Signaling Pathways in Human Blastocyst Development: From Molecular Mechanisms to In Vitro Optimization
Abstract
1. Introduction
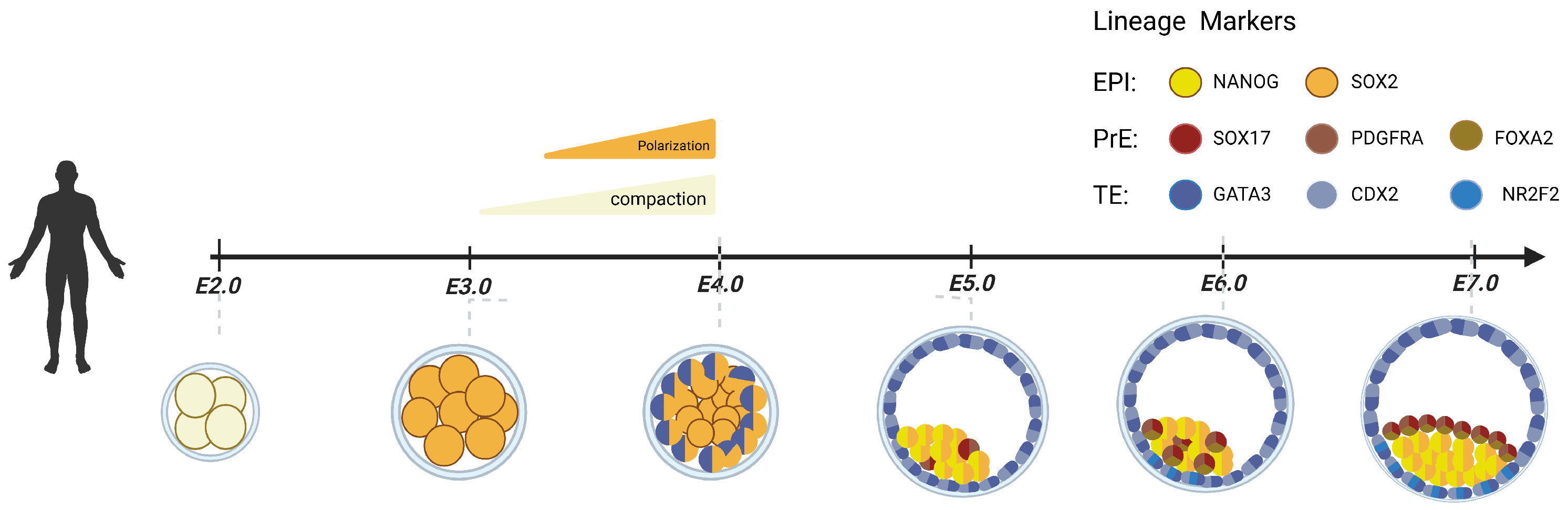
2. Preimplantation Human Embryonic Development
3. Signal Pathway in Human Preimplantation Embryo Development
3.1. Hippo Signaling Pathway: Regulating TE Differentiation
3.2. Wnt Signaling Pathway: Disruption Impairs Trophectoderm Formation
3.3. FGF Signaling Pathway: A Central Driver of Hypoblast Formation
3.4. TGF-β Signaling Pathway: Maintaining EPI Stability and Regulating Apoptosis
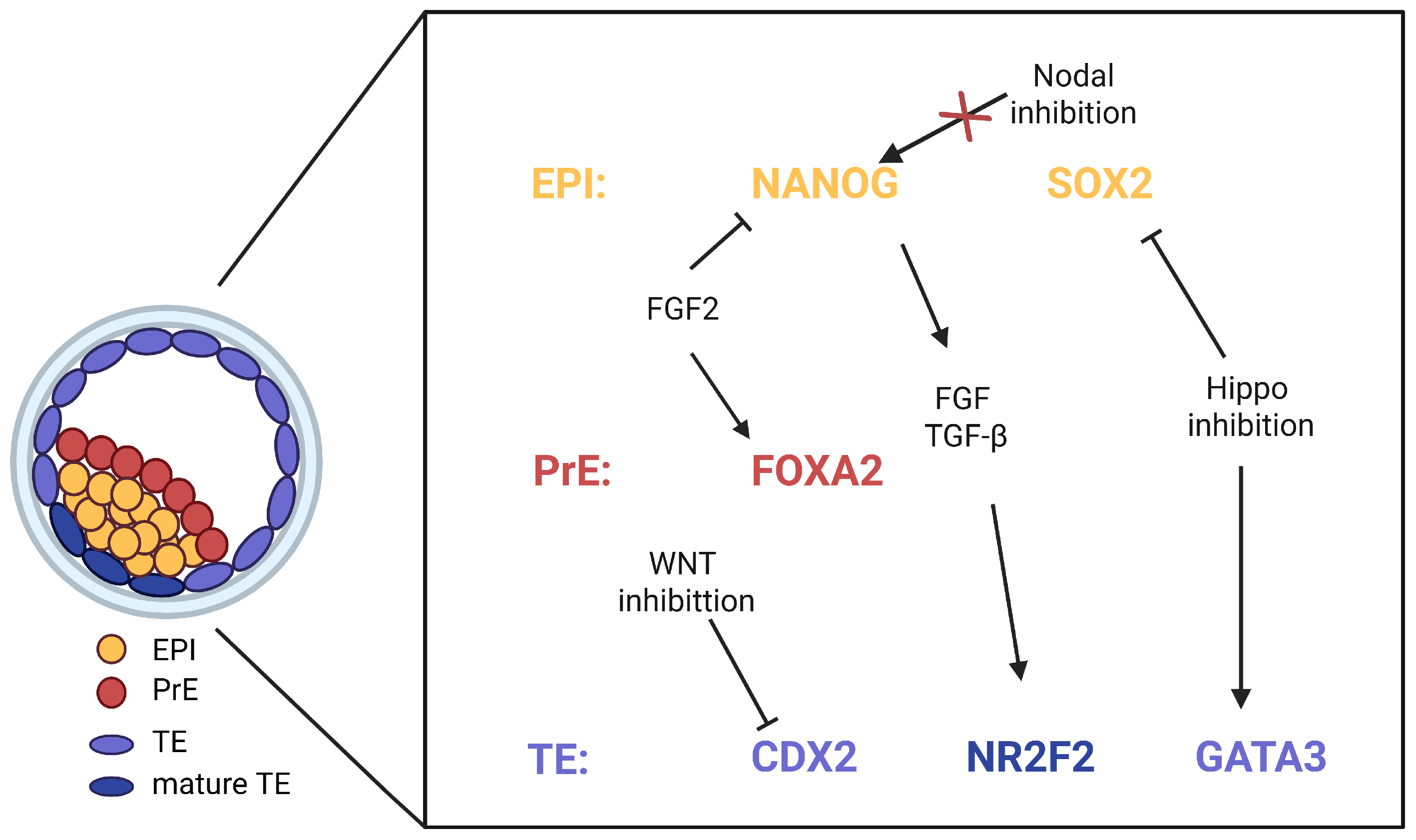
3.5. Signaling Networks in Early Human Embryo Development
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wasilewski, T.; Łukaszewicz-Zając, M.; Wasilewska, J.; Mroczko, B. Biochemistry of infertility. Clin. Chim. Acta 2020, 508, 185–190. [Google Scholar] [CrossRef]
- Inhorn, M.C.; Patrizio, P. Infertility around the globe: New thinking on gender, reproductive technologies and global movements in the 21st century. Hum. Reprod. Update 2015, 21, 411–426. [Google Scholar] [CrossRef]
- Graham, M.E.; Jelin, A.; Hoon, A.H., Jr.; Wilms Floet, A.M.; Levey, E.; Graham, E.M. Assisted reproductive technology: Short- and long-term outcomes. Dev. Med. Child Neurol. 2023, 65, 38–49. [Google Scholar] [CrossRef]
- Fishel, S. First in vitro fertilization baby-this is how it happened. Fertil. Steril. 2018, 110, 5–11. [Google Scholar] [CrossRef]
- Goisis, A.; Håberg, S.E.; Hanevik, H.I.; Magnus, M.C.; Kravdal, Ø. The demographics of assisted reproductive technology births in a Nordic country. Hum. Reprod. 2020, 35, 1441–1450. [Google Scholar] [CrossRef]
- Anagnostopoulou, C.; Maldonado Rosas, I.; Singh, N.; Gugnani, N.; Chockalingham, A.; Singh, K.; Desai, D.; Darbandi, M.; Manoharan, M.; Darbandi, S.; et al. Oocyte quality and embryo selection strategies: A review for the embryologists, by the embryologists. Panminerva Med. 2022, 64, 171–184. [Google Scholar] [CrossRef]
- Nuñez-Calonge, R.; Santamaria, N.; Rubio, T.; Manuel Moreno, J. Making and Selecting the Best Embryo in In vitro Fertilization. Arch. Med. Res. 2024, 55, 103068. [Google Scholar] [CrossRef]
- Gardner, D.K.; Lane, M.; Stevens, J.; Schlenker, T.; Schoolcraft, W.B. Blastocyst score affects implantation and pregnancy outcome: Towards a single blastocyst transfer. Fertil. Steril. 2000, 73, 1155–1158. [Google Scholar] [CrossRef] [PubMed]
- Biondic, S.; Canizo, J.; Vandal, K.; Zhao, C.; Petropoulos, S. Cross-species comparison of mouse and human preimplantation development with an emphasis on lineage specification. Reproduction 2023, 165, R103–R116. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Delgado, E.; Maître, J.L. A theoretical understanding of mammalian preimplantation development. Cells Dev. 2021, 168, 203752. [Google Scholar] [CrossRef] [PubMed]
- Coticchio, G.; Lagalla, C.; Sturmey, R.; Pennetta, F.; Borini, A. The enigmatic morula: Mechanisms of development, cell fate determination, self-correction and implications for ART. Hum. Reprod. Update 2019, 25, 422–438. [Google Scholar] [CrossRef]
- Swain, J.E. Controversies in ART: Can the IVF laboratory influence preimplantation embryo aneuploidy? Reprod. Biomed. Online 2019, 39, 599–607. [Google Scholar] [CrossRef]
- Bari, M.W.; Ishiyama, S.; Matsumoto, S.; Mochizuki, K.; Kishigami, S. From lessons on the long-term effects of the preimplantation environment on later health to a “modified ART-DOHaD” animal model. Reprod. Med. Biol. 2022, 21, e12469. [Google Scholar] [CrossRef]
- Karami, N.; Taei, A.; Eftekhari-Yazdi, P.; Hassani, F. Signaling pathway regulators in preimplantation embryos. J. Mol. Histol. 2024, 56, 57. [Google Scholar] [CrossRef]
- Boroviak, T.; Loos, R.; Lombard, P.; Okahara, J.; Behr, R.; Sasaki, E.; Nichols, J.; Smith, A.; Bertone, P. Lineage-Specific Profiling Delineates the Emergence and Progression of Naive Pluripotency in Mammalian Embryogenesis. Dev. Cell 2015, 35, 366–382. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Z.; Wu, J. Signaling pathways and preimplantation development of mammalian embryos. FEBS J. 2007, 274, 4349–4359. [Google Scholar] [CrossRef]
- Lee, S.H.; Rinaudo, P.F. Metabolic regulation of preimplantation embryo development in vivo and in vitro: Molecular mechanisms and insights. Biochem. Biophys. Res. Commun. 2024, 726, 150256. [Google Scholar] [CrossRef]
- David, L.; Bruneau, A.; Fréour, T.; Rivron, N.; Van de Velde, H. An update on human pre- and peri-implantation development: A blueprint for blastoids. Curr. Opin. Genet. Dev. 2023, 83, 102125. [Google Scholar] [CrossRef] [PubMed]
- Boroviak, T.; Stirparo, G.G.; Dietmann, S.; Hernando-Herraez, I.; Mohammed, H.; Reik, W.; Smith, A.; Sasaki, E.; Nichols, J.; Bertone, P. Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development. Development 2018, 145, dev167833. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.T.; Bonneau, A.R.; Giraldez, A.J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 2014, 30, 581–613. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Yang, M.; Hai, Z.; Fei, X.; Zhu, Y.; Pan, B.; Yang, Q.; Xie, Y.; Cheng, Y.; Xiong, Y.; et al. Maternal Kdm2a-mediated PI3K/Akt signaling and E-cadherin stimulate the morula-to-blastocyst transition revealing crucial roles in early embryonic development. Theriogenology 2023, 209, 60–75. [Google Scholar] [CrossRef]
- Zenker, J.; White, M.D.; Gasnier, M.; Alvarez, Y.D.; Lim, H.Y.G.; Bissiere, S.; Biro, M.; Plachta, N. Expanding Actin Rings Zipper the Mouse Embryo for Blastocyst Formation. Cell 2018, 173, 776–791.e717. [Google Scholar] [CrossRef]
- Chan, C.J.; Costanzo, M.; Ruiz-Herrero, T.; Mönke, G.; Petrie, R.J.; Bergert, M.; Diz-Muñoz, A.; Mahadevan, L.; Hiiragi, T. Hydraulic control of mammalian embryo size and cell fate. Nature 2019, 571, 112–116. [Google Scholar] [CrossRef]
- Ma, M.; Zhang, L.; Liu, Z.; Teng, Y.; Li, M.; Peng, X.; An, L. Effect of blastocyst development on hatching and embryo implantation. Theriogenology 2024, 214, 66–72. [Google Scholar] [CrossRef]
- Shafei, R.A.; Syrkasheva, A.G.; Romanov, A.Y.; Makarova, N.P.; Dolgushina, N.V.; Semenova, M.L. Blastocyst Hatching in Humans. Ontogenez 2017, 48, 8–20. [Google Scholar] [CrossRef]
- Chazaud, C.; Yamanaka, Y. Lineage specification in the mouse preimplantation embryo. Development 2016, 143, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, C.; Krivega, M.; Cauffman, G.; Geens, M.; Van de Velde, H. Totipotency and lineage segregation in the human embryo. Mol. Hum. Reprod. 2014, 20, 599–618. [Google Scholar] [CrossRef] [PubMed]
- Wamaitha, S.E.; Niakan, K.K. Human Pre-gastrulation Development. Curr. Top. Dev. Biol. 2018, 128, 295–338. [Google Scholar] [CrossRef]
- Gerri, C.; McCarthy, A.; Alanis-Lobato, G.; Demtschenko, A.; Bruneau, A.; Loubersac, S.; Fogarty, N.M.E.; Hampshire, D.; Elder, K.; Snell, P.; et al. Initiation of a conserved trophectoderm program in human, cow and mouse embryos. Nature 2020, 587, 443–447. [Google Scholar] [CrossRef]
- Gerri, C.; McCarthy, A.; Mei Scott, G.; Regin, M.; Stamatiadis, P.; Brumm, S.; Simon, C.S.; Lee, J.; Montesinos, C.; Hassitt, C.; et al. A conserved role of the Hippo signalling pathway in initiation of the first lineage specification event across mammals. Development 2023, 150, dev201112. [Google Scholar] [CrossRef] [PubMed]
- Krivega, M.; Essahib, W.; Van de Velde, H. WNT3 and membrane-associated β-catenin regulate trophectoderm lineage differentiation in human blastocysts. Mol. Hum. Reprod. 2015, 21, 711–722. [Google Scholar] [CrossRef]
- Roode, M.; Blair, K.; Snell, P.; Elder, K.; Marchant, S.; Smith, A.; Nichols, J. Human hypoblast formation is not dependent on FGF signalling. Dev. Biol. 2012, 361, 358–363. [Google Scholar] [CrossRef]
- Dattani, A.; Corujo-Simon, E.; Radley, A.; Heydari, T.; Taheriabkenar, Y.; Carlisle, F.; Lin, S.; Liddle, C.; Mill, J.; Zandstra, P.W.; et al. Naive pluripotent stem cell-based models capture FGF-dependent human hypoblast lineage specification. Cell Stem Cell 2024, 31, 1058–1071.e1055. [Google Scholar] [CrossRef]
- Van der Jeught, M.; Heindryckx, B.; O’Leary, T.; Duggal, G.; Ghimire, S.; Lierman, S.; Van Roy, N.; Chuva de Sousa Lopes, S.M.; Deroo, T.; Deforce, D.; et al. Treatment of human embryos with the TGFβ inhibitor SB431542 increases epiblast proliferation and permits successful human embryonic stem cell derivation. Hum. Reprod. 2014, 29, 41–48. [Google Scholar] [CrossRef]
- Brumm, A.S.; McCarthy, A.; Gerri, C.; Fallesen, T.; Woods, L.; McMahon, R.; Papathanasiou, A.; Elder, K.; Snell, P.; Christie, L.; et al. Initiation and maintenance of the pluripotent epiblast in pre-implantation human development is independent of NODAL signaling. Dev. Cell 2025, 60, 174–185.e175. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, C.; Aberkane, A.; Dewandre, D.; Essahib, W.; Sermon, K.; Geens, M.; Verheyen, G.; Tournaye, H.; Van de Velde, H. BMP4 plays a role in apoptosis during human preimplantation development. Mol. Reprod. Dev. 2019, 86, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Harvey, K.F.; Pfleger, C.M.; Hariharan, I.K. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 2003, 114, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, J.; Dong, J.; Pan, D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 2003, 114, 445–456. [Google Scholar] [CrossRef]
- Juan, W.C.; Hong, W. Targeting the Hippo Signaling Pathway for Tissue Regeneration and Cancer Therapy. Genes 2016, 7, 55. [Google Scholar] [CrossRef]
- Yu, F.X.; Zhao, B.; Guan, K.L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015, 163, 811–828. [Google Scholar] [CrossRef]
- Yildirim, E.; Bora, G.; Onel, T.; Talas, N.; Yaba, A. Cell fate determination and Hippo signaling pathway in preimplantation mouse embryo. Cell Tissue Res. 2021, 386, 423–444. [Google Scholar] [CrossRef]
- Leung, C.Y.; Zernicka-Goetz, M. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat. Commun. 2013, 4, 2251. [Google Scholar] [CrossRef]
- Hirate, Y.; Hirahara, S.; Inoue, K.; Suzuki, A.; Alarcon, V.B.; Akimoto, K.; Hirai, T.; Hara, T.; Adachi, M.; Chida, K.; et al. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr. Biol. 2013, 23, 1181–1194. [Google Scholar] [CrossRef]
- Rayon, T.; Menchero, S.; Nieto, A.; Xenopoulos, P.; Crespo, M.; Cockburn, K.; Cañon, S.; Sasaki, H.; Hadjantonakis, A.K.; de la Pompa, J.L.; et al. Notch and hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev. Cell 2014, 30, 410–422. [Google Scholar] [CrossRef]
- Biggins, J.S.; Royer, C.; Watanabe, T.; Srinivas, S. Towards understanding the roles of position and geometry on cell fate decisions during preimplantation development. Semin. Cell Dev. Biol. 2015, 47–48, 74–79. [Google Scholar] [CrossRef]
- Cockburn, K.; Biechele, S.; Garner, J.; Rossant, J. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 2013, 23, 1195–1201. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, N.; Yamamoto, S.; Kiyonari, H.; Sato, H.; Sawada, A.; Ota, M.; Nakao, K.; Sasaki, H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 2008, 125, 270–283. [Google Scholar] [CrossRef]
- Ralston, A.; Cox, B.J.; Nishioka, N.; Sasaki, H.; Chea, E.; Rugg-Gunn, P.; Guo, G.; Robson, P.; Draper, J.S.; Rossant, J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 2010, 137, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Stamatiadis, P.; Cosemans, G.; Boel, A.; Menten, B.; De Sutter, P.; Stoop, D.; Chuva de Sousa Lopes, S.M.; Lluis, F.; Coucke, P.; Heindryckx, B. TEAD4 regulates trophectoderm differentiation upstream of CDX2 in a GATA3-independent manner in the human preimplantation embryo. Hum. Reprod. 2022, 37, 1760–1773. [Google Scholar] [CrossRef] [PubMed]
- Sha, Q.Q.; Zheng, W.; Wu, Y.W.; Li, S.; Guo, L.; Zhang, S.P.; Lin, G.; Ou, X.H.; Fan, H.Y. Dynamics and clinical relevance of maternal mRNA clearance during the oocyte-to-embryo transition in humans. Nat. Commun. 2020, 11, 4917. [Google Scholar] [CrossRef]
- Regin, M.; Essahib, W.; Demtschenko, A.; Dewandre, D.; David, L.; Gerri, C.; Niakan, K.K.; Verheyen, G.; Tournaye, H.; Sterckx, J.; et al. Lineage segregation in human pre-implantation embryos is specified by YAP1 and TEAD1. Hum. Reprod. 2023, 38, 1484–1498. [Google Scholar] [CrossRef]
- Hart, M.; Concordet, J.P.; Lassot, I.; Albert, I.; del los Santos, R.; Durand, H.; Perret, C.; Rubinfeld, B.; Margottin, F.; Benarous, R.; et al. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr. Biol. CB 1999, 9, 207–210. [Google Scholar] [CrossRef]
- Piao, S.; Lee, S.H.; Kim, H.; Yum, S.; Stamos, J.L.; Xu, Y.B.; Lee, S.J.; Lee, J.; Oh, S.; Han, J.K.; et al. Direct Inhibition of GSK3β by the Phosphorylated Cytoplasmic Domain of LRP6 in Wnt/β-Catenin Signaling. PLoS ONE 2008, 3, e4046. [Google Scholar] [CrossRef]
- Behrens, J.; von Kries, J.P.; Kuhl, M.; Bruhn, L.; Wedlich, D.; Grosschedl, R.; Birchmeier, W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 1996, 382, 638–642. [Google Scholar] [CrossRef]
- Garabedian, M.V.; Good, M.C. OptoLRP6 Illuminates Wnt Signaling in Early Embryo Development. J. Mol. Biol. 2021, 433, 167053. [Google Scholar] [CrossRef]
- Huelsken, J.; Vogel, R.; Brinkmann, V.; Erdmann, B.; Birchmeier, C.; Birchmeier, W. Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 2000, 148, 567–578. [Google Scholar] [CrossRef]
- Athanasouli, P.; Balli, M.; De Jaime-Soguero, A.; Boel, A.; Papanikolaou, S.; van der Veer, B.K.; Janiszewski, A.; Vanhessche, T.; Francis, A.; El Laithy, Y.; et al. The Wnt/TCF7L1 transcriptional repressor axis drives primitive endoderm formation by antagonizing naive and formative pluripotency. Nat. Commun. 2023, 14, 1210. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, M.; Andani, M.R.; Hajian, M.; Sanei, N.; Moradi-Hajidavaloo, R.; Mahvash, N.; Jafarpour, F.; Nasr-Esfahani, M.H. Developmental competence of IVF and SCNT goat embryos is improved by inhibition of canonical WNT signaling. PLoS ONE 2023, 18, e0281331. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Enkemann, S.A.; Liang, P.; Hersmus, R.; Zanazzi, C.; Huang, J.; Wu, C.; Chen, Z.; Looijenga, L.H.; Keefe, D.L.; et al. Genome-wide gene expression profiling reveals aberrant MAPK and Wnt signaling pathways associated with early parthenogenesis. J. Mol. Cell. Biol. 2010, 2, 333–344. [Google Scholar] [CrossRef]
- Denicol, A.C.; Dobbs, K.B.; McLean, K.M.; Carambula, S.F.; Loureiro, B.; Hansen, P.J. Canonical WNT signaling regulates development of bovine embryos to the blastocyst stage. Sci. Rep. 2013, 3, 1266. [Google Scholar] [CrossRef] [PubMed]
- de Jaime-Soguero, A.; Abreu de Oliveira, W.A.; Lluis, F. The Pleiotropic Effects of the Canonical Wnt Pathway in Early Development and Pluripotency. Genes 2018, 9, 93. [Google Scholar] [CrossRef]
- Petropoulos, S.; Edsgärd, D.; Reinius, B.; Deng, Q.; Panula, S.P.; Codeluppi, S.; Plaza Reyes, A.; Linnarsson, S.; Sandberg, R.; Lanner, F. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 2016, 165, 1012–1026. [Google Scholar] [CrossRef]
- Jia, Y.; Liu, Y.; Li, Y.; Guan, X.; Xu, J.; Yan, Z.; Liu, K.; Zhang, Y.; Bai, D.; Xiang, J.; et al. Persistent Wnt signaling affects IVF embryo implantation and offspring metabolism. Sci. Bull. 2025, 70, 2297–2311. [Google Scholar] [CrossRef]
- Wang, S.; Leng, L.; Wang, Q.; Gu, Y.; Li, J.; An, Y.; Deng, Q.; Xie, P.; Cheng, C.; Chen, X.; et al. A single-cell transcriptome atlas of human euploid and aneuploid blastocysts. Nat. Genet. 2024, 56, 1468–1481. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Su, N.; Yang, J.; Tan, Q.; Huang, S.; Jin, M.; Ni, Z.; Zhang, B.; Zhang, D.; Luo, F.; et al. FGF/FGFR signaling in health and disease. Signal Transduct. Target. Ther. 2020, 5, 1852–1889. [Google Scholar] [CrossRef]
- Soszyńska, A.; Klimczewska, K.; Suwińska, A. FGF/ERK signaling pathway: How it operates in mammalian preimplantation embryos and embryo-derived stem cells. Int. J. Dev. Biol. 2019, 63, 171–186. [Google Scholar] [CrossRef]
- Frankenberg, S.; Gerbe, F.; Bessonnard, S.; Belville, C.; Pouchin, P.; Bardot, O.; Chazaud, C. Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Dev. Cell 2011, 21, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Goissis, M.D.; Bradshaw, B.; Posfai, E.; Rossant, J. Influence of FGF4 and BMP4 on FGFR2 dynamics during the segregation of epiblast and primitive endoderm cells in the pre-implantation mouse embryo. PLoS ONE 2023, 18, e0279515. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, K.; Wilczak, K.; Szczepańska, K.; Maleszewski, M.; Suwińska, A. Paracrine interactions through FGFR1 and FGFR2 receptors regulate the development of preimplantation mouse chimaeric embryo. Open Biol. 2022, 12, 220193. [Google Scholar] [CrossRef]
- Molotkov, A.; Mazot, P.; Brewer, J.R.; Cinalli, R.M.; Soriano, P. Distinct Requirements for FGFR1 and FGFR2 in Primitive Endoderm Development and Exit from Pluripotency. Dev. Cell 2017, 41, 511–526.e514. [Google Scholar] [CrossRef]
- Wamaitha, S.E.; del Valle, I.; Cho, L.T.; Wei, Y.; Fogarty, N.M.; Blakeley, P.; Sherwood, R.I.; Ji, H.; Niakan, K.K. Gata6 potently initiates reprograming of pluripotent and differentiated cells to extraembryonic endoderm stem cells. Genes Dev. 2015, 29, 1239–1255. [Google Scholar] [CrossRef]
- Krupa, M.; Mazur, E.; Szczepańska, K.; Filimonow, K.; Maleszewski, M.; Suwińska, A. Allocation of inner cells to epiblast vs primitive endoderm in the mouse embryo is biased but not determined by the round of asymmetric divisions (8→16- and 16→32-cells). Dev. Biol. 2014, 385, 136–148. [Google Scholar] [CrossRef]
- Saiz, N.; Williams, K.M.; Seshan, V.E.; Hadjantonakis, A.K. Asynchronous fate decisions by single cells collectively ensure consistent lineage composition in the mouse blastocyst. Nat. Commun. 2016, 7, 13463. [Google Scholar] [CrossRef]
- Saiz, N.; Mora-Bitria, L.; Rahman, S.; George, H.; Herder, J.P.; Garcia-Ojalvo, J.; Hadjantonakis, A.K. Growth-factor-mediated coupling between lineage size and cell fate choice underlies robustness of mammalian development. eLife 2020, 9, e56079. [Google Scholar] [CrossRef]
- Yamanaka, Y.; Lanner, F.; Rossant, J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 2010, 137, 715–724. [Google Scholar] [CrossRef]
- Nichols, J.; Silva, J.; Roode, M.; Smith, A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 2009, 136, 3215–3222. [Google Scholar] [CrossRef] [PubMed]
- Cang, Z.; Wang, Y.; Wang, Q.; Cho, K.W.Y.; Holmes, W.; Nie, Q. A multiscale model via single-cell transcriptomics reveals robust patterning mechanisms during early mammalian embryo development. PLoS Comput. Biol. 2021, 17, e1008571. [Google Scholar] [CrossRef] [PubMed]
- Bontovics, B.; Maraghechi, P.; Lázár, B.; Anand, M.; Németh, K.; Fábián, R.; Vašíček, J.; Makarevich, A.V.; Gócza, E.; Chrenek, P. The effect of dual inhibition of Ras-MEK-ERK and GSK3β pathways on development of in vitro cultured rabbit embryos. Zygote 2020, 28, 183–190. [Google Scholar] [CrossRef]
- Kuijk, E.W.; van Tol, L.T.; Van de Velde, H.; Wubbolts, R.; Welling, M.; Geijsen, N.; Roelen, B.A. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development 2012, 139, 871–882. [Google Scholar] [CrossRef]
- Rodríguez, A.; Allegrucci, C.; Alberio, R. Modulation of pluripotency in the porcine embryo and iPS cells. PLoS ONE 2012, 7, e49079. [Google Scholar] [CrossRef] [PubMed]
- Abdollah, S.; Macias-Silva, M.; Tsukazaki, T.; Hayashi, H.; Attisano, L.; Wrana, J.L. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J. Biol. Chem. 1997, 272, 27678–27685. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.Y.; Chacko, B.M.; Lam, S.S.; de Caestecker, M.P.; Correia, J.J.; Lin, K. Structural basis of Smad1 activation by receptor kinase phosphorylation. Mol. Cell 2001, 8, 1303–1312. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, M.; Liu, F.; Hata, A.; Doody, J.; Massagué, J. The TGF-beta family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes. Dev. 1997, 11, 984–995. [Google Scholar] [CrossRef] [PubMed]
- Pauklin, S.; Vallier, L. Activin/Nodal signalling in stem cells. Development 2015, 142, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Wang, Q. Molecular regulation of Nodal signaling during mesendoderm formation. Acta Biochim. Biophys. Sin. 2018, 50, 74–81. [Google Scholar] [CrossRef]
- Papanayotou, C.; Collignon, J. Activin/Nodal signalling before implantation: Setting the stage for embryo patterning. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130539. [Google Scholar] [CrossRef]
- Granier, C.; Gurchenkov, V.; Perea-Gomez, A.; Camus, A.; Ott, S.; Papanayotou, C.; Iranzo, J.; Moreau, A.; Reid, J.; Koentges, G.; et al. Nodal cis-regulatory elements reveal epiblast and primitive endoderm heterogeneity in the peri-implantation mouse embryo. Dev. Biol. 2011, 349, 350–362. [Google Scholar] [CrossRef]
- Xu, J.; Zheng, Z.; Du, X.; Shi, B.; Wang, J.; Gao, D.; Zhu, Q.; Chen, X.; Han, J. A cytokine screen using CRISPR-Cas9 knock-in reporter pig iPS cells reveals that Activin A regulates NANOG. Stem Cell. Res. Ther. 2020, 11, 67. [Google Scholar] [CrossRef]
- Bayerl, J.; Ayyash, M.; Shani, T.; Manor, Y.S.; Gafni, O.; Massarwa, R.; Kalma, Y.; Aguilera-Castrejon, A.; Zerbib, M.; Amir, H.; et al. Principles of signaling pathway modulation for enhancing human naive pluripotency induction. Cell Stem Cell 2021, 28, 1549–1565.e1512. [Google Scholar] [CrossRef]
- Osnato, A.; Brown, S.; Krueger, C.; Andrews, S.; Collier, A.J.; Nakanoh, S.; Quiroga Londoño, M.; Wesley, B.T.; Muraro, D.; Brumm, A.S.; et al. TGFβ signalling is required to maintain pluripotency of human naïve pluripotent stem cells. eLife 2021, 10, e67259. [Google Scholar] [CrossRef]
- Kiyonari, H.; Kaneko, M.; Abe, S.; Aizawa, S. Three inhibitors of FGF receptor, ERK, and GSK3 establishes germline-competent embryonic stem cells of C57BL/6N mouse strain with high efficiency and stability. Genesis 2010, 48, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Zinski, J.; Tajer, B.; Mullins, M.C. TGF-β Family Signaling in Early Vertebrate Development. Cold Spring Harb. Perspect. Biol. 2018, 10, a033274. [Google Scholar] [CrossRef] [PubMed]
- Malinauskas, T.; Moore, G.; Rudolf, A.F.; Eggington, H.; Belnoue-Davis, H.L.; El Omari, K.; Griffiths, S.C.; Woolley, R.E.; Duman, R.; Wagner, A.; et al. Molecular mechanism of BMP signal control by Twisted gastrulation. Nat. Commun. 2024, 15, 4976. [Google Scholar] [CrossRef]
- Graham, S.J.; Wicher, K.B.; Jedrusik, A.; Guo, G.; Herath, W.; Robson, P.; Zernicka-Goetz, M. BMP signalling regulates the pre-implantation development of extra-embryonic cell lineages in the mouse embryo. Nat. Commun. 2014, 5, 5667. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.M.; Ezashi, T.; Temple, J.; Owen, J.R.; Soncin, F.; Parast, M.M. The role of BMP4 signaling in trophoblast emergence from pluripotency. Cell Mol. Life Sci. 2022, 79, 447. [Google Scholar] [CrossRef]
- Reyes de Mochel, N.S.; Luong, M.; Chiang, M.; Javier, A.L.; Luu, E.; Toshihiko, F.; MacGregor, G.R.; Cinquin, O.; Cho, K.W. BMP signaling is required for cell cleavage in preimplantation-mouse embryos. Dev. Biol. 2015, 397, 45–55. [Google Scholar] [CrossRef]
- Murphy, M.E.; Leu, J.I.; George, D.L. p53 moves to mitochondria: A turn on the path to apoptosis. Cell Cycle 2004, 3, 836–839. [Google Scholar] [CrossRef]
- Spanos, S.; Becker, D.L.; Winston, R.M.; Hardy, K. Anti-apoptotic action of insulin-like growth factor-I during human preimplantation embryo development. Biol. Reprod. 2000, 63, 1413–1420. [Google Scholar] [CrossRef]
- Iyer, D.P.; Khoei, H.H.; van der Weijden, V.A.; Kagawa, H.; Pradhan, S.J.; Novatchkova, M.; McCarthy, A.; Rayon, T.; Simon, C.S.; Dunkel, I.; et al. mTOR activity paces human blastocyst stage developmental progression. Cell 2024, 187, 6566–6583.e6522. [Google Scholar] [CrossRef]
- Song, C.; Xu, F.; Ren, Z.; Zhang, Y.; Meng, Y.; Yang, Y.; Lingadahalli, S.; Cheung, E.; Li, G.; Liu, W.; et al. Elevated Exogenous Pyruvate Potentiates Mesodermal Differentiation through Metabolic Modulation and AMPK/mTOR Pathway in Human Embryonic Stem Cells. Stem Cell Rep. 2019, 13, 338–351. [Google Scholar] [CrossRef]
- Riley, J.K.; Carayannopoulos, M.O.; Wyman, A.H.; Chi, M.; Ratajczak, C.K.; Moley, K.H. The PI3K/Akt pathway is present and functional in the preimplantation mouse embryo. Dev. Biol. 2005, 284, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Gross, V.S.; Hess, M.; Cooper, G.M. Mouse embryonic stem cells and preimplantation embryos require signaling through the phosphatidylinositol 3-kinase pathway to suppress apoptosis. Mol. Reprod. Dev. 2005, 70, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Martyn, I.; Kanno, T.Y.; Ruzo, A.; Siggia, E.D.; Brivanlou, A.H. Self-organization of a human organizer by combined Wnt and Nodal signalling. Nature 2018, 558, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Ben-Haim, N.; Lu, C.; Guzman-Ayala, M.; Pescatore, L.; Mesnard, D.; Bischofberger, M.; Naef, F.; Robertson, E.J.; Constam, D.B. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev. Cell 2006, 11, 313–323. [Google Scholar] [CrossRef]
- Martyn, I.; Brivanlou, A.H.; Siggia, E.D. A wave of WNT signaling balanced by secreted inhibitors controls primitive streak formation in micropattern colonies of human embryonic stem cells. Development 2019, 146, dev172791. [Google Scholar] [CrossRef]
| Small Molecule | Target Pathway | A./I. | Embryo Number | Treatment Duration | Concentration | Blastocyst Development Rate (Control) | ICM Marker | TE Marker | PrE Maeker | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| CRT0276121 | Hippo | A. | 12 | pre-compaction—blastocyst stage | 1.5 μM | 25% (83%) | → | ↓ | - | [29] |
| TRULI | Hippo | I. | 5 | pre-compaction—blastocyst stage | 2.5 μM | 100% (100%) | ↑ | ↓ | - | [30] |
| 1-Azakenpaullone | Wnt/β-catenin | A. | 68 | D 3–D 5/6 | 20 μM | 70% (86%) | → | ↓ | - | [31] |
| Wnt3 | Wnt/β-catenin | A. | 25 | D 3–5/6 | 100 ng/mL | 80% (87%) | → | → | - | |
| Cardamonin | Wnt/β-catenin | I. | 77 | D 3–5/6 | 20 μM | 46% (75%) | → | ↓ | - | |
| PD0325901 | FGF | I. | 3 | D 3–6/7 | 1.0 μM | - | → | - | → | [32] |
| PD0325901 +PD173074 | FGF | I. | 3 | D 3–6/7 | 0.5 μM/100 nM | - | → | - | → | |
| PD173074 | FGF | I. | 11 | D 5–D6/7 | 0.5 μM | - | ↑ | - | ↓ | [33] |
| FGF2 | FGF | A. | 4 | D 5–D6/7 | 250 ng/mL | - | ↓ | - | ↑ | |
| SB431542 | TGF-β/ACTIVIN/Nodal | I. | 64 | D 3–D 6 | 10 μM | 25% (28%) | ↑ | - | → | [34] |
| Activin A | TGF-β/ACTIVIN/Nodal | A. | 44 | D 3–D 6 | 50 ng/mL | 27% (28%) | → | - | → | |
| A8301 | TGF-β/ACTIVIN/Nodal | I. | 7 | D 6–D 7 | 100 μM | - | → | - | → | [35] |
| BMP4 | BMP | A. | - | D 3–D 6 | 100 ng/mL | 17.4% (61.5) | → | → | → | [36] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, Y.; Liu, J.; Li, C.; Hu, Y.; Zhao, S. Signaling Pathways in Human Blastocyst Development: From Molecular Mechanisms to In Vitro Optimization. J. Dev. Biol. 2025, 13, 33. https://doi.org/10.3390/jdb13030033
Jiao Y, Liu J, Li C, Hu Y, Zhao S. Signaling Pathways in Human Blastocyst Development: From Molecular Mechanisms to In Vitro Optimization. Journal of Developmental Biology. 2025; 13(3):33. https://doi.org/10.3390/jdb13030033
Chicago/Turabian StyleJiao, Yan, Jiapeng Liu, Congge Li, Yuexin Hu, and Sanjun Zhao. 2025. "Signaling Pathways in Human Blastocyst Development: From Molecular Mechanisms to In Vitro Optimization" Journal of Developmental Biology 13, no. 3: 33. https://doi.org/10.3390/jdb13030033
APA StyleJiao, Y., Liu, J., Li, C., Hu, Y., & Zhao, S. (2025). Signaling Pathways in Human Blastocyst Development: From Molecular Mechanisms to In Vitro Optimization. Journal of Developmental Biology, 13(3), 33. https://doi.org/10.3390/jdb13030033







