology, 4(2), 19. https://doi.org/10.3390/jdb4020019







