Profilin and Non-Canonical Wnt Signaling: Coordinating Cytoskeletal Dynamics from Development to Disease
Abstract
1. Introduction
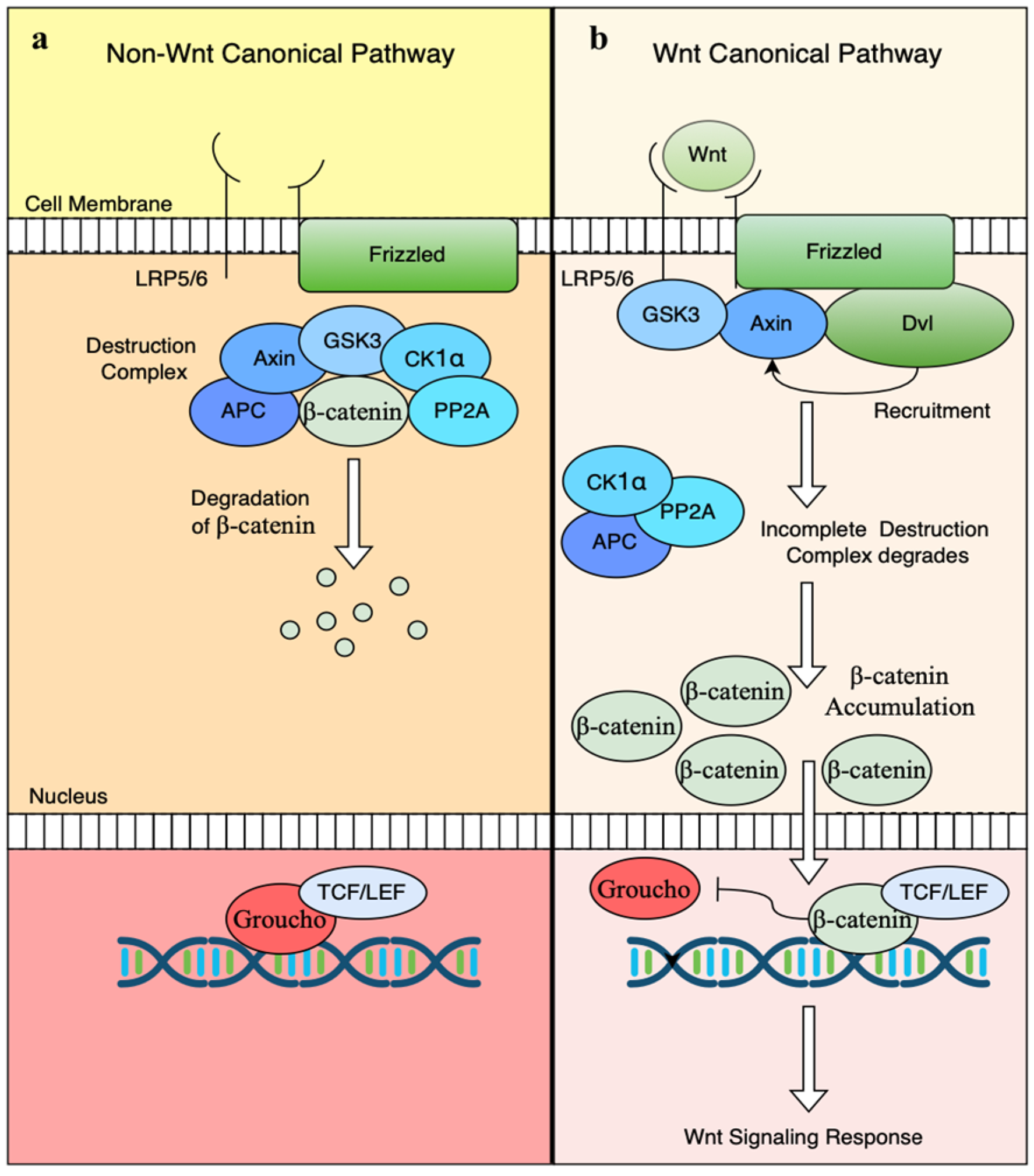
2. Canonical or Wnt/β-Catenin-Dependent Pathway
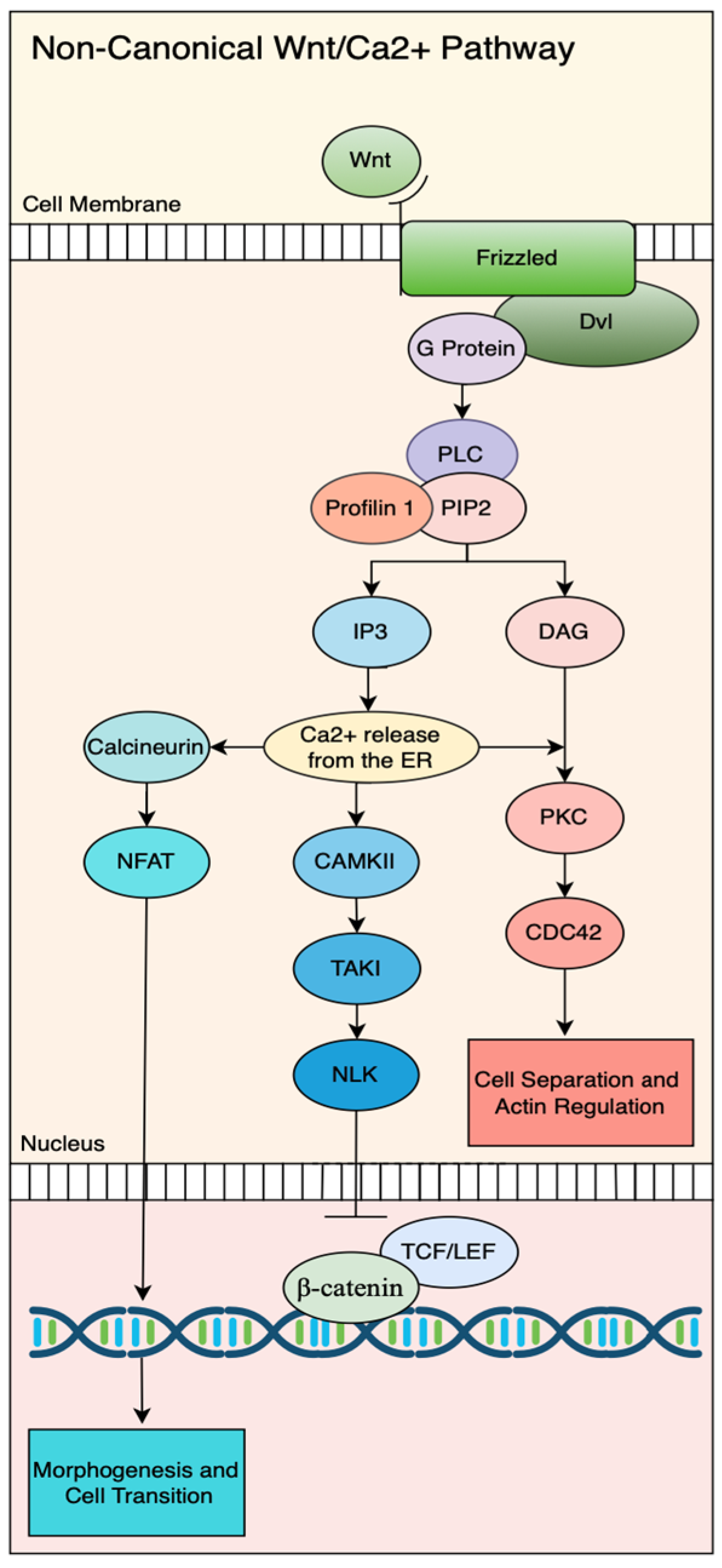
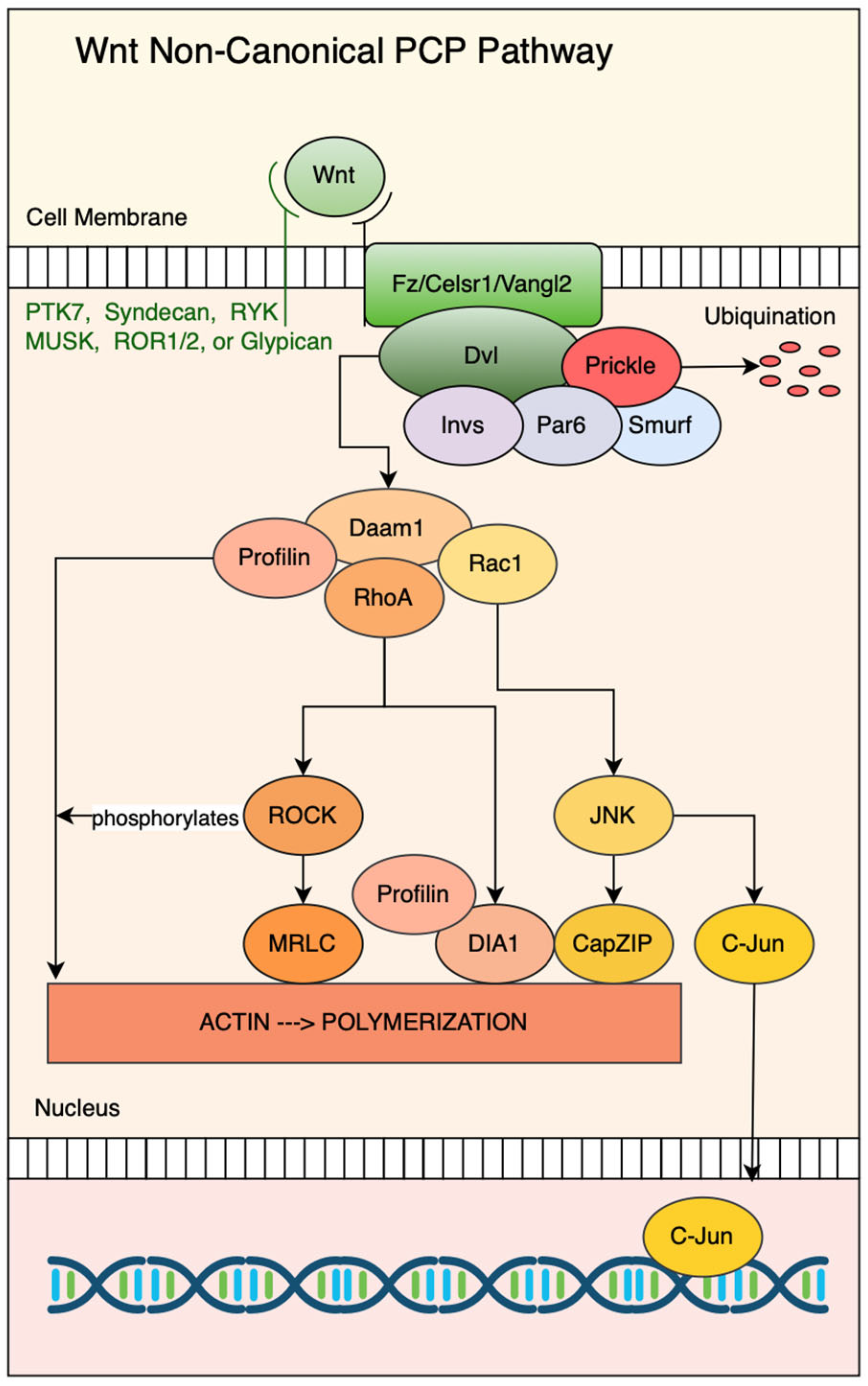
3. Non-Canonical Wnt Signaling Pathway/β-Catenin-Independent Pathway
3.1. Profilin Function in the Non-Canonical Wnt/Ca2+ Pathway
3.2. Profilin as a Key Effector of the Non-Canonical Planar Cell Polarity (PCP) Pathway
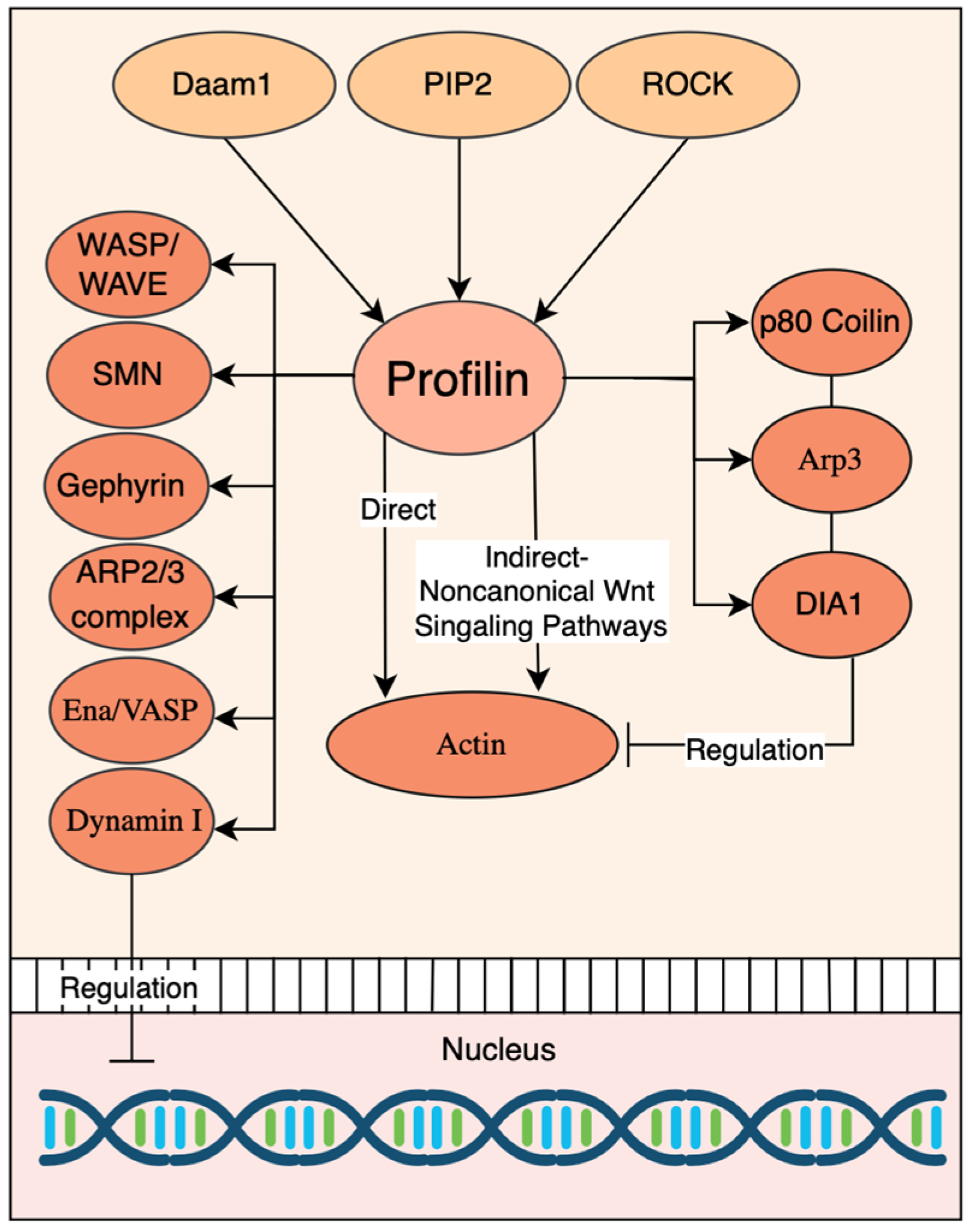
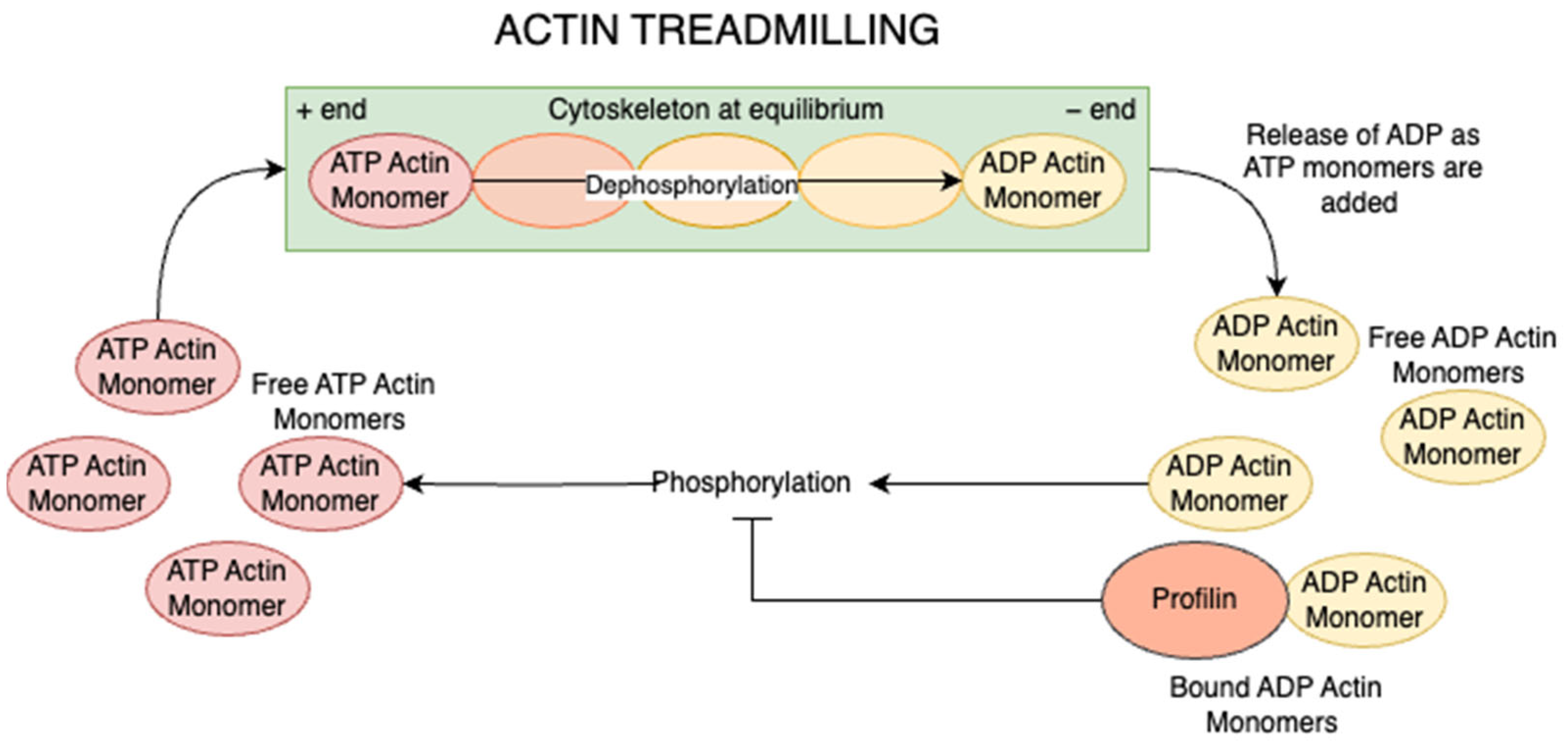
4. Role of Profilin as a Key Effector of Actin Dynamics
5. Expression and Function of Profilin Genes Across Species
5.1. Human Profilins
5.2. Chicken Profilins
5.3. Mouse Profilins
5.4. Bovine Profilins
5.5. Rat Profilins
5.6. Frog (Xenopus) Profilins
5.7. Zebrafish Profilins
6. Dysregulation of Profilin and Its Implications in Disease
6.1. Neurological Disorders
6.1.1. Amyotrophic Lateral Sclerosis (ALS)
6.1.2. Fragile X Syndrome (FXS)
6.1.3. Huntington’s Disease (HD)
6.1.4. Spinal Muscular Atrophy (SMA)
6.2. Cardiovascular and Metabolic Disease
6.3. Cancer Biology
6.4. Clinical Biomarker Potential
7. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Cadigan, K.M.; Nusse, R. Wnt signaling: A common theme in animal development. Genes Dev. 1997, 11, 3286–3305. [Google Scholar] [CrossRef]
- Sharma, R.P.; Chopra, V.L. Effect of the Wingless (wg1) mutation on wing and haltere development in Drosophila melanogaster. Dev. Biol. 1976, 48, 461–465. [Google Scholar] [CrossRef]
- Wodarz, A.; Nusse, R. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 1998, 14, 59–88. [Google Scholar] [CrossRef] [PubMed]
- Rijsewijk, F.; Schuermann, M.; Wagenaar, E.; Parren, P.; Weigel, D.; Nusse, R. The Drosophila homology of the mouse mammary oncogene int-1 is identical to the segment polarity gene wingless. Cell 1987, 50, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Logan, C.Y.; Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.R. The Wnts. Genome Biol. 2002, 3, REVIEWS3001. [Google Scholar]
- Duncan, R.N.; Panahi, S.; Piotrowski, T.; Dorsky, R.I. Identification of Wnt genes expressed in neural progenitor zones during zebrafish brain development. PLoS ONE 2015, 10, e0145810. [Google Scholar] [CrossRef]
- Ruzicka, L.; Howe, D.G.; Ramachandran, S.; Toro, S.; Van Slyke, C.E.; Bradford, Y.M.; Eagle, A.; Fashena, D.; Frazer, K.; Kalita, P.; et al. The Zebrafish Information Network: New support for non-coding genes, richer Gene Ontology annotations and the Alliance of Genome Resources. Nucleic Acids Res. 2019, 47, D867–D873. [Google Scholar] [CrossRef]
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/β-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef]
- Clevers, H.; Nusse, R. Wnt/β-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Uluçkan, Ö.; Jimenez, M.; Karbach, S.; Jeschke, A.; Graña, O.; Keller, J.; Busse, B.; Croxford, A.L.; Finzel, S.; Koenders, M.; et al. Chronic skin inflammation leads to bone loss by IL-17-mediated inhibition of Wnt signaling in osteoblasts. Sci. Transl. Med. 2016, 8, 330ra37. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, Y.; Capdevila, J.; Büscher, D.; Itoh, T.; Esteban, C.R.; Belmonte, J.C.I. WNT signals control FGF-dependent limb initiation and AER induction in the chick embryo. Cell 2001, 104, 891–900. [Google Scholar] [CrossRef]
- Shah, R.; Amador, C.; Chun, S.T.; Ghiam, S.; Saghizadeh, M.; Kramerov, A.A.; Ljubimov, A.V. Non-canonical Wnt signaling in the eye. Prog. Retin. Eye Res. 2023, 95, 101149. [Google Scholar] [CrossRef] [PubMed]
- Vijayaragavan, K.; Szabo, E.; Bossé, M.; Ramos-Mejia, V.; Moon, R.T.; Bhatia, M. Noncanonical Wnt signaling orchestrates early developmental events toward hematopoietic cell fate from human embryonic stem cells. Cell Stem Cell 2009, 4, 248–262. [Google Scholar] [CrossRef]
- Habas, R.; Kato, Y.; He, X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel formin homology protein Daam1. Cell 2001, 107, 843–854. [Google Scholar] [CrossRef]
- Sato, A.; Khadka, D.K.; Liu, W.; Bharti, R.; Runnels, L.W.; Dawid, I.B.; Habas, R. Profilin is an effector for Daam1 in non-canonical Wnt signaling and is required for vertebrate gastrulation. Development 2006, 133, 4219–4231. [Google Scholar] [CrossRef]
- Lai, S.L.; Chan, T.H.; Lin, M.J.; Huang, W.P.; Lou, S.W.; Lee, S.J. Diaphanous-Related Formin 2 and Profilin I Are Required for Gastrulation Cell Movements. PLoS ONE 2008, 3, e3439. [Google Scholar] [CrossRef]
- Khadka, D.K.; Liu, W.; Habas, R. Non-redundant roles for Profilin2 and Profilin1 during vertebrate gastrulation. Dev. Biol. 2009, 332, 396–406. [Google Scholar] [CrossRef]
- Zeng, Y.A.; Nusse, R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell 2010, 6, 568–577. [Google Scholar] [CrossRef]
- Jeong, W.-J.; Yoon, J.; Park, J.C.; Lee, S.H.; Lee, S.H.; Kaduwal, S.; Kim, H.; Yoon, J.B.; Choi, K.Y. Ras stabilization through aberrant activation of Wnt/β-catenin signaling promotes intestinal tumorigenesis. Sci. Signal 2012, 5, ra30. [Google Scholar] [CrossRef]
- Chang, C.H.; Jiang, T.X.; Lin, C.M.; Burrus, L.W.; Chuong, C.M.; Widelitz, R. Distinct Wnt members regulate the hierarchical morphogenesis of skin regions (spinal tract) and individual feathers. Mech. Dev. 2004, 121, 157–171. [Google Scholar] [CrossRef]
- Lickert, H.; Kutsch, S.; Kanzler, B.; Tamai, Y.; Taketo, M.M.; Kemler, R. Formation of multiple hearts in mice following deletion of β-catenin in the embryonic endoderm. Dev. Cell 2002, 3, 171–181. [Google Scholar] [CrossRef]
- Micsenyi, A.; Tan, X.; Sneddon, T.; Luo, J.H.; Michalopoulos, G.K.; Monga, S.P.S. β-Catenin is temporally regulated during normal liver development. Gastroenterology 2004, 126, 1134–1146. [Google Scholar] [CrossRef] [PubMed]
- Sutton, G.; Kelsh, R.N.; Scholpp, S. Review: The role of Wnt/β-catenin signalling in neural crest development in zebrafish. Front. Cell Dev. Biol. 2021, 9, 782445. [Google Scholar] [CrossRef] [PubMed]
- Perugorria, M.J.; Olaizola, P.; Labiano, I.; Esparza-Baquer, A.; Marzioni, M.; Marin, J.J.G.; Bujanda, L.; Banales, J.M. Wnt-β-catenin signalling in liver development, health and disease. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-C.; Tsai, C.-W.; Deak, F.; Rogers, J.; Penuliar, M.; Sung, Y.M.; Maher, J.N.; Fu, Y.; Li, X.; Xu, H.; et al. Deficiency in LRP6-mediated Wnt signaling contributes to synaptic abnormalities and amyloid pathology in Alzheimer’s disease. Neuron 2014, 84, 63–77. [Google Scholar] [CrossRef]
- Singh, S.; Mishra, A.; Mohanbhai, S.J.; Tiwari, V.; Chaturvedi, R.K.; Khurana, S.; Shukla, S. Axin-2 knockdown promotes mitochondrial biogenesis and dopaminergic neurogenesis by regulating Wnt/β-catenin signaling in a rat model of Parkinson’s disease. Free Radic. Biol. Med. 2018, 129, 73–87. [Google Scholar] [CrossRef]
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-Catenin or APC. Science 1997, 275, 1787–1790. [Google Scholar] [CrossRef]
- Hayes, M.J.; Thomas, D.; Emmons, A.; Giordano, T.J.; Kleer, C.G. Genetic changes of Wnt pathway genes are common events in metaplastic carcinomas of the breast. Clin. Cancer Res. 2008, 14, 4038–4045. [Google Scholar] [CrossRef]
- Pan, J.; Fang, S.; Tian, H.; Zhou, C.; Zhao, X.; Tian, H.; He, J.; Shen, W.; Meng, X.; Jin, X. lncRNA JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/β-catenin signaling. Mol. Cancer 2020, 19, 9. [Google Scholar] [CrossRef]
- Takada, R.; Satomi, Y.; Kurata, T.; Ueno, N.; Norioka, S.; Kondoh, H.; Takao, T.; Takada, S. Monounsaturated fatty acid modification of Wnt protein: Its role in Wnt secretion. Dev. Cell 2006, 11, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, J.; Singh, J.; Mahadevan, D. Identification of a frizzled-like cysteine rich domain in the extracellular region of developmental receptor tyrosine kinases. Protein Sci. 1998, 7, 1632–1635. [Google Scholar] [CrossRef] [PubMed]
- Janda, C.Y.; Waghray, D.; Levin, A.M.; Thomas, C.; Garcia, K.C. Structural basis of Wnt recognition by Frizzled. Science 2012, 337, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.K.; Samos, C.H.; Peoples, R.; Perez-Jurado, L.A.; Nusse, R.; Francke, U. A novel human homologue of the Drosophila frizzled Wnt receptor gene binds wingless protein and is in the Williams syndrome deletion at 7q11.23. Hum. Mol. Genet. 1997, 6, 465–472. [Google Scholar] [CrossRef]
- Koike, J.; Takagi, A.; Miwa, T.; Hirai, M.; Terada, M.; Katoh, M. Molecular cloning of Frizzled-10, a novel member of the Frizzled gene family. Biochem. Biophys. Res. Commun. 1999, 262, 39–43. [Google Scholar] [CrossRef]
- He, X.; Semenov, M.; Tamai, K.; Zeng, X. LDL receptor-related proteins 5 and 6 in Wnt/β-catenin signaling: Arrows point the way. Dev. 2004, 131, 1663–1677. [Google Scholar] [CrossRef]
- Wu, G.; Huang, H.; Abreu, J.G.; He, X. Inhibition of GSK3 phosphorylation of β-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PLoS ONE 2009, 4, e4926. [Google Scholar] [CrossRef]
- Beurel, E.; Grieco, S.F.; Jope, R.S. Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol. Ther. 2015, 148, 114–131. [Google Scholar] [CrossRef]
- Tsutsumi, N.; Hwang, S.; Waghray, D.; Hansen, S.; Jude, K.M.; Wang, N.; Miao, Y.; Glassman, C.R.; Caveney, N.A.; Janda, C.Y.; et al. Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination. Proc. Natl. Acad. Sci. USA 2023, 120, e2218238120. [Google Scholar] [CrossRef]
- Veeman, M.T.; Slusarski, D.C.; Kaykas, A.; Louie, S.H.; Moon, R.T. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 2003, 13, 680–685. [Google Scholar] [CrossRef]
- Cadigan, K.M.; Liu, Y.I. Wnt signaling: Complexity at the surface. J. Cell Sci. 2006, 119, 395–402. [Google Scholar] [CrossRef]
- Tu, X.; Joeng, K.S.; Nakayama, K.I.; Nakayama, K.; Rajagopal, J.; Carroll, T.J.; McMahon, A.P.; Long, F. Noncanonical Wnt signaling through G protein-linked PKCδ activation promotes bone formation. Dev. Cell 2007, 12, 113–127. [Google Scholar] [CrossRef]
- He, X.; Saint-Jeannet, J.P.; Wang, Y.; Nathans, J.; Dawid, I.; Varmus, H. A member of the Frizzled protein family mediating axis induction by Wnt-5A. Science 1997, 275, 1652–1654. [Google Scholar] [CrossRef]
- Tao, Q.; Yokota, C.; Puck, H.; Kofron, M.; Birsoy, B.; Yan, D.; Asashima, M.; Wylie, C.C.; Lin, X.; Heasman, J. Maternal Wnt11 activates the canonical Wnt signaling pathway required for axis formation in Xenopus embryos. Cell 2005, 120, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Rao, T.P.; Kühl, M. An updated overview on Wnt signaling pathways. Circ. Res. 2010, 106, 1798–1806. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Bae, Y.H.; Roy, P. Molecular insights on context-specific role of profilin-1 in cell migration. Cell Adhes. Migr. 2012, 6, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, L.; Nyström, L.; Sundkvist, I.; Markey, F.; Lindberg, U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol. 1977, 115, 465–483. [Google Scholar] [CrossRef]
- Pinto-Costa, R.; Sousa, M.M. Profilin as a dual regulator of actin and microtubule dynamics. Cytoskeleton 2020, 77, 76–83. [Google Scholar] [CrossRef]
- MacDonald, B.T.; He, X. Frizzled and LRP5/6 receptors for Wnt/β-catenin signaling. Cold Spring Harb. Perspect. Biol. 2012, 4, a007880. [Google Scholar] [CrossRef]
- Hsu, W.; Zeng, L.; Costantini, F. Identification of a domain of Axin that binds to the serine/threonine protein phosphatase 2A and a self-binding domain. J. Biol. Chem. 1999, 274, 3439–3445. [Google Scholar] [CrossRef]
- Seeling, J.M.; Miller, J.R.; Gil, R.; Moon, R.T.; White, R.; Virshup, D.M. Regulation of β-catenin signaling by the B56 subunit of protein phosphatase 2A. Science 1999, 283, 2089–2091. [Google Scholar] [CrossRef]
- Ratcliffe, M.J.; Itoh, K.; Sokol, S.Y. A positive role for the PP2A catalytic subunit in Wnt signal transduction. J. Biol. Chem. 2000, 275, 35680–35683. [Google Scholar] [CrossRef]
- Yamamoto, H.; Hinoi, T.; Michiue, T.; Fukui, A.; Usui, H.; Janssens, V.; Van Hoof, C.; Goris, J.; Asashima, M.; Kikuchi, A. Inhibition of the Wnt signaling pathway by the PR61 subunit of protein phosphatase 2A. J. Biol. Chem. 2001, 276, 26875–26882. [Google Scholar] [CrossRef]
- Behrens, J.; von Kries, J.P.; Kühl, M.; Bruhn, L.; Wedlich, D.; Grosschedl, R.; Birchmeier, W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature 1996, 382, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, M.; van de Wetering, M.; Oosterwegel, M.; Peterson-Maduro, J.; Godsave, S.; Korinek, V.; Roose, J.; Destrée, O.; Clevers, H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 1996, 86, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Lien, W.H.; Fuchs, E. Wnt some lose some: Transcriptional governance of stem cells by Wnt/β-catenin signaling. Genes Dev. 2014, 28, 1517–1532. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Reid, C.D.; Kessler, D.S. Siamois and Twin are redundant and essential in formation of the Spemann organizer. Dev. Biol. 2011, 352, 367–381. [Google Scholar] [CrossRef]
- Chong, J.M.; Uren, A.; Rubin, J.S.; Speicher, D.W. Disulfide Bond Assignments of Secreted Frizzled-Related Protein-1 Provide Insights about Frizzled Homology and Netrin Modules. J. Biol. Chem. 2002, 277, 5134–5144. [Google Scholar] [CrossRef]
- Malinauskas, T.; Aricescu, A.R.; Lu, W.; Siebold, C.; Jones, E.Y. Modular Mechanism of Wnt Signaling Inhibition by Wnt Inhibitory Factor 1. Nat. Struct. Mol. Biol. 2011, 18, 886–893. [Google Scholar] [CrossRef]
- Claudel, M.; Jouzeau, J.Y.; Cailotto, F. Secreted Frizzled-related proteins (sFRPs) in osteo-articular diseases: Much more than simple antagonists of Wnt signaling? FEBS J. 2019, 286, 4832–4851. [Google Scholar] [CrossRef]
- Semënov, M.; Tamai, K.; He, X. SOST Is a Ligand for LRP5/LRP6 and a Wnt Signaling Inhibitor. J. Biol. Chem. 2005, 280, 26770–26775. [Google Scholar] [CrossRef]
- Zhang, X.; He, X. Methods for Studying Wnt Protein Modifications/Inactivations by Extracellular Enzymes, Tiki and Notum. Methods Mol. Biol. 2016, 1481, 29–38. [Google Scholar]
- Kakugawa, S.; Langton, P.F.; Zebisch, M.; Howell, S.; Chang, H.; Liu, Y.; Feizi, T.; Bineva, G.; Snijders, A.P.; Jones, E.Y.; et al. Notum deacylates Wnts to suppress signalling activity. Nature 2015, 519, 187–192. [Google Scholar] [CrossRef]
- Mao, B.; Wu, W.; Davidson, G.; Marhold, J.; Li, M.; Mechler, B.M.; Delius, H.; Hoppe, D.; Stannek, P.; Walter, C.; et al. Kremen Proteins Are Dickkopf Receptors That Regulate Wnt/β-Catenin Signalling. Nature 2002, 417, 664–667. [Google Scholar] [CrossRef]
- Niida, A.; Hiroko, T.; Kasai, M.; Furukawa, Y.; Nakamura, Y.; Suzuki, Y.; Sugano, S.; Akiyama, T. DKK1, a Negative Regulator of Wnt Signaling, Is a Target of the β-Catenin/TCF Pathway. Oncogene 2004, 23, 8520–8526. [Google Scholar] [CrossRef] [PubMed]
- Pendás-Franco, N.; García, J.M.; Peña, C.; Valle, N.; Pálmer, H.G.; Heinäniemi, M.; Carlberg, C.; Jiménez, B.; Bonilla, F.; Muñoz, A.; et al. DICKKOPF-4 Is Induced by TCF/β-Catenin and Upregulated in Human Colon Cancer, Promotes Tumour Cell Invasion and Angiogenesis and Is Repressed by 1α,25-Dihydroxyvitamin D3. Oncogene 2008, 27, 4467–4477. [Google Scholar] [CrossRef] [PubMed]
- Carmon, K.S.; Gong, X.; Lin, Q.; Thomas, A.; Liu, Q. R-spondins Function as Ligands of the Orphan Receptors LGR4 and LGR5 to Regulate Wnt/β-Catenin Signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 11452–11457. [Google Scholar] [CrossRef] [PubMed]
- de Lau, W.; Barker, N.; Low, T.Y.; Koo, B.K.; Li, V.S.; Teunissen, H.; Kujala, P.; Haegebarth, A.; Peters, P.J.; van de Wetering, M.; et al. Lgr5 Homologues Associate with Wnt Receptors and Mediate R-Spondin Signalling. Nature 2011, 476, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.A.; Kakitani, M.; Zhao, J.; Zhao, J.; Andarmani, S.; Kakitani, M.; Oshima, T.; Binnerts, M.E.; Abo, A.; Tomizuka, K.; et al. R-Spondin Proteins: A Novel Link to β-Catenin Activation. Cell Cycle 2006, 5, 23–26. [Google Scholar] [CrossRef]
- Tocci, J.M.; Felcher, C.M.; García Solá, M.E.; Kordon, E.C. R-spondin-Mediated WNT Signaling Potentiation in Mammary and Breast Cancer Development. IUBMB Life 2020, 72, 1546–1559. [Google Scholar] [CrossRef]
- Hao, H.X.; Xie, Y.; Zhang, Y.; Charlat, O.; Oster, E.; Avello, M.; Lei, H.; Mickanin, C.; Liu, D.; Ruffner, H.; et al. ZNRF3 Promotes Wnt Receptor Turnover in an R-Spondin-Sensitive Manner. Nature 2012, 485, 195–200. [Google Scholar] [CrossRef]
- Koo, B.K.; Spit, M.; Jordens, I.; Low, T.Y.; Stange, D.E.; van de Wetering, M.; van Es, J.H.; Mohammed, S.; Heck, A.J.R.; Maurice, M.M.; et al. Tumour Suppressor RNF43 Is a Stem-Cell E3 Ligase That Induces Endocytosis of Wnt Receptors. Nature 2012, 488, 665–669. [Google Scholar] [CrossRef]
- Foulquier, S.; Daskalopoulos, E.P.; Lluri, G.; Hermans, K.C.M.; Deb, A.; Blankesteijn, W.M. WNT Signaling in Cardiac and Vascular Disease. Pharmacol. Rev. 2018, 70, 68–141. [Google Scholar] [CrossRef]
- Kim, A.; Wagle, M.; Tran, K.; Zhan, X.; Dixon, M.A.; Liu, S.; Gros, D.; Korver, W.; Yonkovich, S.; Tomasevic, N.; et al. R-Spondin Family Members Regulate the Wnt Pathway by a Common Mechanism. Mol. Biol. Cell 2008, 19, 2588. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wang, Y.; Dabdoub, A.; Smallwood, P.M.; Williams, J.; Woods, C.; Kelley, M.W.; Jiang, L.; Tasman, W.; Zhang, K.; et al. Vascular Development in the Retina and Inner Ear: Control by Norrin and Frizzled-4, a High-Affinity Ligand-Receptor Pair. Cell 2004, 116, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Messéant, J.; Dobbertin, A.; Girard, E.; Delers, P.; Manuel, M.; Mangione, F.; Schmitt, A.; Le Denmat, D.; Molgó, J.; Zytnicki, D.; et al. MuSK frizzled-like domain is critical for mammalian neuromuscular junction formation and maintenance. J. Neurosci. 2015, 35, 4926–4941. [Google Scholar] [CrossRef] [PubMed]
- Eubelen, M.; Bostaille, N.; Cabochette, P.; Gauquier, A.; Tebabi, P.; Dumitru, A.C.; Koehler, M.; Gut, P.; Alsteens, D.; Stainier, D.Y.R.; et al. A Molecular Mechanism for Wnt Ligand-Specific Signaling. Science 2018, 361, eaat1178. [Google Scholar] [CrossRef]
- Posokhova, E.; Shukla, A.; Seaman, S.; Volate, S.; Hilton, M.B.; Wu, B.; Morris, H.; Swing, D.A.; Zhou, M.; Zudaire, E.; et al. GPR124 Functions as a WNT7-Specific Coactivator of Canonical β-Catenin Signaling. Cell Rep. 2015, 10, 123–130. [Google Scholar] [CrossRef]
- Vallon, M.; Yuki, K.; Nguyen, T.D.; Chang, J.; Yuan, J.; Siepe, D.; Miao, Y.; Essler, M.; Noda, M.; Garcia, K.C.; et al. A RECK-WNT7 Receptor-Ligand Interaction Enables Isoform-Specific Regulation of Wnt Bioavailability. Proc. Natl. Acad. Sci. USA 2018, 115, E11921–E11930. [Google Scholar] [CrossRef]
- Morkel, M.; Huelsken, J.; Wakamiya, M.; Ding, J.; van de Wetering, M.; Clevers, H.; Taketo, M.M.; Behringer, R.R.; Shen, M.M.; Birchmeier, W. β-Catenin regulates Cripto- and Wnt3-dependent gene expression programs in mouse axis and mesoderm formation. Development 2003, 130, 6283–6294. [Google Scholar] [CrossRef]
- Wei, L.; Roberts, W.; Wang, L.; Yamada, M.; Zhang, S.; Zhao, Z.; Rivkees, S.A.; Schwartz, R.J.; Imanaka-Yoshida, K. Rho kinases play an obligatory role in vertebrate embryonic organogenesis. Development 2001, 128, 2953–2962. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nathans, J. Tissue/planar cell polarity in vertebrates: New insights and new questions. Development 2007, 134, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Veeman, M.T.; Axelrod, J.D.; Moon, R.T. A second canon: Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev. Cell 2003, 5, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Curtin, J.A.; Quint, E.; Tsipouri, V.; Arkell, R.M.; Cattanach, B.; Copp, A.J.; Henderson, D.J.; Spurr, N.; Stanier, P.; Fisher, E.M. Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr. Biol. 2003, 13, 1129–1133. [Google Scholar] [CrossRef]
- Wallingford, J.B.; Habas, R. The developmental biology of Dishevelled: An enigmatic protein governing cell fate and cell polarity. Development 2005, 132, 4421–4436. [Google Scholar] [CrossRef]
- Liu, W.; Komiya, Y.; Mezzacappa, C.; Khadka, D.K.; Runnels, L.W.; Habas, R. MIM regulates vertebrate neural tube closure. Development 2011, 138, 2035–2047. [Google Scholar] [CrossRef]
- Han, C.; Li, J.; Wang, C.; Ouyang, H.; Ding, X.; Liu, Y.; Chen, S.; Luo, L. Wnt5a contributes to the differentiation of human embryonic stem cells into lentoid bodies through the noncanonical Wnt/JNK signaling pathway. Investig. Ophthalmol. Vis. Sci. 2018, 59, 3449–3460. [Google Scholar] [CrossRef]
- Rothe, M.; Kanwal, N.; Dietmann, P.; Seigfried, F.A.; Hempel, A.; Schütz, D.; Reim, D.; Engels, R.; Linnemann, A.; Schmeisser, M.J.; et al. An Epha4/Sipa1l3/Wnt pathway regulates eye development and lens maturation. Development 2017, 144, 321–333. [Google Scholar] [CrossRef]
- Liang, H.; Chen, Q.; Coles, A.H.; Anderson, S.J.; Pihan, G.; Bradley, A.; Gerstein, R.; Jurecic, R.; Jones, S.N. Wnt5a inhibits B cell proliferation and functions as a tumor suppressor in hematopoietic tissue. Cancer Cell 2003, 4, 349–360. [Google Scholar] [CrossRef]
- Kokolus, K.; Nemeth, M.J. Non-canonical Wnt signaling pathways in hematopoiesis. Immunol. Res. 2009, 46, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Carbone, C.; Piro, G.; Gaianigo, N.; Ligorio, F.; Santoro, R.; Merz, V.; Simionato, F.; Zecchetto, C.; Falco, G.; Conti, G.; et al. Adipocytes sustain pancreatic cancer progression through a non-canonical WNT paracrine network inducing ROR2 nuclear shuttling. Int. J. Obes. 2018, 42, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.H.L.; Chari, R.; Lam, A.; Ng, R.T.; Yee, J.; English, J.; Evans, K.G.; Macaulay, C.; Lam, S.; Lam, W.L. Disruption of the non-canonical WNT pathway in lung squamous cell carcinoma. Clin. Med. Oncol. 2008, 2, CMO.S612. [Google Scholar] [CrossRef]
- Gujral, T.S.; Chan, M.; Peshkin, L.; Sorger, P.K.; Kirschner, M.W.; MacBeath, G. A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell 2014, 159, 844–856. [Google Scholar] [CrossRef]
- Kurayoshi, M.; Oue, N.; Yamamoto, H.; Kishida, M.; Inoue, A.; Asahara, T.; Yasui, W.; Kikuchi, A. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res. 2006, 66, 10439–10448. [Google Scholar] [CrossRef]
- Frantzi, M.; Klimou, Z.; Makridakis, M.; Zoidakis, J.; Latosinska, A.; Borràs, D.M.; Janssen, B.; Giannopoulou, I.; Lygirou, V.; Lazaris, A.C.; et al. Silencing of profilin-1 suppresses cell adhesion and tumor growth via predicted alterations in integrin and Ca2+ signaling in T24M-based bladder cancer models. Oncotarget 2016, 7, 70750–70768. [Google Scholar] [CrossRef]
- Gong, B.; Shen, W.; Xiao, W.; Meng, Y.; Meng, A.; Jia, S. The Sec14-like phosphatidylinositol transfer proteins Sec14l3/SEC14L2 act as GTPase proteins to mediate Wnt/Ca2+ signaling. Elife 2017, 6, e26362. [Google Scholar] [CrossRef]
- Kühl, M.; Sheldahl, L.C.; Malbon, C.C.; Moon, R.T. Ca2+/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J. Biol. Chem. 2000, 275, 12701–12711. [Google Scholar] [CrossRef]
- Sheldahl, L.C.; Park, M.; Malbon, C.C.; Moon, R.T. Protein kinase C is differentially stimulated by Wnt and Frizzled homologs in a G-protein-dependent manner. Curr. Biol. 1999, 9, 695–698. [Google Scholar] [CrossRef]
- Penzo-Méndez, A.; Umbhauer, M.; Djiane, A.; Boucaut, J.C.; Riou, J.F. Activation of Gbetagamma signaling downstream of Wnt-11/Xfz7 regulates Cdc42 activity during Xenopus gastrulation. Dev. Biol. 2003, 257, 302–314. [Google Scholar] [CrossRef]
- Courtemanche, N.; Pollard, T.D. Interaction of profilin with the barbed end of actin filaments. Biochemistry 2013, 52, 6456–6466. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt-Clermont, P.J.; Machesky, L.M.; Baldassare, J.J.; Pollard, T.D. The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science 1990, 247, 1575–1578. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt-Clermont, P.J.; Kim, J.W.; Machesky, L.M.; Rhee, S.G.; Pollard, T.D. Regulation of phospholipase C-γ1 by profilin and tyrosine phosphorylation. Science 1991, 251, 1231–1233. [Google Scholar] [CrossRef] [PubMed]
- Ricci, M.M.C.; Orenberg, A.; Ohayon, L.; Gau, D.; Wills, R.C.; Bae, Y.; Das, T.; Koes, D.; Hammond, G.R.V.; Roy, P. Actin-binding protein profilin1 is an important determinant of cellular phosphoinositide control. J. Biol. Chem. 2024, 300, 105583. [Google Scholar] [CrossRef]
- Bae, Y.H.; Ding, Z.; Das, T.; Wells, A.; Gertler, F.; Roy, P. Profilin1 regulates PI(3,4)P2 and lamellipodin accumulation at the leading edge thus influencing motility of MDA-MB-231 cells. Proc. Natl. Acad. Sci. USA 2010, 107, 21547–21552. [Google Scholar] [CrossRef]
- Gau, D.; Ding, Z.; Baty, C.; Roy, P. Fluorescence resonance energy transfer (FRET)-based detection of profilin-VASP interaction. Cell. Mol. Bioeng. 2011, 4, 1–8. [Google Scholar] [CrossRef]
- Zoidakis, J.; Makridakis, M.; Lianidou, E.; Latosinska, A.; Mischak, H.; Vlahou, A. Profilin 1 is a potential biomarker for bladder cancer aggressiveness. Mol. Cell. Proteom. 2012, 11, M111.009449. [Google Scholar] [CrossRef]
- Ding, Z.; Joy, M.; Bhargava, R.; Gunsaulus, M.; Lakshman, N.; Miron-Mendoza, M.; Petroll, M.; Condeelis, J.; Wells, A.; Roy, P. Profilin-1 downregulation has contrasting effects on early vs late steps of breast cancer metastasis. Oncogene 2013, 33, 2065–2074. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, M.; Xu, F.; Jiang, S. Wnt signaling in breast cancer: Biological mechanisms, challenges and opportunities. Mol. Cancer 2020, 19, 165. [Google Scholar] [CrossRef]
- Jenny, A.; Das, G.; Burnett, M.; Mlodzik, M. Diego and Prickle regulate Frizzled planar cell polarity signalling by competing for Dishevelled binding. Nat. Cell Biol. 2005, 7, 691–697. [Google Scholar] [CrossRef]
- Peradziryi, H.; Tolwinski, N.S.; Borchers, A. The many roles of PTK7: A versatile regulator of cell-cell communication. Arch. Biochem. Biophys. 2012, 524, 71–76. [Google Scholar] [CrossRef]
- Berger, H.; Wodarz, A.; Borchers, A. PTK7 Faces the Wnt in Development and Disease. Front. Cell Dev. Biol. 2017, 5, 31. [Google Scholar] [CrossRef]
- Katoh, M.; Katoh, M. Molecular genetics and targeted therapy of WNT-related human diseases (Review). Int. J. Mol. Med. 2017, 40, 587–606. [Google Scholar] [CrossRef] [PubMed]
- Bovolenta, P.; Rodriguez, J.; Esteve, P. Frizzled/RYK mediated signalling in axon guidance. Development 2006, 133, 4399–4408. [Google Scholar] [CrossRef] [PubMed]
- Pataki, C.A.; Couchman, J.R.; Brábek, J. Wnt signaling cascades and the roles of syndecan proteoglycans. J. Histochem. Cytochem. 2015, 63, 465–480. [Google Scholar] [CrossRef] [PubMed]
- de Moraes, G.F.A.; Listik, E.; Justo, G.Z.; Vicente, C.M.; Toma, L. The Glypican proteoglycans show intrinsic interactions with Wnt-3a in human prostate cancer cells that are not always associated with cascade activation. BMC Mol. Cell Biol. 2021, 22, 26. [Google Scholar] [CrossRef]
- Miller, M.A.; Steele, R.E. Lemon encodes an unusual receptor protein-tyrosine kinase expressed during gametogenesis in Hydra. Dev. Biol. 2000, 224, 286–298. [Google Scholar] [CrossRef]
- Kroiher, M.; Miller, M.A.; Steele, R.E. Deceiving appearances: Signaling by “dead” and “fractured” receptor protein-tyrosine kinases. Bioessays 2001, 23, 69–76. [Google Scholar] [CrossRef]
- Shnitsar, I.; Borchers, A. PTK7 recruits dsh to regulate neural crest migration. Development 2008, 135, 4015–4024. [Google Scholar] [CrossRef]
- Puppo, F.; Thomé, V.; Lhoumeau, A.C.; Cibois, M.; Gangar, A.; Lembo, F.; Belotti, E.; Marchetto, S.; Lécine, P.; Prébet, T.; et al. Protein tyrosine kinase 7 has a conserved role in Wnt/β-catenin canonical signalling. EMBO Rep. 2011, 12, 43–49. [Google Scholar] [CrossRef]
- Wehner, P.; Shnitsar, I.; Urlaub, H.; Borchers, A. RACK1 is a novel interaction partner of PTK7 that is required for neural tube closure. Development 2011, 138, 1321–1327. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.; Naito, M.; Daulat, A.; Angers, S.; Ciruna, B. Ptk7 promotes non-canonical Wnt/PCP-mediated morphogenesis and inhibits Wnt/β-catenin-dependent cell fate decisions during vertebrate development. Development 2013, 140, 1807–1818. [Google Scholar] [CrossRef] [PubMed]
- Petrova, I.M.; Malessy, M.J.; Verhaagen, J.; Fradkin, L.G.; Noordermeer, J.N. Wnt Signaling through the Ror Receptor in the Nervous System. Mol. Neurobiol. 2014, 49, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Green, J.L.; Inoue, T.; Sternberg, P.W. The C. elegans ROR receptor tyrosine kinase, CAM-1, non-autonomously inhibits the Wnt pathway. Development 2007, 134, 4053–4062. [Google Scholar] [CrossRef]
- Green, J.; Nusse, R.; van Amerongen, R. The role of Ryk and Ror receptor tyrosine kinases in Wnt signal transduction. Cold Spring Harb. Perspect. Biol. 2014, 6, a009175. [Google Scholar] [CrossRef]
- Oishi, I.; Suzuki, H.; Onishi, N.; Takada, R.; Kani, S.; Ohkawara, B.; Koshida, I.; Suzuki, K.; Yamada, G.; Schwabe, G.C.; et al. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells 2003, 8, 645–654. [Google Scholar] [CrossRef]
- Schambony, A.; Wedlich, D. Wnt-5A/Ror2 regulate expression of XPAPC through an alternative noncanonical signaling pathway. Dev. Cell 2007, 12, 779–792. [Google Scholar] [CrossRef]
- Bafico, A.; Gazit, A.; Pramila, T.; Finch, P.W.; Yaniv, A.; Aaronson, S.A. Interaction of frizzled related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J. Biol. Chem. 1999, 274, 16180–16187. [Google Scholar] [CrossRef]
- Kawano, Y.; Kypta, R. Secreted antagonists of the Wnt signalling pathway. J. Cell Sci. 2003, 116, 2627–2634. [Google Scholar] [CrossRef]
- Alberts, A.S. Diaphanous-related Formin homology proteins. Curr. Biol. 2002, 12, R796. [Google Scholar] [CrossRef]
- Wallar, B.J.; Alberts, A.S. The formins: Active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 2003, 13, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Higgs, H.N. Formin proteins: A domain-based approach. Trends Biochem. Sci. 2005, 30, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, M.; Pruyne, D.; Amberg, D.C.; Boone, C.; Bretscher, A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 2002, 4, 32–41. [Google Scholar] [CrossRef]
- Frazier, J.A.; Field, C.M. Actin cytoskeleton: Are FH proteins local organizers? Curr. Biol. 1997, 7, R414–R417. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Severson, A.F.; Baillie, D.L.; Bowerman, B. A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr. Biol. 2002, 12, 2066–2075. [Google Scholar] [CrossRef]
- Habas, R.; Dawid, I.B.; He, X. Coactivation of Rac and Rho by Wnt/Frizzled Signaling Is Required for Vertebrate Gastrulation. Genes Dev. 2003, 17, 295–309. [Google Scholar] [CrossRef]
- Peng, Y.; Chen, Z.; Chen, Y.; Li, S.; Jiang, Y.; Yang, H.; Wu, C.; You, F.; Zheng, C.; Zhu, J.; et al. ROCK isoforms differentially modulate cancer cell motility by mechanosensing the substrate stiffness. Acta Biomater. 2019, 88, 86–101. [Google Scholar] [CrossRef]
- Eyers, C.E.; McNeill, H.; Knebel, A.; Morrice, N.; Arthur, S.J.; Cuenda, A.; Cohen, P. The phosphorylation of CapZ-interacting protein (CapZIP) by stress-activated protein kinases triggers its dissociation from CapZ. Biochem. J. 2005, 389, 127–135. [Google Scholar] [CrossRef]
- Witke, W.; Podtelejnikov, A.V.; Di Nardo, A.; Sutherland, J.D.; Gurniak, C.B.; Dotti, C.; Mann, M. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998, 17, 967–976. [Google Scholar] [CrossRef]
- Da Silva, J.S.; Medina, M.; Zuliani, C.; Di Nardo, A.; Witke, W.; Dotti, C.G. RhoA/ROCK regulation of neuritogenesis via profilin IIa–mediated control of actin stability. J. Cell Biol. 2003, 162, 1267–1279. [Google Scholar] [CrossRef]
- Korenbaum, E.; Nordberg, P.; Björkegren-Sjögren, C.; Schutt, C.E.; Lindberg, U.; Karlsson, R. The role of profilin in actin polymerization and nucleotide exchange. Biochemistry 1998, 37, 9274–9283. [Google Scholar] [CrossRef]
- Reinhard, M.; Giehl, K.; Abel, K.; Haffner, C.; Jarchau, T.; Hoppe, V.; Jockusch, B.M.; Walter, U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995, 14, 1583–1589. [Google Scholar] [CrossRef]
- Nejedla, M.; Sadi, S.; Sulimenko, V.; de Almeida, F.N.; Blom, H.; Draber, P.; Aspenström, P.; Karlsson, R. Profilin connects actin assembly with microtubule dynamics. Mol. Biol. Cell 2016, 27, 2381–2393. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, R.; Dráber, P. Profilin-A master coordinator of actin and microtubule organization in mammalian cells. J. Cell Physiol. 2021, 236, 7256–7265. [Google Scholar] [PubMed]
- Goode, B.L.; Eck, M.J. Mechanism and Function of Formins in Control of Actin Assembly. Annu. Rev. Biochem. 2007, 76, 593–627. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.S.; Pollard, T.D. The role of the FH1 domain and profilin in formin mediated actin-filament elongation and nucleation. Curr. Biol. 2008, 18, 9–19. [Google Scholar] [CrossRef]
- Bartolini, F.; Ramalingam, N.; Gundersen, G.G. Actin-capping protein promotes microtubule stability by antagonizing the actin activity of mDia1. Mol. Biol. Cell 2012, 23, 3871–3882. [Google Scholar] [CrossRef]
- Moustafa-Bayoumi, M.; Alhaj, M.A.; El-Sayed, O.; Wisel, S.; Chotani, M.A.; Abouelnaga, Z.A.; Hassona, M.D.H.; Rigatto, K.; Morris, M.; Nuovo, G.; et al. Vascular hypertrophy and hypertension caused by transgenic overexpression of profilin 1. J. Biol. Chem. 2007, 282, 37632–37639. [Google Scholar] [CrossRef]
- Stritt, S.; Birkholz, I.; Beck, S.; Sorrentino, S.; Sapra, K.T.; Viaud, J.; Heck, J.; Gaits-Iacovoni, F.; Schulze, H.; Du, X.; et al. Profilin 1–Mediated Cytoskeletal Rearrangements Regulate Integrin Function in Mouse Platelets. Blood Adv. 2018, 2, 1040–1045. [Google Scholar] [CrossRef]
- Kim, A.S.; Kakalis, L.T.; Abdul-Manan, N.; Liu, G.A.; Rosen, M.K. Autoinhibition and activation mechanisms of the Wiskott–Aldrich syndrome protein. Nature 2000, 404, 151–158. [Google Scholar] [CrossRef]
- Yang, C.; Huang, M.; DeBiasio, J.; Pring, M.; Joyce, M.; Miki, H.; Takenawa, T.; Zigmond, S.H. Profilin enhances Cdc42-induced nucleation of actin polymerization. J. Cell Biol. 2000, 150, 1001–1012. [Google Scholar] [CrossRef]
- Pimm, M.L.; Liu, X.; Tuli, F.; Heritz, J.; Lojko, A.; Henty-Ridilla, J.L. Visualizing molecules of functional human profilin. eLife 2022, 11, e76485. [Google Scholar] [CrossRef] [PubMed]
- Read, T.A.; Cisterna, B.A.; Skruber, K.; Ahmadieh, S.; Liu, T.M.; Vitriol, J.A.; Shi, Y.; Black, J.B.; Butler, M.T.; Lindamood, H.L.; et al. The actin binding protein profilin 1 localizes inside mitochondria and is critical for their function. EMBO Rep. 2024, 25, 3240–3262. [Google Scholar] [CrossRef] [PubMed]
- Ferron, F.; Rebowski, G.; Lee, S.H.; Dominguez, R. Structural basis for the recruitment of profilin–actin complexes during filament elongation by Ena/VASP. EMBO J. 2007, 26, 4597–4606. [Google Scholar] [CrossRef] [PubMed]
- Mammoto, A.; Sasaki, T.; Asakura, T.; Hotta, I.; Imamura, H.; Takahashi, K.; Matsuura, Y.; Shirao, T.; Takai, Y. Interactions of drebrin and gephyrin with profilin. Biochem. Biophys. Res. Commun. 1998, 243, 86–89. [Google Scholar] [CrossRef]
- Rawe, V.Y.; Payne, C.; Schatten, G. Profilin and actin-related proteins regulate microfilament dynamics during early mammalian embryogenesis. Hum. Reprod. 2006, 21, 1143–1153. [Google Scholar] [CrossRef]
- Jockusch, B.M.; Murk, K.; Rothkegel, M. The profile of profilins. Rev. Physiol. Biochem. Pharmacol. 2007, 159, 131–149. [Google Scholar]
- Liu, X.; Pimm, M.L.; Haarer, B.; Brawner, A.T.; Henty-Ridilla, J.L. Biochemical characterization of actin assembly mechanisms with ALS-associated profilin variants. Eur. J. Cell Biol. 2022, 101, 151212. [Google Scholar] [CrossRef]
- Gupta, C.M.; Ambaru, B.; Bajaj, R. Emerging functions of actins and actin binding proteins in trypanosomatids. Front. Cell Dev. Biol. 2020, 8, 587685. [Google Scholar] [CrossRef]
- Pollard, T.D. Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a018226. [Google Scholar] [CrossRef]
- Zhang, W.; Cao, Y.; Li, W.; Wang, W. Allostery and molecular stripping mechanism in profilin regulated actin filament growth. New J. Phys. 2021, 23, 123010. [Google Scholar] [CrossRef]
- Davey, R.J.; Moens, P.D. Profilin: Many facets of a small protein. Biophys. Rev. 2020, 12, 827–849. [Google Scholar] [CrossRef]
- Rebowski, G.; Boczkowska, M.; Drazic, A.; Ree, R.; Goris, M.; Arnesen, T.; Dominguez, R. Mechanism of actin N-terminal acetylation. Sci. Adv. 2020, 6, eaay8793. [Google Scholar] [CrossRef]
- Honoré, B.; Madsen, P.; Andersen, A.H.; Leffers, H. Cloning and expression of a novel human profilin variant, profilin II. FEBS Lett. 1993, 330, 151–155. [Google Scholar] [CrossRef]
- Ding, Z.; Lambrechts, A.; Parepally, M.; Roy, P. Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J. Cell Sci. 2006, 119, 4127–4137. [Google Scholar] [CrossRef]
- Schoppmeyer, R.; Zhao, R.; Cheng, H.; Hamed, M.; Liu, C.; Zhou, X.; Schwarz, E.C.; Zhou, Y.; Knörck, A.; Schwär, G.; et al. Human profilin 1 is a negative regulator of CTL mediated cell-killing and migration. Eur. J. Immunol. 2017, 47, 1562–1572. [Google Scholar] [CrossRef]
- Teyssou, E.; Chartier, L.; Roussel, D.; Perera, N.D.; Nemazanyy, I.; Langui, D.; Albert, M.; Larmonier, T.; Saker, S.; Salachas, F.; et al. The Amyotrophic Lateral Sclerosis M114T PFN1 Mutation Deregulates Alternative Autophagy Pathways and Mitochondrial Homeostasis. Int. J. Mol. Sci. 2022, 23, 5694. [Google Scholar] [CrossRef]
- Lambrechts, A.; Braun, A.; Jonckheere, V.; Aszodi, A.; Lanier, L.M.; Robbens, J.; Van Colen, I.; Vandekerckhove, J.; Fässler, R.; Ampe, C. Profilin II is alternatively spliced, resulting in profilin isoforms that are differentially expressed and have distinct biochemical properties. Mol. Cell. Biol. 2000, 20, 8209–8219. [Google Scholar] [CrossRef][Green Version]
- Li, Z.; Huo, X.; Chen, K.; Yang, F.; Tan, W.; Zhang, Q.; Yu, H.; Li, C.; Zhou, D.; Chen, H.; et al. Profilin 2 and endothelial exosomal profilin 2 promote angiogenesis and myocardial infarction repair in mice. Front. Cardiovasc. Med. 2022, 9, 781753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, W.; Yan, J.; Zhou, K.; Wan, B.; Shi, P.; Chen, Y.; He, S.; Li, D. Loss of profilin 2 contributes to enhanced epithelial–mesenchymal transition and metastasis of colorectal cancer. Int. J. Oncol. 2018, 53, 1118–1128. [Google Scholar] [CrossRef] [PubMed]
- Hu, E.; Chen, Z.; Fredrickson, T.; Zhu, Y. Molecular cloning and characterization of profilin-3: A novel cytoskeleton-associated gene expressed in rat kidney and testes. Exp. Nephrol. 2001, 9, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Obermann, H.; Raabe, I.; Balvers, M.; Brunswig, B.; Schulze, W.; Kirchhoff, C. Novel testis-expressed profilin IV associated with acrosome biogenesis and spermatid elongation. Mol. Hum. Reprod. 2005, 11, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Murk, K.; Ornaghi, M.; Schiweck, J. Profilin isoforms in health and disease—All the same but different. Front. Cell Dev. Biol. 2021, 9, 681122. [Google Scholar] [CrossRef] [PubMed]
- Behnen, M.; Murk, K.; Kursula, P.; Cappallo-Obermann, H.; Rothkegel, M.; Kierszenbaum, A.L.; Kirchhoff, C. Testis-expressed profilins 3 and 4 show distinct functional characteristics and localize in the acroplaxome-manchette complex in spermatids. BMC Cell Biol. 2009, 10, 34. [Google Scholar] [CrossRef]
- Murk, K.; Buchmeier, S.; Jockusch, B.M.; Rothkegel, M. In birds, profilin-2a is ubiquitously expressed and contributes to actin-based motility. J. Cell Sci. 2009, 122, 957–964. [Google Scholar] [CrossRef]
- Witke, W.; Sutherland, J.D.; Sharpe, A.; Arai, M.; Kwiatkowski, D.J. Profilin I is essential for cell survival and cell division in early mouse development. Proc. Natl. Acad. Sci. USA 2001, 98, 3832–3836. [Google Scholar] [CrossRef]
- Pilo Boyl, P.P.; Di Nardo, A.; Mulle, C.; Sassoè-Pognetto, M.; Panzanelli, P.; Mele, A.; Kneussel, M.; Costantini, V.; Perlas, E.; Massimi, M.; et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 2007, 26, 2991–3002. [Google Scholar] [CrossRef]
- Di Nardo, A.D.; Gareus, R.; Kwiatkowski, D.; Witke, W. Alternative splicing of the mouse profilin II gene generates functionally different profilin isoforms. J. Cell Sci. 2000, 113, 3795–3803. [Google Scholar] [CrossRef]
- Umer, N.; Arévalo, L.; Phadke, S.; Lohanadan, K.; Kirfel, G.; Sons, D.; Sofia, D.; Witke, W.; Schorle, H. Loss of Profilin3 impairs spermiogenesis by affecting acrosome biogenesis, autophagy, manchette development and mitochondrial organization. Front. Cell Dev. Biol. 2021, 9, 749559. [Google Scholar] [CrossRef]
- Umer, N.; Phadke, S.; Shakeri, F.; Arevalo, L.; Lohanadan, K.; Kirfel, G.; Sylvester, M.; Buness, A.; Schorle, H. PFN4 is required for manchette development and acrosome biogenesis during mouse spermiogenesis. Development 2022, 149, dev200499. [Google Scholar] [CrossRef]
- Lambrechts, A.; van Damme, J.; Goethals, M.; Vandekerckhove, J.; Ampe, C. Purification and characterization of bovine profilin II. Actin, poly(L-proline) and inositolphospholipid binding. Eur. J. Biochem. 1995, 230, 281–286. [Google Scholar] [CrossRef]
- Polet, D.; Lambrechts, A.; Vandepoele, K.; Vandekerckhove, J.; Ampe, C. On the origin and evolution of vertebrate and viral profilins. FEBS Lett. 2007, 581, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Romeo, G.; Frangioni, J.V.; Kazlauskas, A. Profilin acts downstream of LDL to mediate diabetic endothelial cell dysfunction. FASEB J. 2004, 18, 725–727. [Google Scholar] [CrossRef] [PubMed]
- Merlotti, D.; Materozzi, M.; Bianciardi, S.; Guarnieri, V.; Rendina, D.; Volterrani, L.; Bellan, C.; Mingiano, C.; Picchioni, T.; Frosali, A.; et al. Mutation of PFN1 gene in an early onset, poly-ostotic Paget-like disease. J. Clin. Endocrinol. Metab. 2020, 105, dgaa252. [Google Scholar] [CrossRef] [PubMed]
- Scotto di Carlo, F.; Pazzaglia, L.; Esposito, T.; Gianfrancesco, F. The loss of profilin 1 causes early onset Paget’s disease of bone. J. Bone Miner. Res. 2020, 35, 1387–1398. [Google Scholar] [CrossRef]
- Nölle, A.; Zeug, A.; van Bergeijk, J.; Tönges, L.; Gerhard, R.; Brinkmann, H.; Al Rayes, S.; Hensel, N.; Schill, Y.; Apkhazava, D.; et al. The spinal muscular atrophy disease protein SMN is linked to the rho-kinase pathway via profilin. Hum. Mol. Genet. 2011, 20, 4865–4878. [Google Scholar] [CrossRef]
- Wu, C.H.; Fallini, C.; Ticozzi, N.; Keagle, P.J.; Sapp, P.C.; Piotrowska, K.; Lowe, P.; Koppers, M.; McKenna-Yasek, D.; Baron, D.M.; et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 2012, 488, 499–503. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, Z.Z.; Huang, R.; Chen, K.; Song, W.; Zhao, B.; Chen, X.; Yang, Y.; Yuan, L.; Shang, H.F. PFN1 mutations are rare in Han Chinese populations with amyotrophic lateral sclerosis. Neurobiol. Aging 2013, 34, 1922.e1–1922.e5. [Google Scholar] [CrossRef]
- Ingre, C.; Landers, J.E.; Rizik, N.; Volk, A.E.; Akimoto, C.; Birve, A.; Hubers, A.; Keagle, P.J.; Piotrowska, K.; Press, R.; et al. A novel phosphorylation site mutation in profilin 1 revealed in a large screen of US, Nordic, and German amyotrophic lateral sclerosis/frontotemporal dementia cohorts. Neurobiol. Aging 2013, 34, 1708.e1–1708.e6. [Google Scholar] [CrossRef]
- Smith, B.N.; Vance, C.; Scotter, E.L.; Troakes, C.; Wong, C.H.; Topp, S.; Maekawa, S.; King, A.; Mitchell, J.C.; Lund, K.; et al. Novel mutations support a role for Profilin 1 in the pathogenesis of ALS. Neurobiol. Aging 2015, 36, 1602.e17–1602.e27. [Google Scholar] [CrossRef]
- Alkam, D.; Feldman, E.Z.; Singh, A.; Kiaei, M. Profilin1 biology and its mutation, actin(g) in disease. Cell. Mol. Life Sci. 2017, 74, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Danielson, E.W.; Qiao, T.; Metterville, J.; Brown, R.H., Jr.; Landers, J.E.; Xu, Z. Mutant PFN1 causes ALS phenotypes and progressive motor neuron degeneration in mice by a gain of toxicity. Proc. Natl. Acad. Sci. USA 2016, 113, E6209–E6218. [Google Scholar] [CrossRef] [PubMed]
- Fil, D.; DeLoach, A.; Yadav, S.; Alkam, D.; MacNicol, M.; Singh, A.; Compadre, C.M.; Goellner, J.J.; O’Brien, C.A.; Fahmi, T.; et al. Mutant Profilin1 transgenic mice recapitulate cardinal features of motor neuron disease. Hum. Mol. Genet. 2017, 26, 686–701. [Google Scholar] [CrossRef] [PubMed]
- Brettle, M.; Stefen, H.; Djordjevic, A.; Fok, S.Y.Y.; Chan, J.W.; van Hummel, A.; van der Hoven, J.; Przybyla, M.; Volkerling, A.; Ke, Y.D.; et al. Developmental expression of mutant PFN1 in motor neurons impacts neuronal growth and motor performance of young and adult mice. Front. Mol. Neurosci. 2019, 12, 231. [Google Scholar] [CrossRef]
- Boopathy, S.; Silvas, T.V.; Tischbein, M.; Jansen, S.; Shandilya, S.M.; Zitzewitz, J.A.; Landers, J.E.; Goode, B.L.; Schiffer, C.A.; Bosco, D.A. Structural basis for mutation-induced destabilization of profilin 1 in ALS. Proc. Natl. Acad. Sci. USA 2015, 112, 7984–7989. [Google Scholar] [CrossRef]
- Freischmidt, A.; Schopflin, M.; Feiler, M.S.; Fleck, A.K.; Ludolph, A.C.; Weishaupt, J.H. Profilin 1 with the amyotrophic lateral sclerosis associated mutation T109M displays unaltered actin binding and does not affect the actin cytoskeleton. BMC Neurosci. 2015, 16, 77. [Google Scholar] [CrossRef]
- Del Poggetto, E.; Gori, L.; Chiti, F. Biophysical analysis of three novel profilin-1 variants associated with amyotrophic lateral sclerosis indicates a correlation between their aggregation propensity and the structural features of their globular state. Biol. Chem. 2016, 397, 927–937. [Google Scholar] [CrossRef]
- Del Poggetto, E.; Toto, A.; Aloise, C.; Di Piro, F.; Gori, L.; Malatesta, F.; Gianni, S.; Chiti, F.; Bemporad, F. Stability of an aggregation-prone partially folded state of human profilin-1 correlates with aggregation propensity. J. Biol. Chem. 2018, 293, 10303–10313. [Google Scholar] [CrossRef]
- Schmidt, E.J.; Funes, S.; McKeon, J.E.; Morgan, B.R.; Boopathy, S.; O’Connor, L.C.; Bilsel, O.; Massi, F.; Jegou, A.; Bosco, D.A. ALS-linked PFN1 variants exhibit loss and gain of functions in the context of formin-induced actin polymerization. Proc. Natl. Acad. Sci. USA 2021, 118, e2024605118. [Google Scholar] [CrossRef]
- Bassell, G.J.; Warren, S.T. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron 2008, 60, 201–214. [Google Scholar] [CrossRef]
- Reeve, S.P.; Bassetto, L.; Genova, G.K.; Kleyner, Y.; Leyssen, M.; Jackson, F.R.; Hassan, B.A. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr. Biol. 2005, 15, 1156–1163. [Google Scholar] [CrossRef]
- Tessier, C.R.; Broadie, K. Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development 2008, 135, 1547–1557. [Google Scholar] [CrossRef]
- Saffary, R.; Xie, Z. FMRP regulates the nuclear export of NXF1 mRNA via N6-methyladenosine-dependent pathway. J. Cell Biol. 2011, 194, 339–345. [Google Scholar]
- Michaelsen-Preusse, K.; Zessin, S.; Grigoryan, G.; Feederle, R.; Eliava, M.; Dityatev, A.; Gottmann, K. Neuronal profilin isoforms are differently involved in spine development and synapse formation. J. Neurosci. 2016, 36, 6097–6109. [Google Scholar]
- Scharkowski, F.; Frotscher, M.; Lutz, D.; Korte, M.; Winter, D.; Krieglstein, K.; Schneider, M. Environmental enrichment induces synaptic and cellular changes in the hippocampus that are associated with reduced anxiety and improved spatial memory. Front. Neurosci. 2018, 12, 20. [Google Scholar]
- Zuccato, C.; Valenza, M.; Cattaneo, E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev. 2010, 90, 905–981. [Google Scholar] [CrossRef] [PubMed]
- Saudou, F.; Humbert, S. The biology of huntingtin. Neuron 2016, 89, 910–926. [Google Scholar] [CrossRef]
- Shao, J.; Diamond, M.I. Protein phosphatase 1 dephosphorylates profilin-1 at Ser-137. PLoS One 2012, 7, e32802. [Google Scholar] [CrossRef]
- Posey, A.E.; Ruff, K.M.; Harmon, T.S.; Crick, S.L.; Li, A.; Diamond, M.I.; Shorter, J.; Pappu, R.V. Profilin reduces aggregation and phase separation of huntingtin N-terminal fragments by preferentially binding to soluble monomers and oligomers. J. Biol. Chem. 2018, 293, 3734–3746. [Google Scholar] [CrossRef]
- Ceccon, A.; Tugarinov, V.; Ghirlando, R.; Clore, G.M. Abrogation of prenucleation, transient oligomerization of the Huntingtin exon 1 protein by human profilin I. Proc. Natl. Acad. Sci. USA 2020, 117, 5844–5852. [Google Scholar] [CrossRef]
- Bauer, P.O.; Wong, H.K.; Oyama, F.; Goswami, A.; Okuno, M.; Kino, Y.; Ross, C.A.; Nukina, N. Inhibition of Rho kinases enhances the degradation of mutant huntingtin. J. Biol. Chem. 2009, 284, 13153–13164. [Google Scholar] [CrossRef]
- Li, M.; Yasumura, D.; Ma, A.A.K.; Matthes, M.T.; Yang, H.; Nielson, G.; Hsu, J.; Ozaki, M.; Yoshida, T.; Chiba, K.; et al. Intravitreal administration of HA-1077, a ROCK inhibitor, improves retinal function in a mouse model of Huntington disease. PLoS One 2013, 8, e56026. [Google Scholar] [CrossRef] [PubMed]
- Burnett, B.G.; Andrews, J.; Ranganathan, S.; Fischbeck, K.H.; Di Prospero, N.A. Expression of expanded polyglutamine targets profilin for degradation and alters actin dynamics. Neurobiol. Dis. 2008, 30, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Giesemann, T.; Rathke-Hartlieb, S.; Rothkegel, M.; Bartsch, J.W.; Buchmeier, S.; Jockusch, B.M.; Jockusch, H. A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with SMN in nuclear gems. J. Biol. Chem. 1999, 274, 37908–37914. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Lambrechts, A.; Hao, L.T.; Le, T.T.; Sewry, C.A.; Ampe, C.; Burghes, A.H.; Morris, G.E. A role for complexes of survival of motor neurons (SMN) protein with gemins and profilin in neurite-like cytoplasmic extensions of cultured nerve cells. Exp. Cell Res. 2005, 309, 185–197. [Google Scholar] [CrossRef]
- Walter, L.M.; Rademacher, S.; Pich, A.; Claus, P. Profilin 2 regulates actin rod assembly in neuronal cells. Sci. Rep. 2021, 11, 10287. [Google Scholar] [CrossRef]
- Pimm, M.L.; Hotaling, J.; Henty-Ridilla, J.L. Profilin choreographs actin and microtubules in cells and cancer. Int. Rev. Cell Mol. Biol. 2020, 355, 155–204. [Google Scholar]
- Jiang, C.; Veon, W.; Li, H.; Hallows, K.R.; Roy, P. Epithelial morphological reversion drives Profilin-1-induced elevation of p27(kip1) in mesenchymal triple-negative human breast cancer cells through AMP-activated protein kinase activation. Cell Cycle 2015, 14, 2914–2923. [Google Scholar] [CrossRef]
- Cao, Q.; Liu, Y.; Wu, Y.; Hu, C.; Sun, L.; Wang, J.; Li, C.; Guo, M.; Liu, X.; Lv, J.; et al. Profilin 2 promotes growth, metastasis, and angiogenesis of small cell lung cancer through cancer-derived exosomes. Aging 2020, 12, 25981–25999. [Google Scholar] [CrossRef]
- Özkan, S.; Aksan, A.; Fıratlıgil, F.B.; Kurt, D.; Sucu, S.; Coşkun, A.; Yücel, K.Y.; Çağlar, A.T.; Üstün, Y.E. Profilin-1 levels in preeclampsia: Associations with disease and adverse neonatal outcomes. Placenta 2025, 159, 140–145. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Gao, H.; Zhao, S.; Ji, X.; Liu, X.; You, B.; Li, X.; Qiu, J. Profilin-1 promotes the development of hypertension-induced artery remodeling. J. Histochem. Cytochem. 2014, 62, 298–307. [Google Scholar] [CrossRef]
- Erdem, E.; Yadigaroglu, M.; Güzel, M.; Gülbüz, L.; Yilman, M.; Ocak, M.; Aksu, E.A.; Görgün, S.; Yücel, M. A new biomarker in the diagnosis and prognosis of pulmonary thromboembolism: Serum profilin-1. Heliyon 2024, 10, e23693. [Google Scholar] [CrossRef]
| Organism | Gene(s) | Expression Pattern | Known Functions | References |
|---|---|---|---|---|
| Human | PFN1 | Ubiquitous | Cytoskeleton organization, endothelial migration, mitochondrial homeostasis, ALS, CTL regulation | [164] |
| PFN2a/b | Broad; enriched in brain | Tumor suppression, angiogenesis post-MI | [168,169,170] | |
| PFN3 | Kidney, testis | Spermatogenesis; lacks actin-binding | [171,172,173] | |
| PFN4 | Testis-specific | Sperm maturation, PIP-binding, diagnostic marker | [172,174] | |
| Mouse | PFN1 | Ubiquitous | Embryonic survival, | [176] |
| PFN2a/b | Neuronal | Synaptic plasticity, actin regulation, behavior | [177,178] | |
| PFN3 | Kidney, testis | Manchette development, sperm morphology | [179] | |
| PFN4 | Testis-specific | Manchette and acrosome formation, male fertility | [180] | |
| Chicken | PFN1 | Declines post-embryo | Less critical for adhesion/motility | [175] |
| PFN2a | Embryonic and adult | Cell adhesion and locomotion | [175] | |
| Bovine | PFN1, PFN2 | PFN1: spleen, PFN2: brain; early embryo | Required for cleavage and blastocyst formation; PFN2 strongly inhibits actin polymerization | [156,168] |
| Rat | PFN1, PFN3, PFN4 | Testis (PFN3, PFN4) | Sperm development (acroplaxome, manchette) | [174] |
| Xenopus | PFN1 | Gastrulation stage | Blastopore closure | [17] |
| PFN2 | Only isoform | Neural tube closure, convergent extension | [19] | |
| PFN3 | Not identified | - | - | |
| PFN4 | Conserved structure | Presumed role in spermatogenesis | [182] | |
| Zebrafish | PFN1 (zpfn1) | Early development | Gastrulation, epiboly, zDia2 interaction | [18] |
| PFN2 (zpfn2) | Weak phenotype | Minor role, no synergy with zDia2 | [18] | |
| PFN3, PFN4, PFN2a/b | Predicted, uncharacterized | Expression and function unknown | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alam, S.; Duncan, D.; Hasan, S. Profilin and Non-Canonical Wnt Signaling: Coordinating Cytoskeletal Dynamics from Development to Disease. J. Dev. Biol. 2025, 13, 31. https://doi.org/10.3390/jdb13030031
Alam S, Duncan D, Hasan S. Profilin and Non-Canonical Wnt Signaling: Coordinating Cytoskeletal Dynamics from Development to Disease. Journal of Developmental Biology. 2025; 13(3):31. https://doi.org/10.3390/jdb13030031
Chicago/Turabian StyleAlam, Samira, Danielle Duncan, and Sharmin Hasan. 2025. "Profilin and Non-Canonical Wnt Signaling: Coordinating Cytoskeletal Dynamics from Development to Disease" Journal of Developmental Biology 13, no. 3: 31. https://doi.org/10.3390/jdb13030031
APA StyleAlam, S., Duncan, D., & Hasan, S. (2025). Profilin and Non-Canonical Wnt Signaling: Coordinating Cytoskeletal Dynamics from Development to Disease. Journal of Developmental Biology, 13(3), 31. https://doi.org/10.3390/jdb13030031






