Developmental Impacts of Epigenetics and Metabolism in COVID-19
Abstract
1. Introduction
2. SARS-CoV-2 Induces Metabolic Reprogramming and Epigenetic Changes
3. COVID-19 Is Associated with Accelerated Epigenetic Aging and Hence Development
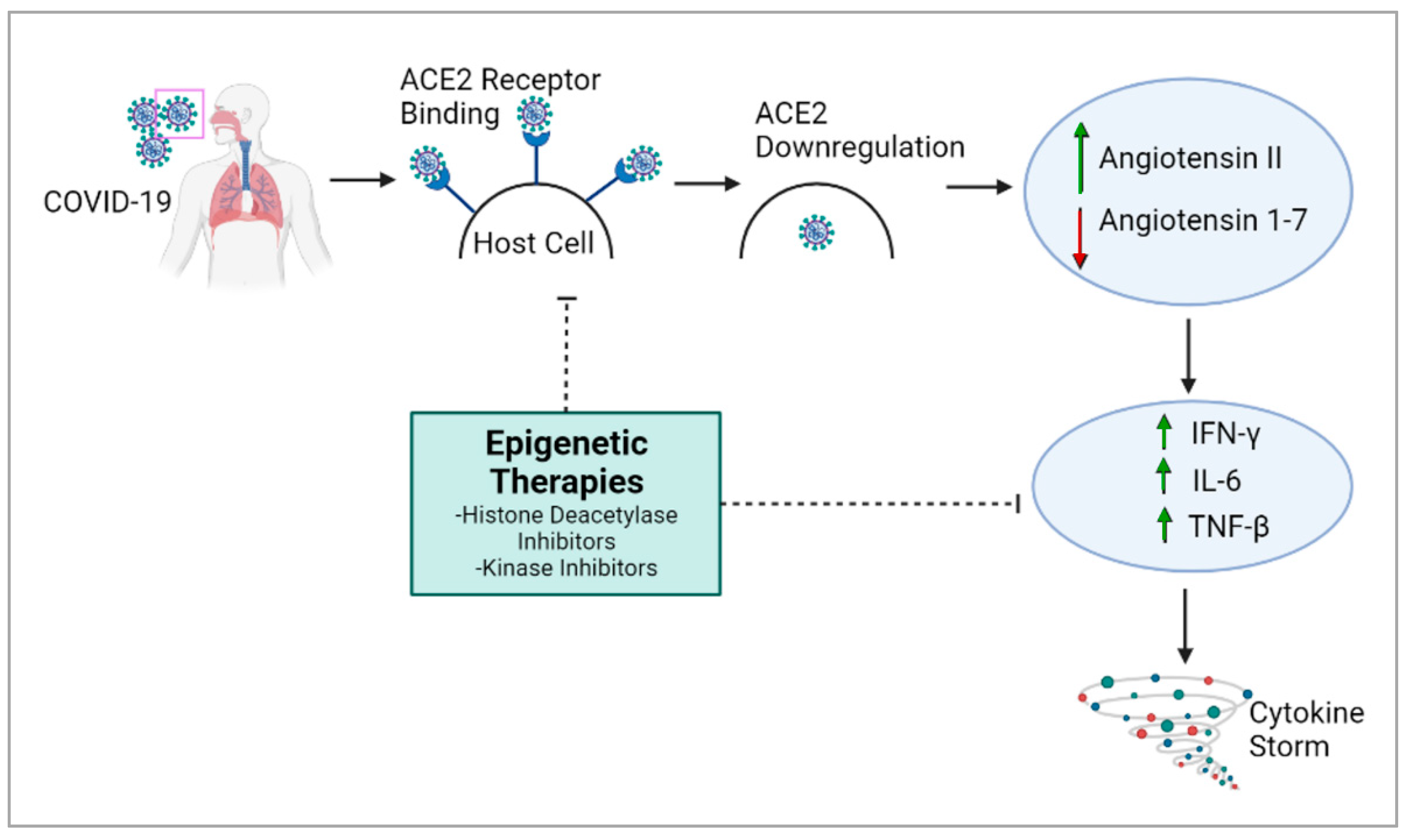
4. Epigenetic Regulation of Viral Pathogenicity Suggests Epi-Drugs as a Therapeutic Approach against COVID-19
5. Epigenetic Therapies May Help to Mitigate COVID-19 Severity
6. Vitamin D Has a Plausible Protective Effect against COVID-19
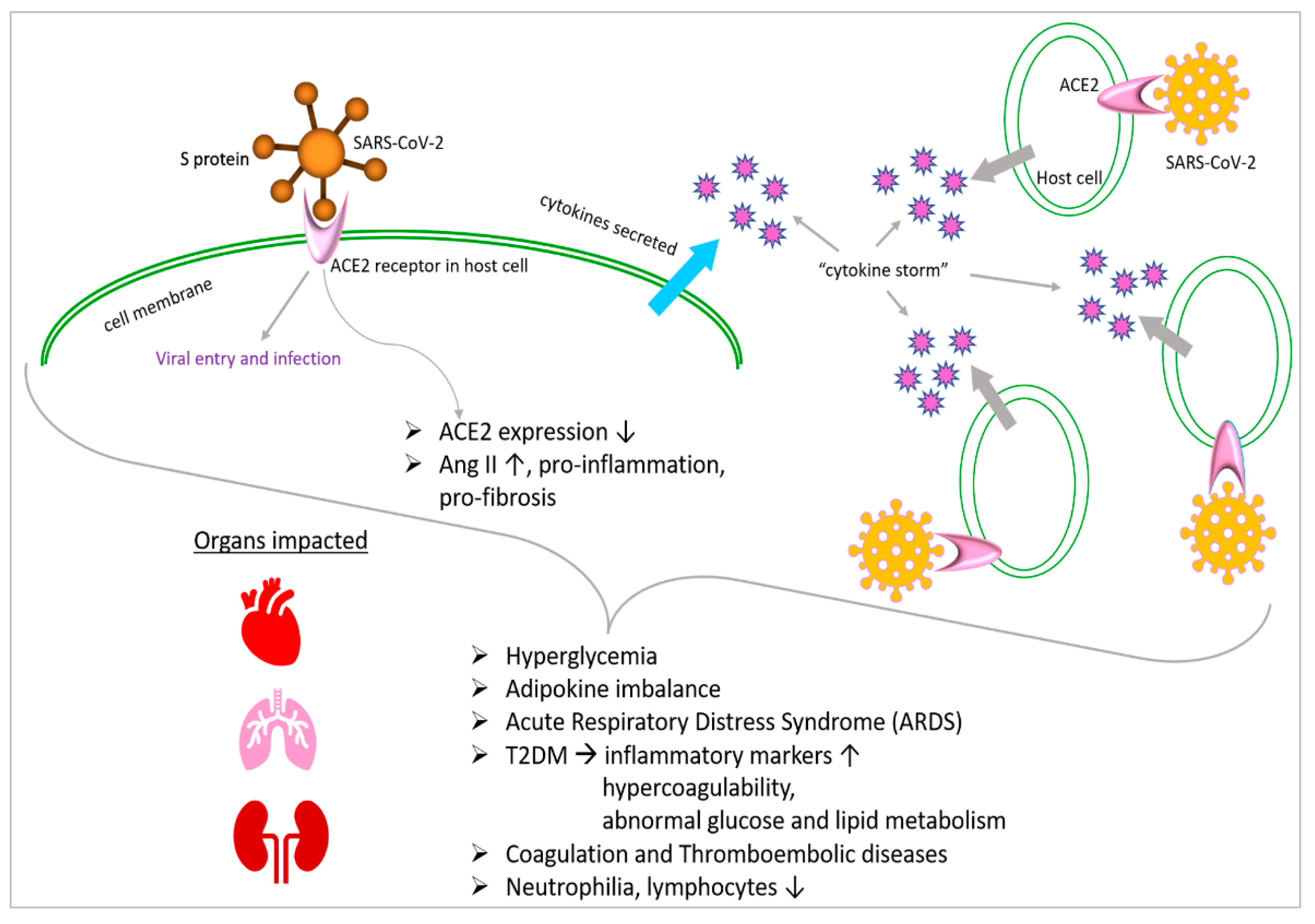
7. Overview of Metabolic Abnormalities Associated with COVID-19
8. Abnormal Metabolism and Diabetes Are Often Manifested in COVID-19
9. Hyperglycemia Is Associated with COVID-19 Severity
10. COVID-19 Affects Adipokines with Impacts on Glucose Metabolism
11. COVID-19 Affects Metabolism through Interactions with Angiotensin
12. Metabolism-Related Therapeutics Could Be Promising against COVID-19
13. Inflammatory Immune Response in Diabetic COVID-19 Patients Is Deleterious
14. Immunometabolic Phenotyping Reveals T Cell and Myeloid Cell Populations Unique to Severe COVID-19
15. Hyperglycemia Associated with COVID-19 Impacts Blood Coagulation
16. Discussion
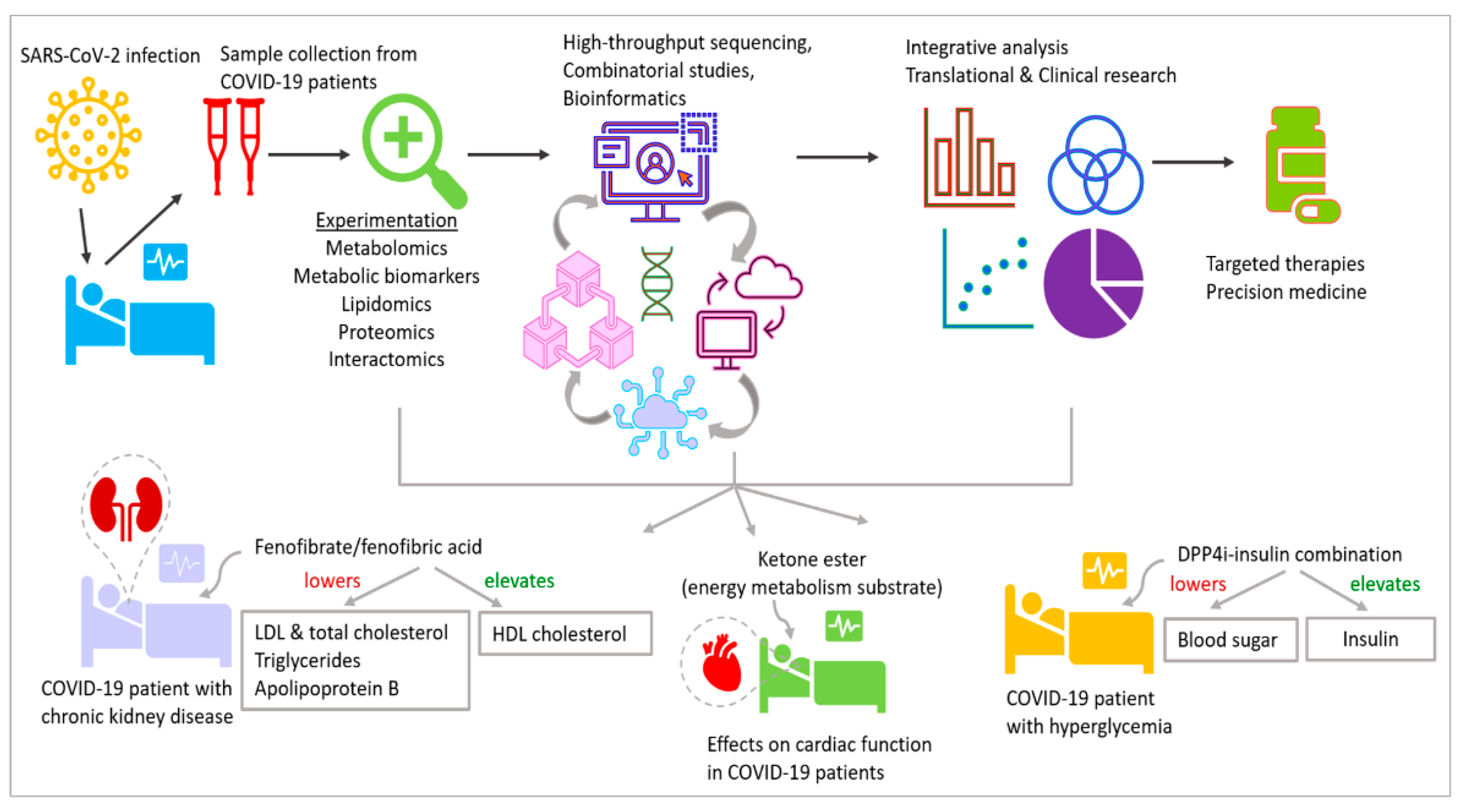
17. Future Perspectives: Potential Aims and Experimental Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spinney, L. How pandemics shape social evolution. Nature 2019, 574, 324–326. [Google Scholar] [CrossRef]
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, J.; Rao, P.; Poojara, L.; Goswami, D.; Acharya, D.; Patel, S.K.; Rawal, R.M. Unravelling the antifungal mode of action of curcumin by potential inhibition of CYP51B: A computational study validated in vitro on mucormycosis agent, Rhizopus oryzae. Arch. Biochem. Biophys. 2021, 712, 109048. [Google Scholar] [CrossRef] [PubMed]
- Bornstein, S.R.; Dalan, R.; Hopkins, D.; Mingrone, G.; Boehm, B.O. Endocrine and metabolic link to coronavirus infection. Nat. Rev. Endocrinol. 2020, 16, 297–298. [Google Scholar] [CrossRef]
- Cao, X.; Li, W.; Wang, T.; Ran, D.; Davalos, V.; Planas-Serra, L.; Pujol, A.; Esteller, M.; Wang, X.; Yu, H. Accelerated biological aging in COVID-19 patients. Nat. Commun. 2022, 13, 2135. [Google Scholar] [CrossRef] [PubMed]
- Arif, T.; Amsalem, Z.; Shoshan-Barmatz, V. Metabolic Reprograming Via Silencing of Mitochondrial VDAC1 Expression Encourages Differentiation of Cancer Cells. Mol. Ther. Nucleic Acids 2019, 17, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Santorelli, L.; Caterino, M.; Costanzo, M. Dynamic Interactomics by Cross-Linking Mass Spectrometry: Mapping the Daily Cell Life in Postgenomic Era. OMICS 2022, 26, 633–649. [Google Scholar] [CrossRef]
- Mar, D.; Babenko, I.M.; Zhang, R.; Noble, W.S.; Denisenko, O.; Vaisar, T.; Bomsztyk, K. A High-Throughput PIXUL-Matrix-Based Toolbox to Profile Frozen and Formalin-Fixed Paraffin-Embedded Tissues Multiomes. Lab. Investig. 2024, 104, 100282. [Google Scholar] [CrossRef]
- Caterino, M.; Costanzo, M.; Fedele, R.; Cevenini, A.; Gelzo, M.; Di Minno, A.; Andolfo, I.; Capasso, M.; Russo, R.; Annunziata, A.; et al. The Serum Metabolome of Moderate and Severe COVID-19 Patients Reflects Possible Liver Alterations Involving Carbon and Nitrogen Metabolism. Int. J. Mol. Sci. 2021, 22, 9548. [Google Scholar] [CrossRef]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef]
- Bonow, R.O.; Fonarow, G.C.; O’Gara, P.T.; Yancy, C.W. Association of Coronavirus Disease 2019 (COVID-19) With Myocardial Injury and Mortality. JAMA Cardiol. 2020, 5, 751–753. [Google Scholar] [CrossRef]
- Hamming, I.; Timens, W.; Bulthuis, M.L.; Lely, A.T.; Navis, G.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Li, S.; Ma, F.; Yokota, T.; Garcia, G., Jr.; Palermo, A.; Wang, Y.; Farrell, C.; Wang, Y.C.; Wu, R.; Zhou, Z.; et al. Metabolic reprogramming and epigenetic changes of vital organs in SARS-CoV-2-induced systemic toxicity. JCI Insight 2021, 6, r145027. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Sen, R. Insights into Cardiovascular Defects and Cardiac Epigenome in the Context of COVID-19. Epigenomes 2022, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Mueller, A.L.; McNamara, M.S.; Sinclair, D.A. Why does COVID-19 disproportionately affect older people? Aging 2020, 12, 9959–9981. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.T.; Hanage, W.P.; Owusu-Boaitey, N.; Cochran, K.B.; Walsh, S.P.; Meyerowitz-Katz, G. Assessing the age specificity of infection fatality rates for COVID-19: Systematic review, meta-analysis, and public policy implications. Eur. J. Epidemiol. 2020, 35, 1123–1138. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Hayek, S.S.; Wang, W.; Chan, L.; Mathews, K.S.; Melamed, M.L.; Brenner, S.K.; Leonberg-Yoo, A.; Schenck, E.J.; Radbel, J.; et al. Factors Associated With Death in Critically Ill Patients With Coronavirus Disease 2019 in the US. JAMA Intern. Med. 2020, 180, 1436–1447. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.J.; Pang, A.P.S.; Dody, K.; Mudd, P.A.; Patterson, B.K.; Seethamraju, H.; Bram, Y.; Peluso, M.J.; Torres, L.; Iyer, N.S.; et al. Genome-wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID-19. J. Leukoc. Biol. 2021, 110, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Hazelton, W.D.; Luebeck, G.E.; Grady, W.M. Epigenetic Aging: More Than Just a Clock When It Comes to Cancer. Cancer Res. 2020, 80, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, J.P.; Mishra, N.; Tran, F.; Bahmer, T.; Best, L.; Blase, J.I.; Bordoni, D.; Franzenburg, J.; Geisen, U.; Josephs-Spaulding, J.; et al. Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19. Immunity 2020, 53, 1296–1314.e9. [Google Scholar] [CrossRef]
- Ogata, A.F.; Maley, A.M.; Wu, C.; Gilboa, T.; Norman, M.; Lazarovits, R.; Mao, C.P.; Newton, G.; Chang, M.; Nguyen, K.; et al. Ultra-Sensitive Serial Profiling of SARS-CoV-2 Antigens and Antibodies in Plasma to Understand Disease Progression in COVID-19 Patients with Severe Disease. Clin. Chem. 2020, 66, 1562–1572. [Google Scholar] [CrossRef] [PubMed]
- Mollica, V.; Rizzo, A.; Massari, F. The pivotal role of TMPRSS2 in coronavirus disease 2019 and prostate cancer. Future Oncol 2020, 16, 2029–2033. [Google Scholar] [CrossRef] [PubMed]
- Beyerstedt, S.; Casaro, E.B.; Rangel, E.B. COVID-19: Angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 905–919. [Google Scholar] [CrossRef] [PubMed]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, J.; Zhang, D.; Xu, Z.; Ji, J.; Wen, C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020, 11, 1708. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Vaishak, K.; Deka, D.; Radhakrishnan, A.K.; Paul, S.; Shanmugam, P.; Daniel, A.P.; Pathak, S.; Duttaroy, A.K.; Banerjee, A. Epigenetic perspectives associated with COVID-19 infection and related cytokine storm: An updated review. Infection 2023, 51, 1603–1618. [Google Scholar] [CrossRef] [PubMed]
- Kgatle, M.M.; Lawal, I.O.; Mashabela, G.; Boshomane, T.M.G.; Koatale, P.C.; Mahasha, P.W.; Ndlovu, H.; Vorster, M.; Rodrigues, H.G.; Zeevaart, J.R.; et al. COVID-19 Is a Multi-Organ Aggressor: Epigenetic and Clinical Marks. Front. Immunol. 2021, 12, 752380. [Google Scholar] [CrossRef] [PubMed]
- AbdelHamid, S.G.; Refaat, A.A.; Benjamin, A.M.; Elmawardy, L.A.; Elgendy, L.A.; Manolly, M.M.; Elmaksoud, N.A.; Sherif, N.; Hamdy, N.M. Deciphering epigenetic(s) role in modulating susceptibility to and severity of COVID-19 infection and/or outcome: A systematic rapid review. Environ. Sci. Pollut. Res. Int. 2021, 28, 54209–54221. [Google Scholar] [CrossRef]
- Gracia-Ramos, A.E. Is the ACE2 Overexpression a Risk Factor for COVID-19 Infection? Arch. Med. Res. 2020, 51, 345–346. [Google Scholar] [CrossRef]
- Takahashi, Y.; Hayakawa, A.; Sano, R.; Fukuda, H.; Harada, M.; Kubo, R.; Okawa, T.; Kominato, Y. Histone deacetylase inhibitors suppress ACE2 and ABO simultaneously, suggesting a preventive potential against COVID-19. Sci. Rep. 2021, 11, 3379. [Google Scholar] [CrossRef]
- Yasmin, R.; Siraj, S.; Hassan, A.; Khan, A.R.; Abbasi, R.; Ahmad, N. Epigenetic regulation of inflammatory cytokines and associated genes in human malignancies. Mediat. Inflamm. 2015, 2015, 201703. [Google Scholar] [CrossRef]
- Patra, S.K.; Szyf, M. Epigenetic perspectives of COVID-19: Virus infection to disease progression and therapeutic control. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166527. [Google Scholar] [CrossRef]
- Li, Y.; Li, H.; Zhou, L. EZH2-mediated H3K27me3 inhibits ACE2 expression. Biochem. Biophys. Res. Commun. 2020, 526, 947–952. [Google Scholar] [CrossRef]
- Foolchand, A.; Mazaleni, S.; Ghazi, T.; Chuturgoon, A.A. A Review: Highlighting the Links between Epigenetics, COVID-19 Infection, and Vitamin D. Int. J. Mol. Sci. 2022, 23, 12292. [Google Scholar] [CrossRef]
- Essalmani, R.; Jain, J.; Susan-Resiga, D.; Andreo, U.; Evagelidis, A.; Derbali, R.M.; Huynh, D.N.; Dallaire, F.; Laporte, M.; Delpal, A.; et al. Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity. J. Virol. 2022, 96, e0012822. [Google Scholar] [CrossRef]
- de Abajo, F.J.; Rodriguez-Martin, S.; Lerma, V.; Mejia-Abril, G.; Aguilar, M.; Garcia-Luque, A.; Laredo, L.; Laosa, O.; Centeno-Soto, G.A.; Angeles Galvez, M.; et al. Use of renin-angiotensin-aldosterone system inhibitors and risk of COVID-19 requiring admission to hospital: A case-population study. Lancet 2020, 395, 1705–1714. [Google Scholar] [CrossRef]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef]
- Hassan, F.U.; Rehman, M.S.; Khan, M.S.; Ali, M.A.; Javed, A.; Nawaz, A.; Yang, C. Curcumin as an Alternative Epigenetic Modulator: Mechanism of Action and Potential Effects. Front. Genet. 2019, 10, 514. [Google Scholar] [CrossRef] [PubMed]
- Sfera, A.; Bullock, K.; Price, A.; Inderias, L.; Osorio, C. Ferrosenescence: The iron age of neurodegeneration? Mech. Ageing Dev. 2018, 174, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Sfera, A.; Fayard, L.; Osorio, C.; Price, A. Epigenetic interventions for brain rejuvenation: Anchoring age-related transposons. Neural Regen. Res. 2018, 13, 635–636. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.A.; Norton, R. Vitamin D and respiratory health. Clin. Exp. Immunol. 2009, 158, 20–25. [Google Scholar] [CrossRef]
- Lau, F.H.; Majumder, R.; Torabi, R.; Saeg, F.; Hoffman, R.; Cirillo, J.D.; Greiffenstein, P. Vitamin D insufficiency is prevalent in severe COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Tan, C.W.; Ho, L.P.; Kalimuddin, S.; Cherng, B.P.Z.; Teh, Y.E.; Thien, S.Y.; Wong, H.M.; Tern, P.J.W.; Chandran, M.; Chay, J.W.M.; et al. Cohort study to evaluate the effect of vitamin D, magnesium, and vitamin B(12) in combination on progression to severe outcomes in older patients with coronavirus (COVID-19). Nutrition 2020, 79–80, 111017. [Google Scholar] [CrossRef]
- Montefusco, L.; Ben Nasr, M.; D’Addio, F.; Loretelli, C.; Rossi, A.; Pastore, I.; Daniele, G.; Abdelsalam, A.; Maestroni, A.; Dell’Acqua, M.; et al. Acute and long-term disruption of glycometabolic control after SARS-CoV-2 infection. Nat. Metab. 2021, 3, 774–785. [Google Scholar] [CrossRef]
- Stefan, N. Metabolic disorders, COVID-19 and vaccine-breakthrough infections. Nat. Rev. Endocrinol. 2022, 18, 75–76. [Google Scholar] [CrossRef]
- Stefan, N.; Birkenfeld, A.L.; Schulze, M.B. Global pandemics interconnected—Obesity, impaired metabolic health and COVID-19. Nat. Rev. Endocrinol. 2021, 17, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Brosh-Nissimov, T.; Orenbuch-Harroch, E.; Chowers, M.; Elbaz, M.; Nesher, L.; Stein, M.; Maor, Y.; Cohen, R.; Hussein, K.; Weinberger, M.; et al. BNT162b2 vaccine breakthrough: Clinical characteristics of 152 fully vaccinated hospitalized COVID-19 patients in Israel. Clin. Microbiol. Infect. 2021, 27, 1652–1657. [Google Scholar] [CrossRef] [PubMed]
- Maestre-Muniz, M.M.; Arias, A.; Mata-Vazquez, E.; Martin-Toledano, M.; Lopez-Larramona, G.; Ruiz-Chicote, A.M.; Nieto-Sandoval, B.; Lucendo, A.J. Long-Term Outcomes of Patients with Coronavirus Disease 2019 at One Year after Hospital Discharge. J. Clin. Med. 2021, 10, 2945. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, R.K.; Kashour, T.; Hamid, Q.; Halwani, R.; Tleyjeh, I.M. Unraveling the Mystery Surrounding Post-Acute Sequelae of COVID-19. Front. Immunol. 2021, 12, 686029. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Caterino, M.; Fedele, R.; Cevenini, A.; Pontillo, M.; Barra, L.; Ruoppolo, M. COVIDomics: The Proteomic and Metabolomic Signatures of COVID-19. Int. J. Mol. Sci. 2022, 23, 2414. [Google Scholar] [CrossRef] [PubMed]
- Holman, N.; Knighton, P.; Kar, P.; O’Keefe, J.; Curley, M.; Weaver, A.; Barron, E.; Bakhai, C.; Khunti, K.; Wareham, N.J.; et al. Risk factors for COVID-19-related mortality in people with type 1 and type 2 diabetes in England: A population-based cohort study. Lancet Diabetes Endocrinol. 2020, 8, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Grasselli, G.; Zangrillo, A.; Zanella, A.; Antonelli, M.; Cabrini, L.; Castelli, A.; Cereda, D.; Coluccello, A.; Foti, G.; Fumagalli, R.; et al. Baseline Characteristics and Outcomes of 1591 Patients Infected With SARS-CoV-2 Admitted to ICUs of the Lombardy Region, Italy. JAMA 2020, 323, 1574–1581. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zheng, Y.; Gou, X.; Pu, K.; Chen, Z.; Guo, Q.; Ji, R.; Wang, H.; Wang, Y.; Zhou, Y. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: A systematic review and meta-analysis. Int. J. Infect. Dis. 2020, 94, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; She, Z.G.; Cheng, X.; Qin, J.J.; Zhang, X.J.; Cai, J.; Lei, F.; Wang, H.; Xie, J.; Wang, W.; et al. Association of Blood Glucose Control and Outcomes in Patients with COVID-19 and Pre-existing Type 2 Diabetes. Cell Metab. 2020, 31, 1068–1077.e1063. [Google Scholar] [CrossRef] [PubMed]
- Barron, E.; Bakhai, C.; Kar, P.; Weaver, A.; Bradley, D.; Ismail, H.; Knighton, P.; Holman, N.; Khunti, K.; Sattar, N.; et al. Associations of type 1 and type 2 diabetes with COVID-19-related mortality in England: A whole-population study. Lancet Diabetes Endocrinol. 2020, 8, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Bae, J.H.; Kwon, H.S.; Nauck, M.A. COVID-19 and diabetes mellitus: From pathophysiology to clinical management. Nat. Rev. Endocrinol. 2021, 17, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Seidu, S.; Gillies, C.; Zaccardi, F.; Kunutsor, S.K.; Hartmann-Boyce, J.; Yates, T.; Singh, A.K.; Davies, M.J.; Khunti, K. The impact of obesity on severe disease and mortality in people with SARS-CoV-2: A systematic review and meta-analysis. Endocrinol. Diabetes Metab. 2021, 4, e00176. [Google Scholar] [CrossRef] [PubMed]
- Khunti, K.; Kosiborod, M.; Ray, K.K. Legacy benefits of blood glucose, blood pressure and lipid control in individuals with diabetes and cardiovascular disease: Time to overcome multifactorial therapeutic inertia? Diabetes Obes. Metab. 2018, 20, 1337–1341. [Google Scholar] [CrossRef]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Mabrey, F.L.; Morrell, E.D.; Wurfel, M.M. TLRs in COVID-19: How they drive immunopathology and the rationale for modulation. Innate Immun. 2021, 27, 503–513. [Google Scholar] [CrossRef]
- Totura, A.L.; Whitmore, A.; Agnihothram, S.; Schafer, A.; Katze, M.G.; Heise, M.T.; Baric, R.S. Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. mBio 2015, 6, e00638-15. [Google Scholar] [CrossRef]
- Iacobellis, G. COVID-19 and diabetes: Can DPP4 inhibition play a role? Diabetes Res. Clin. Pract. 2020, 162, 108125. [Google Scholar] [CrossRef]
- Kokic Males, V. Letter to the editor in response to the article “COVID-19 and diabetes: Can DPP4 inhibition play a role?”. Diabetes Res. Clin. Pract. 2020, 163, 108163. [Google Scholar] [CrossRef]
- Morin, N. Response to COVID-19 and diabetes: Can DPP4 inhibition play a role?—GLP-1 might play one too. Diabetes Res. Clin. Pract. 2020, 164, 108160. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Li, M.; Dong, Y.; Zhou, H.; Zhang, Z.; Tian, C.; Qin, R.; Wang, H.; Shen, Y.; Du, K.; et al. Diabetes is a risk factor for the progression and prognosis of COVID-19. Diabetes Metab. Res. Rev. 2020, 36, e3319. [Google Scholar] [CrossRef] [PubMed]
- Blanke, C.D. In response: Diabetes is a risk factor for the progression and prognosis of COVID-19. Diabetes Metab. Res. Rev. 2020, 36, e3331. [Google Scholar] [CrossRef] [PubMed]
- Fadini, G.P.; Morieri, M.L.; Longato, E.; Avogaro, A. Prevalence and impact of diabetes among people infected with SARS-CoV-2. J. Endocrinol. Investig. 2020, 43, 867–869. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, Q.; Yang, G.; Lin, C.; Zhang, Y.; Yang, P. Weight and prognosis for influenza A(H1N1)pdm09 infection during the pandemic period between 2009 and 2011: A systematic review of observational studies with meta-analysis. Infect. Dis. 2016, 48, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.J.; Baranauskas, M.N.; Fly, A.D. Considerations for Obesity, Vitamin D, and Physical Activity Amid the COVID-19 Pandemic. Obesity 2020, 28, 1176–1177. [Google Scholar] [CrossRef] [PubMed]
- Remuzzi, A.; Remuzzi, G. COVID-19 and Italy: What next? Lancet 2020, 395, 1225–1228. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, S.; Yu, M.; Wang, K.; Tao, Y.; Zhou, Y.; Shi, J.; Zhou, M.; Wu, B.; Yang, Z.; et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J. Allergy Clin. Immunol. 2020, 146, 110–118. [Google Scholar] [CrossRef]
- Hulme, K.D.; Gallo, L.A.; Short, K.R. Influenza Virus and Glycemic Variability in Diabetes: A Killer Combination? Front. Microbiol. 2017, 8, 861. [Google Scholar] [CrossRef]
- Wysocki, J.; Ye, M.; Soler, M.J.; Gurley, S.B.; Xiao, H.D.; Bernstein, K.E.; Coffman, T.M.; Chen, S.; Batlle, D. ACE and ACE2 activity in diabetic mice. Diabetes 2006, 55, 2132–2139. [Google Scholar] [CrossRef]
- Longato, E.; Di Camillo, B.; Sparacino, G.; Saccavini, C.; Avogaro, A.; Fadini, G.P. Diabetes diagnosis from administrative claims and estimation of the true prevalence of diabetes among 4.2 million individuals of the Veneto region (North East Italy). Nutr. Metab. Cardiovasc. Dis. 2020, 30, 84–91. [Google Scholar] [CrossRef]
- Stegenga, M.E.; van der Crabben, S.N.; Blumer, R.M.; Levi, M.; Meijers, J.C.; Serlie, M.J.; Tanck, M.W.; Sauerwein, H.P.; van der Poll, T. Hyperglycemia enhances coagulation and reduces neutrophil degranulation, whereas hyperinsulinemia inhibits fibrinolysis during human endotoxemia. Blood 2008, 112, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Berbudi, A.; Rahmadika, N.; Tjahjadi, A.I.; Ruslami, R. Type 2 Diabetes and its Impact on the Immune System. Curr. Diabetes Rev. 2020, 16, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef] [PubMed]
- Jafar, N.; Edriss, H.; Nugent, K. The Effect of Short-Term Hyperglycemia on the Innate Immune System. Am. J. Med. Sci. 2016, 351, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Yang, F.; Zhu, X.; Wang, M.; Sun, Z.; Zhao, T.; Yang, X.; Zou, Y. Analysis of clinical features and pulmonary CT features of coronavirus disease 2019 (COVID-19) patients with diabetes mellitus. Endokrynol. Pol. 2020, 71, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Popov, D.; Simionescu, M. Alterations of lung structure in experimental diabetes, and diabetes associated with hyperlipidaemia in hamsters. Eur. Respir. J. 1997, 10, 1850–1858. [Google Scholar] [CrossRef]
- Exley, M.A.; Hand, L.; O’Shea, D.; Lynch, L. Interplay between the immune system and adipose tissue in obesity. J. Endocrinol. 2014, 223, R41–R48. [Google Scholar] [CrossRef]
- Polito, R.; Nigro, E.; Messina, A.; Monaco, M.L.; Monda, V.; Scudiero, O.; Cibelli, G.; Valenzano, A.; Picciocchi, E.; Zammit, C.; et al. Adiponectin and Orexin-A as a Potential Immunity Link Between Adipose Tissue and Central Nervous System. Front. Physiol. 2018, 9, 982. [Google Scholar] [CrossRef] [PubMed]
- Nigro, E.; Stiuso, P.; Matera, M.G.; Monaco, M.L.; Caraglia, M.; Maniscalco, M.; Perrotta, F.; Mazzarella, G.; Daniele, A.; Bianco, A. The anti-proliferative effects of adiponectin on human lung adenocarcinoma A549 cells and oxidative stress involvement. Pulm. Pharmacol. Ther. 2019, 55, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, A.; Nigro, E.; Polito, R.; Monaco, M.L.; Scudiero, O.; Mormile, I.; Cesoni Marcelli, A.; Capasso, M.; Habetswallner, F.; Genovese, A.; et al. Total and High Molecular Weight Adiponectin Expression Is Decreased in Patients with Common Variable Immunodeficiency: Correlation with Ig Replacement Therapy. Front. Immunol. 2017, 8, 895. [Google Scholar] [CrossRef] [PubMed]
- Tipnis, S.R.; Hooper, N.M.; Hyde, R.; Karran, E.; Christie, G.; Turner, A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000, 275, 33238–33243. [Google Scholar] [CrossRef] [PubMed]
- Donoghue, M.; Hsieh, F.; Baronas, E.; Godbout, K.; Gosselin, M.; Stagliano, N.; Donovan, M.; Woolf, B.; Robison, K.; Jeyaseelan, R.; et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000, 87, E1–E9. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Gaddam, R.R.; Chambers, S.; Bhatia, M. ACE and ACE2 in inflammation: A tale of two enzymes. Inflamm. Allergy Drug Targets 2014, 13, 224–234. [Google Scholar] [CrossRef]
- Hemnes, A.R.; Rathinasabapathy, A.; Austin, E.A.; Brittain, E.L.; Carrier, E.J.; Chen, X.; Fessel, J.P.; Fike, C.D.; Fong, P.; Fortune, N.; et al. A potential therapeutic role for angiotensin-converting enzyme 2 in human pulmonary arterial hypertension. Eur. Respir. J. 2018, 51, 1702638. [Google Scholar] [CrossRef]
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W.; et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005, 11, 875–879. [Google Scholar] [CrossRef]
- Imai, Y.; Kuba, K.; Rao, S.; Huan, Y.; Guo, F.; Guan, B.; Yang, P.; Sarao, R.; Wada, T.; Leong-Poi, H.; et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 2005, 436, 112–116. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Y.; Zhang, C.; Huang, F.; Wang, F.; Yuan, J.; Wang, Z.; Li, J.; Li, J.; Feng, C.; et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020, 63, 364–374. [Google Scholar] [CrossRef]
- Dikalova, A.; Clempus, R.; Lassegue, B.; Cheng, G.; McCoy, J.; Dikalov, S.; San Martin, A.; Lyle, A.; Weber, D.S.; Weiss, D.; et al. Nox1 overexpression potentiates angiotensin II-induced hypertension and vascular smooth muscle hypertrophy in transgenic mice. Circulation 2005, 112, 2668–2676. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.S.; Vinh, A.; McCarthy, C.A.; Gaspari, T.A.; Widdop, R.E. AT2 receptors: Functional relevance in cardiovascular disease. Pharmacol. Ther. 2008, 120, 292–316. [Google Scholar] [CrossRef]
- Dikalov, S.I.; Ungvari, Z. Role of mitochondrial oxidative stress in hypertension. Am. J. Physiol. Heart Circ. Physiol. 2013, 305, H1417–H1427. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.; Li, Y.; Jiang, Z.; Deng, G.; Li, M.; Liu, X.; Wang, Y. A ROS-dependent and Caspase-3-mediated apoptosis in sheep bronchial epithelial cells in response to Mycoplasma Ovipneumoniae infections. Vet. Immunol. Immunopathol. 2017, 187, 55–63. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, M.; Jiang, H.; Hao, J.; Zhang, Q.; Zhu, Q.; Saren, G.; Zhang, Y.; Meng, X.; Yue, X. Angiotensin II upregulates endothelial lipase expression via the NF-kappa B and MAPK signaling pathways. PLoS ONE 2014, 9, e107634. [Google Scholar] [CrossRef]
- Kawahara, K.; Hohjoh, H.; Inazumi, T.; Tsuchiya, S.; Sugimoto, Y. Prostaglandin E2-induced inflammation: Relevance of prostaglandin E receptors. Biochim. Biophys. Acta 2015, 1851, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Keidar, S.; Gamliel-Lazarovich, A.; Kaplan, M.; Pavlotzky, E.; Hamoud, S.; Hayek, T.; Karry, R.; Abassi, Z. Mineralocorticoid receptor blocker increases angiotensin-converting enzyme 2 activity in congestive heart failure patients. Circ. Res. 2005, 97, 946–953. [Google Scholar] [CrossRef]
- Ferrario, C.M.; Jessup, J.; Chappell, M.C.; Averill, D.B.; Brosnihan, K.B.; Tallant, E.A.; Diz, D.I.; Gallagher, P.E. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation 2005, 111, 2605–2610. [Google Scholar] [CrossRef]
- Yang, G.; Tan, Z.; Zhou, L.; Yang, M.; Peng, L.; Liu, J.; Cai, J.; Yang, R.; Han, J.; Huang, Y.; et al. Effects of Angiotensin II Receptor Blockers and ACE (Angiotensin-Converting Enzyme) Inhibitors on Virus Infection, Inflammatory Status, and Clinical Outcomes in Patients With COVID-19 and Hypertension: A Single-Center Retrospective Study. Hypertension 2020, 76, 51–58. [Google Scholar] [CrossRef]
- Zhong, J.C.; Ye, J.Y.; Jin, H.Y.; Yu, X.; Yu, H.M.; Zhu, D.L.; Gao, P.J.; Huang, D.Y.; Shuster, M.; Loibner, H.; et al. Telmisartan attenuates aortic hypertrophy in hypertensive rats by the modulation of ACE2 and profilin-1 expression. Regul. Pept. 2011, 166, 90–97. [Google Scholar] [CrossRef]
- Furuhashi, M.; Moniwa, N.; Mita, T.; Fuseya, T.; Ishimura, S.; Ohno, K.; Shibata, S.; Tanaka, M.; Watanabe, Y.; Akasaka, H.; et al. Urinary angiotensin-converting enzyme 2 in hypertensive patients may be increased by olmesartan, an angiotensin II receptor blocker. Am. J. Hypertens. 2015, 28, 15–21. [Google Scholar] [CrossRef]
- Perrotta, F.; Matera, M.G.; Cazzola, M.; Bianco, A. Severe respiratory SARS-CoV2 infection: Does ACE2 receptor matter? Respir. Med. 2020, 168, 105996. [Google Scholar] [CrossRef] [PubMed]
- Ali, R.M.; Al-Shorbagy, M.Y.; Helmy, M.W.; El-Abhar, H.S. Role of Wnt4/beta-catenin, Ang II/TGFbeta, ACE2, NF-kappaB, and IL-18 in attenuating renal ischemia/reperfusion-induced injury in rats treated with Vit D and pioglitazone. Eur. J. Pharmacol. 2018, 831, 68–76. [Google Scholar] [CrossRef]
- Zhang, W.; Li, C.; Liu, B.; Wu, R.; Zou, N.; Xu, Y.Z.; Yang, Y.Y.; Zhang, F.; Zhou, H.M.; Wan, K.Q.; et al. Pioglitazone upregulates hepatic angiotensin converting enzyme 2 expression in rats with steatohepatitis. Ann. Hepatol. 2013, 12, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Darwish, I.; Mubareka, S.; Liles, W.C. Immunomodulatory therapy for severe influenza. Expert. Rev. Anti Infect. Ther. 2011, 9, 807–822. [Google Scholar] [CrossRef]
- Drucker, D.J. Coronavirus Infections and Type 2 Diabetes-Shared Pathways with Therapeutic Implications. Endocr. Rev. 2020, 41, bnaa011. [Google Scholar] [CrossRef]
- Sestan, M.; Marinovic, S.; Kavazovic, I.; Cekinovic, D.; Wueest, S.; Turk Wensveen, T.; Brizic, I.; Jonjic, S.; Konrad, D.; Wensveen, F.M.; et al. Virus-Induced Interferon-gamma Causes Insulin Resistance in Skeletal Muscle and Derails Glycemic Control in Obesity. Immunity 2018, 49, 164–177.e6. [Google Scholar] [CrossRef] [PubMed]
- Rao Kondapally Seshasai, S.; Kaptoge, S.; Thompson, A.; Di Angelantonio, E.; Gao, P.; Sarwar, N.; Whincup, P.H.; Mukamal, K.J.; Gillum, R.F.; Holme, I.; et al. Diabetes mellitus, fasting glucose, and risk of cause-specific death. N. Engl. J. Med. 2011, 364, 829–841. [Google Scholar] [CrossRef]
- van Crevel, R.; van de Vijver, S.; Moore, D.A.J. The global diabetes epidemic: What does it mean for infectious diseases in tropical countries? Lancet Diabetes Endocrinol. 2017, 5, 457–468. [Google Scholar] [CrossRef]
- Hodgson, K.; Morris, J.; Bridson, T.; Govan, B.; Rush, C.; Ketheesan, N. Immunological mechanisms contributing to the double burden of diabetes and intracellular bacterial infections. Immunology 2015, 144, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Boccia, M.; Aronne, L.; Celia, B.; Mazzeo, G.; Ceparano, M.; D’Agnano, V.; Parrella, R.; Valente, T.; Bianco, A.; Perrotta, F. COVID-19 and coagulative axis: Review of emerging aspects in a novel disease. Monaldi Arch. Chest Dis. 2020, 90, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Shah, B.R.; Hux, J.E. Quantifying the risk of infectious diseases for people with diabetes. Diabetes Care 2003, 26, 510–513. [Google Scholar] [CrossRef] [PubMed]
- Muniyappa, R.; Gubbi, S. COVID-19 pandemic, coronaviruses, and diabetes mellitus. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E736–E741. [Google Scholar] [CrossRef] [PubMed]
- Kuri-Cervantes, L.; Pampena, M.B.; Meng, W.; Rosenfeld, A.M.; Ittner, C.A.G.; Weisman, A.R.; Agyekum, R.S.; Mathew, D.; Baxter, A.E.; Vella, L.A.; et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020, 5, eabd7114. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.A.; Cascino, K.; Ordonez, A.A.; Zhou, W.; Vaghasia, A.; Hamacher-Brady, A.; Brady, N.R.; Sun, I.H.; Wang, R.; Rosenberg, A.Z.; et al. Metabolic programs define dysfunctional immune responses in severe COVID-19 patients. Cell Rep. 2021, 34, 108863. [Google Scholar] [CrossRef] [PubMed]
- Shoshan-Barmatz, V.; Shteinfer-Kuzmine, A.; Verma, A. VDAC1 at the Intersection of Cell Metabolism, Apoptosis, and Diseases. Biomolecules 2020, 10, 1485. [Google Scholar] [CrossRef] [PubMed]
- Henson, S.M.; Akbar, A.N. KLRG1--more than a marker for T cell senescence. Age 2009, 31, 285–291. [Google Scholar] [CrossRef]
- Kim, S.J.; Mehta, H.H.; Wan, J.; Kuehnemann, C.; Chen, J.; Hu, J.F.; Hoffman, A.R.; Cohen, P. Mitochondrial peptides modulate mitochondrial function during cellular senescence. Aging 2018, 10, 1239–1256. [Google Scholar] [CrossRef]
- Gu, R.; Mao, T.; Lu, Q.; Tianjiao Su, T.; Wang, J. Myeloid dysregulation and therapeutic intervention in COVID-19. Semin. Immunol. 2021, 55, 101524. [Google Scholar] [CrossRef]
- Terpos, E.; Ntanasis-Stathopoulos, I.; Elalamy, I.; Kastritis, E.; Sergentanis, T.N.; Politou, M.; Psaltopoulou, T.; Gerotziafas, G.; Dimopoulos, M.A. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020, 95, 834–847. [Google Scholar] [CrossRef]
- Liu, B.; Li, M.; Zhou, Z.; Guan, X.; Xiang, Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J. Autoimmun. 2020, 111, 102452. [Google Scholar] [CrossRef]
- Herold, T.; Jurinovic, V.; Arnreich, C.; Lipworth, B.J.; Hellmuth, J.C.; von Bergwelt-Baildon, M.; Klein, M.; Weinberger, T. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J. Allergy Clin. Immunol. 2020, 146, 128–136.e124. [Google Scholar] [CrossRef]
- Perrotta, F.; Nigro, E.; Mollica, M.; Costigliola, A.; D’Agnano, V.; Daniele, A.; Bianco, A.; Guerra, G. Pulmonary Hypertension and Obesity: Focus on Adiponectin. Int. J. Mol. Sci. 2019, 20, 912. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, S.; Singh, U.; Jialal, I. The evolving role of C-reactive protein in atherothrombosis. Clin. Chem. 2009, 55, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Fay, W.P. Linking inflammation and thrombosis: Role of C-reactive protein. World J. Cardiol. 2010, 2, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Nogami, K.; Muraki, I.; Imano, H.; Iso, H. Risk of disseminated intravascular coagulation in patients with type 2 diabetes mellitus: Retrospective cohort study. BMJ Open 2017, 7, e013894. [Google Scholar] [CrossRef] [PubMed]
- Kollias, A.; Kyriakoulis, K.G.; Dimakakos, E.; Poulakou, G.; Stergiou, G.S.; Syrigos, K. Thromboembolic risk and anticoagulant therapy in COVID-19 patients: Emerging evidence and call for action. Br. J. Haematol. 2020, 189, 846–847. [Google Scholar] [CrossRef] [PubMed]
- Senn, J.J.; Klover, P.J.; Nowak, I.A.; Mooney, R.A. Interleukin-6 induces cellular insulin resistance in hepatocytes. Diabetes 2002, 51, 3391–3399. [Google Scholar] [CrossRef]
- Klover, P.J.; Zimmers, T.A.; Koniaris, L.G.; Mooney, R.A. Chronic exposure to interleukin-6 causes hepatic insulin resistance in mice. Diabetes 2003, 52, 2784–2789. [Google Scholar] [CrossRef]
- Plomgaard, P.; Bouzakri, K.; Krogh-Madsen, R.; Mittendorfer, B.; Zierath, J.R.; Pedersen, B.K. Tumor necrosis factor-alpha induces skeletal muscle insulin resistance in healthy human subjects via inhibition of Akt substrate 160 phosphorylation. Diabetes 2005, 54, 2939–2945. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Blocking interleukin-1beta in acute and chronic autoinflammatory diseases. J. Intern. Med. 2011, 269, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.J.; To, K.K.; Li, C.; Lau, C.C.; Poon, V.K.; Chan, C.C.; Zheng, B.J.; Hung, I.F.; Lam, K.S.; Xu, A.; et al. Leptin mediates the pathogenesis of severe 2009 pandemic influenza A(H1N1) infection associated with cytokine dysregulation in mice with diet-induced obesity. J. Infect. Dis. 2013, 207, 1270–1280. [Google Scholar] [CrossRef]
- Hou, N.; Luo, J.D. Leptin and cardiovascular diseases. Clin. Exp. Pharmacol. Physiol. 2011, 38, 905–913. [Google Scholar] [CrossRef]
- Karlsson, E.A.; Hertz, T.; Johnson, C.; Mehle, A.; Krammer, F.; Schultz-Cherry, S. Obesity Outweighs Protection Conferred by Adjuvanted Influenza Vaccination. mBio 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Lima-Martinez, M.M.; Carrera Boada, C.; Madera-Silva, M.D.; Marin, W.; Contreras, M. COVID-19 and diabetes: A bidirectional relationship. Clin. Investig. Arterioscler. 2021, 33, 151–157. [Google Scholar] [CrossRef]
- Steenblock, C.; Schwarz, P.E.H.; Ludwig, B.; Linkermann, A.; Zimmet, P.; Kulebyakin, K.; Tkachuk, V.A.; Markov, A.G.; Lehnert, H.; de Angelis, M.H.; et al. COVID-19 and metabolic disease: Mechanisms and clinical management. Lancet Diabetes Endocrinol. 2021, 9, 786–798. [Google Scholar] [CrossRef] [PubMed]
- Khunti, K.; Davies, M.J.; Kosiborod, M.N.; Nauck, M.A. Long COVID—Metabolic risk factors and novel therapeutic management. Nat. Rev. Endocrinol. 2021, 17, 379–380. [Google Scholar] [CrossRef] [PubMed]
- Nigro, E.; Perrotta, F.; Polito, R.; D’Agnano, V.; Scialo, F.; Bianco, A.; Daniele, A. Metabolic Perturbations and Severe COVID-19 Disease: Implication of Molecular Pathways. Int. J. Endocrinol. 2020, 2020, 8896536. [Google Scholar] [CrossRef]
- Sen, R. High-throughput approaches of diagnosis and therapies for COVID-19: Antibody panels, proteomics and metabolomics. Future Drug Discov. 2021, 3, FDD55. [Google Scholar] [CrossRef]
- Appiasie, D.; Guerra, D.J.; Tanguay, K.; Jelinek, S.; Guerra, D.D.; Sen, R. “Multiomics” Approaches to Understand and Treat COVID-19: Mass Spectrometry and Next-Generation Sequencing. BioChem 2021, 1, 210–237. [Google Scholar] [CrossRef]
- Sen, R.; Garbati, M.; Bryant, K.; Lu, Y. Epigenetic mechanisms influencing COVID-19. Genome 2021, 64, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Domingo, I.K.; Latif, A.; Bhavsar, A.P. Pro-Inflammatory Signalling PRRopels Cisplatin-Induced Toxicity. Int. J. Mol. Sci. 2022, 23, 7227. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Q.; Zhao, Y.; Fu, J.; Su, J. Tumor Cells Transmit Drug Resistance via Cisplatin-Induced Extracellular Vesicles. Int. J. Mol. Sci. 2023, 24, 12347. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Rodriguez, M.G.; Ayala-Sumuano, J.T.; Garcia-Morales, L.; Zamudio-Meza, H.; Perez-Yepez, E.A.; Meza, I. IL-1beta Inflammatory Cytokine-Induced TP63 Isoform ∆NP63alpha Signaling Cascade Contributes to Cisplatin Resistance in Human Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 27. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.J.; Lim, J.Y.; Son, M.K.; Ahn, J.H.; Song, K.H.; Lee, H.J.; Kim, S.; Cho, E.H.; Chung, J.Y.; Cho, H.; et al. TRPV1 inhibition overcomes cisplatin resistance by blocking autophagy-mediated hyperactivation of EGFR signaling pathway. Nat. Commun. 2023, 14, 2691. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Kang, J.; Hwang, J.S.; Li, J.; Boese, A.C.; Wang, X.; Yang, L.; Boggon, T.J.; Chen, G.Z.; Saba, N.F.; et al. Cisplatin-mediated activation of glucocorticoid receptor induces platinum resistance via MAST1. Nat. Commun. 2021, 12, 4960. [Google Scholar] [CrossRef] [PubMed]
- Laurino, S.; Brancaccio, M.; Angrisano, T.; Calice, G.; Russi, S.; Mazzone, P.; Di Paola, G.; Aieta, M.; Grieco, V.; Bianchino, G.; et al. Role of IL-6/STAT3 Axis in Resistance to Cisplatin in Gastric Cancers. Biomedicines 2023, 11, 694. [Google Scholar] [CrossRef]
- Group, R.C. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet 2021, 397, 1637–1645. [Google Scholar] [CrossRef]
- Hill, J.A.; Menon, M.P.; Dhanireddy, S.; Wurfel, M.M.; Green, M.; Jain, R.; Chan, J.D.; Huang, J.; Bethune, D.; Turtle, C.; et al. Tocilizumab in hospitalized patients with COVID-19: Clinical outcomes, inflammatory marker kinetics, and safety. J. Med. Virol. 2021, 93, 2270–2280. [Google Scholar] [CrossRef] [PubMed]
- Sarhan, N.M.; Warda, A.E.A.; Ibrahim, H.S.G.; Schaalan, M.F.; Fathy, S.M. Evaluation of infliximab/tocilizumab versus tocilizumab among COVID-19 patients with cytokine storm syndrome. Sci. Rep. 2023, 13, 6456. [Google Scholar] [CrossRef] [PubMed]
- Szabo, R.; Petrisor, C.; Bodolea, C.; Dobre, V.; Tranca, S.; Clichici, S.; Szabo, I.; Melinte, R.M.; Mocan, T. Effects of Tocilizumab on Inflammation and Iron Metabolism in Critically Ill Patients with COVID-19. Pharmaceutics 2023, 15, 646. [Google Scholar] [CrossRef]
- Duran-Mendez, A.; Aguilar-Arroyo, A.D.; Vivanco-Gomez, E.; Nieto-Ortega, E.; Perez-Ortega, D.; Jimenez-Perez, C.; Hernandez-Skewes, K.Y.; Montiel-Bravo, G.; Roque-Reyes, O.J.; Romero-Lechuga, F.; et al. Tocilizumab reduces COVID-19 mortality and pathology in a dose and timing-dependent fashion: A multi-centric study. Sci. Rep. 2021, 11, 19728. [Google Scholar] [CrossRef]
| NCT * | Intervention/Aim | Clinical Trial Description |
|---|---|---|
| NCT04411563 | Predicting prognosis markers | Quantification of circulating epigenetic factors, e.g., microRNAs, profiling of DNA methylation |
| NCT04859894 | Correlating symptoms and physiology of COVID-19 patients with epigenetics | Studying epigenetic alterations and DNA methylation patterns |
| NCT04939155 | Assess effects of SARS-CoV-2 infection and vaccination | Analyze epigenomes and DNA methylation pre-/post-infection and vaccination |
| NCT04517396 | Fenofibrate/fenofibric acid. Lowers elevated LDL and total cholesterol, triglycerides, apolipoprotein B. Increases HDL cholesterol | Fenofibrate in chronic kidney disease to improve clinical outcomes in COVID-19 |
| NCT04542213 | Dipeptidyl peptidase 4 inhibitor (DPP4i). Elevates insulin, lowers blood sugar | DPP4i–insulin combination for metabolic control and prognosis in hospitalized patients with SARS-CoV-2 and hyperglycemia |
| NCT04573764 | d-β-hydroxybutyrate-(R)-1,3 butanediol monoester. Ketone body, maintains ATP yield at starvation, more efficient substrate for energy metabolism than glucose | Acute effects of oral ketone ester on cardiac function in COVID-19 patients |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naik, N.; Patel, M.; Sen, R. Developmental Impacts of Epigenetics and Metabolism in COVID-19. J. Dev. Biol. 2024, 12, 9. https://doi.org/10.3390/jdb12010009
Naik N, Patel M, Sen R. Developmental Impacts of Epigenetics and Metabolism in COVID-19. Journal of Developmental Biology. 2024; 12(1):9. https://doi.org/10.3390/jdb12010009
Chicago/Turabian StyleNaik, Noopur, Mansi Patel, and Rwik Sen. 2024. "Developmental Impacts of Epigenetics and Metabolism in COVID-19" Journal of Developmental Biology 12, no. 1: 9. https://doi.org/10.3390/jdb12010009
APA StyleNaik, N., Patel, M., & Sen, R. (2024). Developmental Impacts of Epigenetics and Metabolism in COVID-19. Journal of Developmental Biology, 12(1), 9. https://doi.org/10.3390/jdb12010009






