Phase Separation as a Driver of Stem Cell Organization and Function during Development
Abstract
1. Introduction
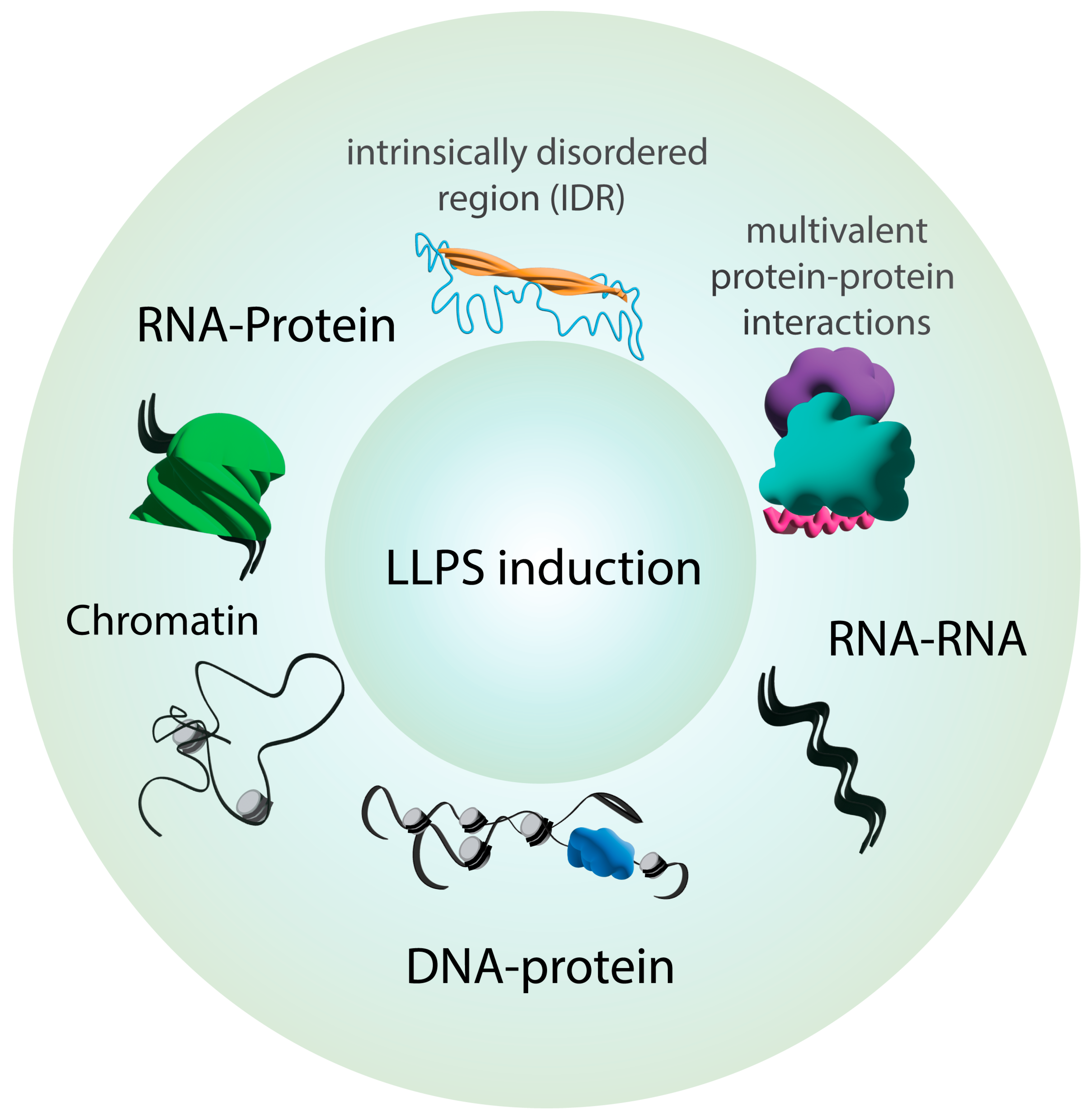
2. Key Principles of Phase Separation in Cellular Organization
3. Phase Separation Drives the Formation of Biological Condensates Essential to Development
4. Phase Separation in Chromatin Organization and Gene Expression
5. Phase Separation in Developmental Signal Transduction Pathways
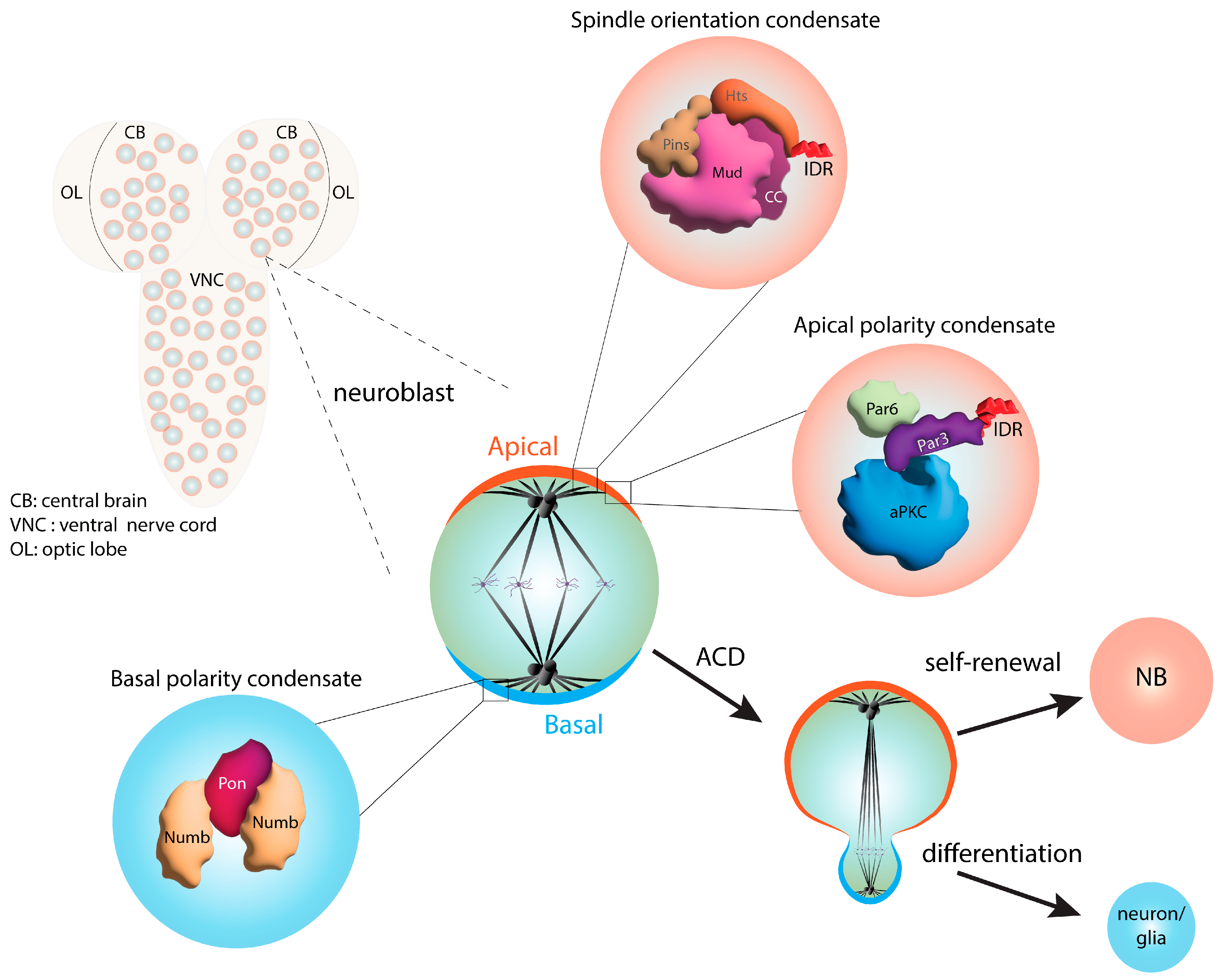
6. Phase Separation in Asymmetric Cell Division
7. Phase Separation in Disease
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef] [PubMed]
- Bracha, D.; Walls, M.T.; Brangwynne, C.P. Probing and engineering liquid-phase organelles. Nat. Biotechnol. 2019, 37, 1435–1445. [Google Scholar] [CrossRef] [PubMed]
- Dignon, G.L.; Best, R.B.; Mittal, J. Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu. Rev. Phys. Chem. 2020, 71, 53–75. [Google Scholar] [CrossRef] [PubMed]
- Rhine, K.; Vidaurre, V.; Myong, S. RNA Droplets. Annu. Rev. Biophys. 2020, 49, 247–265. [Google Scholar] [CrossRef] [PubMed]
- Sabari, B.R.; Dall’Agnese, A.; Young, R.A. Biomolecular Condensates in the Nucleus. Trends Biochem. Sci. 2020, 45, 961–977. [Google Scholar] [CrossRef] [PubMed]
- Chong, S.; Mir, M. Towards Decoding the Sequence-Based Grammar Governing the Functions of Intrinsically Disordered Protein Regions. J. Mol. Biol. 2021, 433, 166724. [Google Scholar] [CrossRef]
- Ruff, K.M.; Choi, Y.H.; Cox, D.; Ormsby, A.R.; Myung, Y.; Ascher, D.B.; Radford, S.E.; Pappu, R.V.; Hatters, D.M. Sequence grammar underlying the unfolding and phase separation of globular proteins. Mol. Cell 2022, 82, 3193–3208.e3198. [Google Scholar] [CrossRef]
- Wang, J.; Choi, J.M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174, 688–699.e616. [Google Scholar] [CrossRef]
- Dvir, S.; Argoetti, A.; Lesnik, C.; Roytblat, M.; Shriki, K.; Amit, M.; Hashimshony, T.; Mandel-Gutfreund, Y. Uncovering the RNA-binding protein landscape in the pluripotency network of human embryonic stem cells. Cell Rep. 2021, 35, 109198. [Google Scholar] [CrossRef]
- Kwon, S.C.; Yi, H.; Eichelbaum, K.; Föhr, S.; Fischer, B.; You, K.T.; Castello, A.; Krijgsveld, J.; Hentze, M.W.; Kim, V.N. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Parra, A.S.; Johnston, C.A. Emerging Roles of RNA-Binding Proteins in Neurodevelopment. J. Dev. Biol. 2022, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Fox, R.G.; Park, F.D.; Koechlein, C.S.; Kritzik, M.; Reya, T. Musashi signaling in stem cells and cancer. Annu. Rev. Cell Dev. Biol. 2015, 31, 249–267. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.C.; Huang, J.R. Musashi-1: An Example of How Polyalanine Tracts Contribute to Self-Association in the Intrinsically Disordered Regions of RNA-Binding Proteins. Int. J. Mol. Sci. 2020, 21, 2289. [Google Scholar] [CrossRef] [PubMed]
- Chiu, S.H.; Ho, W.L.; Sun, Y.C.; Kuo, J.C.; Huang, J.R. Phase separation driven by interchangeable properties in the intrinsically disordered regions of protein paralogs. Commun. Biol. 2022, 5, 400. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Tian, G.G.; Wang, Q.; Liu, M.; He, L.; Li, S.; Wu, J. YTHDF1 phase separation triggers the fate transition of spermatogonial stem cells by activating the IκB-NF-κB-CCND1 axis. Cell Rep. 2023, 42, 112403. [Google Scholar] [CrossRef] [PubMed]
- Case, L.B.; Ditlev, J.A.; Rosen, M.K. Regulation of Transmembrane Signaling by Phase Separation. Annu. Rev. Biophys. 2019, 48, 465–494. [Google Scholar] [CrossRef]
- Feng, Z.; Caballe, A.; Wainman, A.; Johnson, S.; Haensele, A.F.M.; Cottee, M.A.; Conduit, P.T.; Lea, S.M.; Raff, J.W. Structural Basis for Mitotic Centrosome Assembly in Flies. Cell 2017, 169, 1078–1089.e1013. [Google Scholar] [CrossRef]
- Parra, A.S.; Moezzi, C.A.; Johnston, C.A. Drosophila Adducin facilitates phase separation and function of a conserved spindle orientation complex. Front. Cell Dev. Biol. 2023, 11, 1220529. [Google Scholar] [CrossRef]
- Woodruff, J.B.; Ferreira Gomes, B.; Widlund, P.O.; Mahamid, J.; Honigmann, A.; Hyman, A.A. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 2017, 169, 1066–1077.e1010. [Google Scholar] [CrossRef]
- Zeng, M.; Shang, Y.; Araki, Y.; Guo, T.; Huganir, R.L.; Zhang, M. Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity. Cell 2016, 166, 1163–1175.e1112. [Google Scholar] [CrossRef]
- Beutel, O.; Maraspini, R.; Pombo-García, K.; Martin-Lemaitre, C.; Honigmann, A. Phase Separation of Zonula Occludens Proteins Drives Formation of Tight Junctions. Cell 2019, 179, 923–936.e911. [Google Scholar] [CrossRef]
- Case, L.B.; Zhang, X.; Ditlev, J.A.; Rosen, M.K. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 2019, 363, 1093–1097. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Banjade, S.; Cheng, H.C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Chen, X.; Wu, X.; Zhang, M. Formation of biological condensates via phase separation: Characteristics, analytical methods, and physiological implications. J. Biol. Chem. 2019, 294, 14823–14835. [Google Scholar] [CrossRef] [PubMed]
- Harmon, T.S.; Holehouse, A.S.; Rosen, M.K.; Pappu, R.V. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. eLife 2017, 6, e30294. [Google Scholar] [CrossRef] [PubMed]
- López-Palacios, T.P.; Andersen, J.L. Kinase regulation by liquid-liquid phase separation. Trends Cell Biol. 2023, 33, 649–666. [Google Scholar] [CrossRef]
- Luo, Y.Y.; Wu, J.J.; Li, Y.M. Regulation of liquid-liquid phase separation with focus on post-translational modifications. Chem. Commun. 2021, 57, 13275–13287. [Google Scholar] [CrossRef]
- Dodson, A.E.; Kennedy, S. Germ Granules Coordinate RNA-Based Epigenetic Inheritance Pathways. Dev. Cell 2019, 50, 704–715.e704. [Google Scholar] [CrossRef]
- So, C.; Cheng, S.; Schuh, M. Phase Separation during Germline Development. Trends Cell Biol. 2021, 31, 254–268. [Google Scholar] [CrossRef]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Jülicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef]
- Putnam, A.; Cassani, M.; Smith, J.; Seydoux, G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 2019, 26, 220–226. [Google Scholar] [CrossRef]
- Smith, J.; Calidas, D.; Schmidt, H.; Lu, T.; Rasoloson, D.; Seydoux, G. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. eLife 2016, 5, e21337. [Google Scholar] [CrossRef]
- Wang, J.T.; Smith, J.; Chen, B.C.; Schmidt, H.; Rasoloson, D.; Paix, A.; Lambrus, B.G.; Calidas, D.; Betzig, E.; Seydoux, G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife 2014, 3, e04591. [Google Scholar] [CrossRef]
- Trcek, T.; Lehmann, R. Germ granules in Drosophila. Traffic 2019, 20, 650–660. [Google Scholar] [CrossRef]
- Niepielko, M.G.; Eagle, W.V.I.; Gavis, E.R. Stochastic Seeding Coupled with mRNA Self-Recruitment Generates Heterogeneous Drosophila Germ Granules. Curr. Biol. 2018, 28, 1872–1881.e1873. [Google Scholar] [CrossRef]
- Trcek, T.; Grosch, M.; York, A.; Shroff, H.; Lionnet, T.; Lehmann, R. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun. 2015, 6, 7962. [Google Scholar] [CrossRef]
- Strom, A.R.; Brangwynne, C.P. The liquid nucleome—Phase transitions in the nucleus at a glance. J. Cell Sci. 2019, 132, 235093. [Google Scholar] [CrossRef]
- Boisvert, F.M.; van Koningsbruggen, S.; Navascués, J.; Lamond, A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007, 8, 574–585. [Google Scholar] [CrossRef]
- Vilchez, D.; Simic, M.S.; Dillin, A. Proteostasis and aging of stem cells. Trends Cell Biol. 2014, 24, 161–170. [Google Scholar] [CrossRef]
- Gupta, S.; Santoro, R. Regulation and Roles of the Nucleolus in Embryonic Stem Cells: From Ribosome Biogenesis to Genome Organization. Stem Cell Rep. 2020, 15, 1206–1219. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Lee, H.; Han, K.; Kim, S.C.; Choi, Y.; Park, S.W.; Bak, G.; Lee, Y.; Choi, J.K.; Kim, T.K.; et al. SET7/9 methylation of the pluripotency factor LIN28A is a nucleolar localization mechanism that blocks let-7 biogenesis in human ESCs. Cell Stem Cell 2014, 15, 735–749. [Google Scholar] [CrossRef] [PubMed]
- Savić, N.; Bär, D.; Leone, S.; Frommel, S.C.; Weber, F.A.; Vollenweider, E.; Ferrari, E.; Ziegler, U.; Kaech, A.; Shakhova, O.; et al. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 2014, 15, 720–734. [Google Scholar] [CrossRef]
- Brangwynne, C.P.; Mitchison, T.J.; Hyman, A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 4334–4339. [Google Scholar] [CrossRef]
- Mitrea, D.M.; Cika, J.A.; Guy, C.S.; Ban, D.; Banerjee, P.R.; Stanley, C.B.; Nourse, A.; Deniz, A.A.; Kriwacki, R.W. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 2016, 5, e13571. [Google Scholar] [CrossRef]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef]
- Esgleas, M.; Falk, S.; Forné, I.; Thiry, M.; Najas, S.; Zhang, S.; Mas-Sanchez, A.; Geerlof, A.; Niessing, D.; Wang, Z.; et al. Trnp1 organizes diverse nuclear membrane-less compartments in neural stem cells. Embo J. 2020, 39, e103373. [Google Scholar] [CrossRef]
- Dash, S.; Lamb, M.C.; Lange, J.J.; McKinney, M.C.; Tsuchiya, D.; Guo, F.; Zhao, X.; Corbin, T.J.; Kirkman, M.; Delventhal, K.; et al. rRNA transcription is integral to phase separation and maintenance of nucleolar structure. PLoS Genet. 2023, 19, e1010854. [Google Scholar] [CrossRef]
- Mensah, M.A.; Niskanen, H.; Magalhaes, A.P.; Basu, S.; Kircher, M.; Sczakiel, H.L.; Reiter, A.M.V.; Elsner, J.; Meinecke, P.; Biskup, S.; et al. Aberrant phase separation and nucleolar dysfunction in rare genetic diseases. Nature 2023, 614, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Da Costa-Nunes, J.A.; Noordermeer, D. TADs: Dynamic structures to create stable regulatory functions. Curr. Opin. Struct. Biol. 2023, 81, 102622. [Google Scholar] [CrossRef]
- Tena, J.J.; Santos-Pereira, J.M. Topologically Associating Domains and Regulatory Landscapes in Development, Evolution and Disease. Front. Cell Dev. Biol. 2021, 9, 702787. [Google Scholar] [CrossRef]
- Gorkin, D.U.; Leung, D.; Ren, B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell 2014, 14, 762–775. [Google Scholar] [CrossRef]
- Rajderkar, S.; Barozzi, I.; Zhu, Y.; Hu, R.; Zhang, Y.; Li, B.; Alcaina Caro, A.; Fukuda-Yuzawa, Y.; Kelman, G.; Akeza, A.; et al. Topologically associating domain boundaries are required for normal genome function. Commun. Biol. 2023, 6, 435. [Google Scholar] [CrossRef]
- Mohana, G.; Dorier, J.; Li, X.; Mouginot, M.; Smith, R.C.; Malek, H.; Leleu, M.; Rodriguez, D.; Khadka, J.; Rosa, P.; et al. Chromosome-level organization of the regulatory genome in the Drosophila nervous system. Cell 2023, 186, 3826–3844.e3826. [Google Scholar] [CrossRef]
- Palikyras, S.; Papantonis, A. Modes of phase separation affecting chromatin regulation. Open Biol. 2019, 9, 190167. [Google Scholar] [CrossRef]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A Phase Separation Model for Transcriptional Control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef]
- Kotha, S.R.; Staller, M.V. Clusters of acidic and hydrophobic residues can predict acidic transcriptional activation domains from protein sequence. Genetics 2023, 225, iyad131. [Google Scholar] [CrossRef]
- Már, M.; Nitsenko, K.; Heidarsson, P.O. Multifunctional Intrinsically Disordered Regions in Transcription Factors. Chemistry 2023, 29, e202203369. [Google Scholar] [CrossRef]
- Mann, R.; Notani, D. Transcription factor condensates and signaling driven transcription. Nucleus 2023, 14, 2205758. [Google Scholar] [CrossRef]
- Cho, W.K.; Spille, J.H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef]
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175, 1842–1855.e1816. [Google Scholar] [CrossRef]
- Sharma, R.; Choi, K.J.; Quan, M.D.; Sharma, S.; Sankaran, B.; Park, H.; LaGrone, A.; Kim, J.J.; MacKenzie, K.R.; Ferreon, A.C.M.; et al. Liquid condensation of reprogramming factor KLF4 with DNA provides a mechanism for chromatin organization. Nat. Commun. 2021, 12, 5579. [Google Scholar] [CrossRef]
- Kuang, J.; Zhai, Z.; Li, P.; Shi, R.; Guo, W.; Yao, Y.; Guo, J.; Zhao, G.; He, J.; Xu, S.; et al. SS18 regulates pluripotent-somatic transition through phase separation. Nat. Commun. 2021, 12, 4090. [Google Scholar] [CrossRef]
- Eissenberg, J.C.; Elgin, S.C. The HP1 protein family: Getting a grip on chromatin. Curr. Opin. Genet. Dev. 2000, 10, 204–210. [Google Scholar] [CrossRef]
- Mattout, A.; Aaronson, Y.; Sailaja, B.S.; Raghu Ram, E.V.; Harikumar, A.; Mallm, J.P.; Sim, K.H.; Nissim-Rafinia, M.; Supper, E.; Singh, P.B.; et al. Heterochromatin Protein 1β (HP1β) has distinct functions and distinct nuclear distribution in pluripotent versus differentiated cells. Genome Biol. 2015, 16, 213. [Google Scholar] [CrossRef]
- Zeng, A.; Li, Y.Q.; Wang, C.; Han, X.S.; Li, G.; Wang, J.Y.; Li, D.S.; Qin, Y.W.; Shi, Y.; Brewer, G.; et al. Heterochromatin protein 1 promotes self-renewal and triggers regenerative proliferation in adult stem cells. J. Cell Biol. 2013, 201, 409–425. [Google Scholar] [CrossRef]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef]
- Keenen, M.M.; Brown, D.; Brennan, L.D.; Renger, R.; Khoo, H.; Carlson, C.R.; Huang, B.; Grill, S.W.; Narlikar, G.J.; Redding, S. HP1 proteins compact DNA into mechanically and positionally stable phase separated domains. eLife 2021, 10, e64563. [Google Scholar] [CrossRef]
- Sanulli, S.; Trnka, M.J.; Dharmarajan, V.; Tibble, R.W.; Pascal, B.D.; Burlingame, A.L.; Griffin, P.R.; Gross, J.D.; Narlikar, G.J. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 2019, 575, 390–394. [Google Scholar] [CrossRef]
- Gibson, B.A.; Doolittle, L.K.; Schneider, M.W.G.; Jensen, L.E.; Gamarra, N.; Henry, L.; Gerlich, D.W.; Redding, S.; Rosen, M.K. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019, 179, 470–484.e421. [Google Scholar] [CrossRef]
- Mehravar, M.; Ghaemimanesh, F.; Poursani, E.M. An Overview on the Complexity of OCT4: At the Level of DNA, RNA and Protein. Stem Cell Rev. Rep. 2021, 17, 1121–1136. [Google Scholar] [CrossRef]
- Wang, J.; Yu, H.; Ma, Q.; Zeng, P.; Wu, D.; Hou, Y.; Liu, X.; Jia, L.; Sun, J.; Chen, Y.; et al. Phase separation of OCT4 controls TAD reorganization to promote cell fate transitions. Cell Stem Cell 2021, 28, 1868–1883.e1811. [Google Scholar] [CrossRef]
- Clevers, H. Wnt/beta-catenin signaling in development and disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef]
- Nong, J.; Kang, K.; Shi, Q.; Zhu, X.; Tao, Q.; Chen, Y.G. Phase separation of Axin organizes the β-catenin destruction complex. J. Cell Biol. 2021, 220, e202012112. [Google Scholar] [CrossRef]
- Esposito, M.; Fang, C.; Cook, K.C.; Park, N.; Wei, Y.; Spadazzi, C.; Bracha, D.; Gunaratna, R.T.; Laevsky, G.; DeCoste, C.J.; et al. TGF-β-induced DACT1 biomolecular condensates repress Wnt signalling to promote bone metastasis. Nat. Cell Biol. 2021, 23, 257–267. [Google Scholar] [CrossRef]
- Zheng, Y.; Pan, D. The Hippo Signaling Pathway in Development and Disease. Dev. Cell 2019, 50, 264–282. [Google Scholar] [CrossRef]
- Cai, D.; Feliciano, D.; Dong, P.; Flores, E.; Gruebele, M.; Porat-Shliom, N.; Sukenik, S.; Liu, Z.; Lippincott-Schwartz, J. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol. 2019, 21, 1578–1589. [Google Scholar] [CrossRef]
- Lu, Y.; Wu, T.; Gutman, O.; Lu, H.; Zhou, Q.; Henis, Y.I.; Luo, K. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat. Cell Biol. 2020, 22, 453–464. [Google Scholar] [CrossRef]
- Wang, L.; Choi, K.; Su, T.; Li, B.; Wu, X.; Zhang, R.; Driskill, J.H.; Li, H.; Lei, H.; Guo, P.; et al. Multiphase coalescence mediates Hippo pathway activation. Cell 2022, 185, 4376–4393.e4318. [Google Scholar] [CrossRef] [PubMed]
- Bonello, T.T.; Cai, D.; Fletcher, G.C.; Wiengartner, K.; Pengilly, V.; Lange, K.S.; Liu, Z.; Lippincott-Schwartz, J.; Kavran, J.M.; Thompson, B.J. Phase separation of Hippo signalling complexes. Embo J. 2023, 42, e112863. [Google Scholar] [CrossRef] [PubMed]
- Zhuge, R.; Wang, C.; Wang, J.; Yu, S.; Liao, L.; Zheng, X. hCINAP regulates the differentiation of embryonic stem cells by regulating NEDD4 liquid-liquid phase-separation-mediated YAP1 activation. Cell Rep. 2023, 42, 111935. [Google Scholar] [CrossRef] [PubMed]
- Coutu, D.L.; Galipeau, J. Roles of FGF signaling in stem cell self-renewal, senescence and aging. Aging 2011, 3, 920–933. [Google Scholar] [CrossRef] [PubMed]
- Mossahebi-Mohammadi, M.; Quan, M.; Zhang, J.S.; Li, X. FGF Signaling Pathway: A Key Regulator of Stem Cell Pluripotency. Front. Cell Dev. Biol. 2020, 8, 79. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Zhou, F.; Zhao, T.; Zhao, H.; Wang, X.; Chen, L.; Li, J.P.; Luo, S.Z. Phase separation on cell surface facilitates bFGF signal transduction with heparan sulphate. Nat. Commun. 2022, 13, 1112. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Suen, K.M.; Jeffrey, P.A.; Wieteska, L.; Lidster, J.A.; Bao, P.; Curd, A.P.; Stainthorp, A.; Seiler, C.; Koss, H.; et al. Receptor tyrosine kinases regulate signal transduction through a liquid-liquid phase separated state. Mol. Cell 2022, 82, 1089–1106.e1012. [Google Scholar] [CrossRef]
- Weaver, C.J.; Patel, A.L.; Shvartsman, S.Y.; Levine, M.S.; Treen, N. ERK signaling dissolves ERF repression condensates in living embryos. Proc. Natl. Acad. Sci. USA 2022, 119, e2119187119. [Google Scholar] [CrossRef]
- Treen, N.; Chavarria, E.; Weaver, C.J.; Brangwynne, C.P.; Levine, M. An FGF timer for zygotic genome activation. Genes. Dev. 2023, 37, 80–85. [Google Scholar] [CrossRef]
- Dewey, E.B.; Taylor, D.T.; Johnston, C.A. Cell Fate Decision Making through Oriented Cell Division. J. Dev. Biol. 2015, 3, 129–157. [Google Scholar] [CrossRef]
- di Pietro, F.; Echard, A.; Morin, X. Regulation of mitotic spindle orientation: An integrated view. EMBO Rep. 2016, 17, 1106–1130. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Wang, H. Re-visiting the principles of apicobasal polarity in Drosophila neural stem cells. Dev. Biol. 2022, 484, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Rothenberg, M.; Jan, L.Y.; Jan, Y.N. Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell 1998, 95, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Tu, Y.; Yang, Y.; Liu, Z.; Zeng, M.; Xu, H.; Long, J.; Zhang, M.; Cai, Y.; Wen, W. Basal condensation of Numb and Pon complex via phase transition during Drosophila neuroblast asymmetric division. Nat. Commun. 2018, 9, 737. [Google Scholar] [CrossRef] [PubMed]
- Drummond, M.L.; Prehoda, K.E. Molecular Control of Atypical Protein Kinase C: Tipping the Balance between Self-Renewal and Differentiation. J. Mol. Biol. 2016, 428, 1455–1464. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, Y.; Gu, A.; Xu, J.; Mao, Y.; Lu, H.; Hu, W.; Lei, Q.Y.; Li, Z.; Zhang, M.; et al. Par complex cluster formation mediated by phase separation. Nat. Commun. 2020, 11, 2266. [Google Scholar] [CrossRef]
- Xu, J.; Deng, X.; Gu, A.; Cai, Y.; Huang, Y.; Zhang, W.; Zhang, Y.; Wen, W.; Xie, Y. Ccdc85c-Par3 condensates couple cell polarity with Notch to control neural progenitor proliferation. Cell Rep. 2023, 42, 112677. [Google Scholar] [CrossRef]
- Culurgioni, S.; Alfieri, A.; Pendolino, V.; Laddomada, F.; Mapelli, M. Inscuteable and NuMA proteins bind competitively to Leu-Gly-Asn repeat-enriched protein (LGN) during asymmetric cell divisions. Proc. Natl. Acad. Sci. USA 2011, 108, 20998–21003. [Google Scholar] [CrossRef]
- Mauser, J.F.; Prehoda, K.E. Inscuteable regulates the Pins-Mud spindle orientation pathway. PLoS ONE 2012, 7, e29611. [Google Scholar] [CrossRef]
- Zhu, J.; Wen, W.; Zheng, Z.; Shang, Y.; Wei, Z.; Xiao, Z.; Pan, Z.; Du, Q.; Wang, W.; Zhang, M. LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and Gαi/LGN/NuMA pathways. Mol. Cell 2011, 43, 418–431. [Google Scholar] [CrossRef]
- Guo, L.; Kim, H.J.; Wang, H.; Monaghan, J.; Freyermuth, F.; Sung, J.C.; O’Donovan, K.; Fare, C.M.; Diaz, Z.; Singh, N.; et al. Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell 2018, 173, 677–692.e620. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Lee, H.O.; Jawerth, L.; Maharana, S.; Jahnel, M.; Hein, M.Y.; Stoynov, S.; Mahamid, J.; Saha, S.; Franzmann, T.M.; et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015, 162, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.P.; Martyniak, B.; Canning, A.J.; Lei, Y.; Colicino, E.G.; Cosgrove, M.S.; Hehnly, H.; Castaneda, C.A. ALS-Linked Mutations Affect UBQLN2 Oligomerization and Phase Separation in a Position- and Amino Acid-Dependent Manner. Structure 2019, 27, 937–951.e935. [Google Scholar] [CrossRef] [PubMed]
- Hjerpe, R.; Bett, J.S.; Keuss, M.J.; Solovyova, A.; McWilliams, T.G.; Johnson, C.; Sahu, I.; Varghese, J.; Wood, N.; Wightman, M.; et al. UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell 2016, 166, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Cai, C.; Ruan, B.; Hao, M.; Yeo, S.K.; Haas, M.; Yang, F.; Zhang, X.; Guan, J.L. Regulation of RB1CC1/FIP200 stability and autophagy function by CREBBP-mediated acetylation in an intrinsically disordered region. Autophagy 2023, 19, 1662–1677. [Google Scholar] [CrossRef]
- Chen, J.; He, H.J.; Ye, Q.; Feng, F.; Wang, W.W.; Gu, Y.; Han, R.; Xie, C. Defective Autophagy and Mitophagy in Alzheimer’s Disease: Mechanisms and Translational Implications. Mol. Neurobiol. 2021, 58, 5289–5302. [Google Scholar] [CrossRef]
- Tran, M.; Reddy, P.H. Defective Autophagy and Mitophagy in Aging and Alzheimer’s Disease. Front. Neurosci. 2020, 14, 612757. [Google Scholar] [CrossRef]
- Gu, S.; Xu, M.; Chen, L.; Shi, X.; Luo, S.Z. A liquid-to-solid phase transition of Cu/Zn superoxide dismutase 1 initiated by oxidation and disease mutation. J. Biol. Chem. 2023, 299, 102857. [Google Scholar] [CrossRef]
- Zhu, G.; Xie, J.; Kong, W.; Xie, J.; Li, Y.; Du, L.; Zheng, Q.; Sun, L.; Guan, M.; Li, H.; et al. Phase Separation of Disease-Associated SHP2 Mutants Underlies MAPK Hyperactivation. Cell 2020, 183, 490–502.e418. [Google Scholar] [CrossRef]
- Flotho, C.; Valcamonica, S.; Mach-Pascual, S.; Schmahl, G.; Corral, L.; Ritterbach, J.; Hasle, H.; Arico, M.; Biondi, A.; Niemeyer, C.M. RAS mutations and clonality analysis in children with juvenile myelomonocytic leukemia (JMML). Leukemia 1999, 13, 32–37. [Google Scholar] [CrossRef][Green Version]
- Chen, J.; Wang, Y.; Wang, C.; Hu, J.F.; Li, W. LncRNA Functions as a New Emerging Epigenetic Factor in Determining the Fate of Stem Cells. Front. Genet. 2020, 11, 277. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Han, H.; Liu, G.P.; Ma, Y.X.; Pan, R.L.; Sang, L.J.; Li, R.H.; Yang, L.J.; Marks, J.R.; Wang, W.; et al. LncRNA wires up Hippo and Hedgehog signaling to reprogramme glucose metabolism. EMBO J. 2017, 36, 3325–3335. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jiang, T.; Jia, Y.; Zou, J.; Wang, X.; Gu, W. LncRNA MALAT1/miR-181a-5p affects the proliferation and adhesion of myeloma cells via regulation of Hippo-YAP signaling pathway. Cell Cycle 2019, 18, 2509–2523. [Google Scholar] [CrossRef] [PubMed]
- Li, R.H.; Tian, T.; Ge, Q.W.; He, X.Y.; Shi, C.Y.; Li, J.H.; Zhang, Z.; Liu, F.Z.; Sang, L.J.; Yang, Z.Z.; et al. A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid-liquid phase separation to promote oncogenic YAP signaling. Cell Res. 2021, 31, 1088–1105. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Luo, H.; Yee, P.P.; Zhang, L.; Liu, Z.; Zheng, H.; Zhang, L.; Anderson, B.; Tang, M.; Huang, S.; et al. Paraspeckle Protein NONO Promotes TAZ Phase Separation in the Nucleus to Drive the Oncogenic Transcriptional Program. Adv. Sci. 2021, 8, e2102653. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Li, L.; Deng, T.; Liu, Y.; Ling, N.; Qiu, S.; Zhang, L.; Peng, B.; Xiong, W.; Cao, L.; et al. NONO and tumorigenesis: More than splicing. J. Cell Mol. Med. 2020, 24, 4368–4376. [Google Scholar] [CrossRef]
- Zhang, S.; Cooper, J.A.; Chong, Y.S.; Naveed, A.; Mayoh, C.; Jayatilleke, N.; Liu, T.; Amos, S.; Kobelke, S.; Marshall, A.C.; et al. NONO enhances mRNA processing of super-enhancer-associated GATA2 and HAND2 genes in neuroblastoma. EMBO Rep. 2023, 24, e54977. [Google Scholar] [CrossRef]
- Zhu, G.; Xie, J.; Fu, Z.; Wang, M.; Zhang, Q.; He, H.; Chen, Z.; Guo, X.; Zhu, J. Pharmacological inhibition of SRC-1 phase separation suppresses YAP oncogenic transcription activity. Cell Res. 2021, 31, 1028–1031. [Google Scholar] [CrossRef]
- Mehta, S.; Zhang, J. Liquid-liquid phase separation drives cellular function and dysfunction in cancer. Nat. Rev. Cancer 2022, 22, 239–252. [Google Scholar] [CrossRef]
- Tong, X.; Tang, R.; Xu, J.; Wang, W.; Zhao, Y.; Yu, X.; Shi, S. Liquid-liquid phase separation in tumor biology. Signal Transduct. Target. Ther. 2022, 7, 221. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parra, A.S.; Johnston, C.A. Phase Separation as a Driver of Stem Cell Organization and Function during Development. J. Dev. Biol. 2023, 11, 45. https://doi.org/10.3390/jdb11040045
Parra AS, Johnston CA. Phase Separation as a Driver of Stem Cell Organization and Function during Development. Journal of Developmental Biology. 2023; 11(4):45. https://doi.org/10.3390/jdb11040045
Chicago/Turabian StyleParra, Amalia S., and Christopher A. Johnston. 2023. "Phase Separation as a Driver of Stem Cell Organization and Function during Development" Journal of Developmental Biology 11, no. 4: 45. https://doi.org/10.3390/jdb11040045
APA StyleParra, A. S., & Johnston, C. A. (2023). Phase Separation as a Driver of Stem Cell Organization and Function during Development. Journal of Developmental Biology, 11(4), 45. https://doi.org/10.3390/jdb11040045




