Abstract
The COMMD (copper metabolism MURR1 domain containing) family includes ten structurally conserved proteins (COMMD1 to COMMD10) in eukaryotic multicellular organisms that are involved in a diverse array of cellular and physiological processes, including endosomal trafficking, copper homeostasis, and cholesterol metabolism, among others. To understand the role of COMMD10 in embryonic development, we used Commd10Tg(Vav1-icre)A2Kio/J mice, where the Vav1-cre transgene is integrated into an intron of the Commd10 gene, creating a functional knockout of Commd10 in homozygous mice. Breeding heterozygous mice produced no COMMD10-deficient (Commd10Null) offspring, suggesting that COMMD10 is required for embryogenesis. Analysis of Commd10Null embryos demonstrated that they displayed stalled development by embryonic day 8.5 (E8.5). Transcriptome analysis revealed that numerous neural crest-specific gene markers had lower expression in mutant versus wild-type (WT) embryos. Specifically, Commd10Null embryos displayed significantly lower expression levels of a number of transcription factors, including a major regulator of the neural crest, Sox10. Moreover, several cytokines/growth factors involved in early embryonic neurogenesis were also lower in mutant embryos. On the other hand, Commd10Null embryos demonstrated higher expression of genes involved in tissue remodeling and regression processes. Taken together, our findings show that Commd10Null embryos die by day E8.5 due to COMMD10-dependent neural crest failure, revealing a new and critical role for COMMD10 in neural development.
1. Introduction
Endosomes are intracellular lipid bilayer organelles that regulate the trafficking of biological cargo between the plasma membrane and other subcellular compartments, including the trans-Golgi network and lysosomes. Following endocytosis, transmembrane proteins undergo sorting to be recycled back to the cell surface or sent for degradation in lysosomes. Cell surface recycling is essential for membrane receptor maintenance and is executed by two distinct protein complexes: Retromer and Retriever (reviewed in [1]). Each of these recycling complexes associates with other multi-protein structures, such as the Wiscott-Aldrich and Scar Homolog (WASH) complex and the COMMD/CCDC93/CCDC22 (CCC) complex [2,3]. Mutations in these multi-protein complexes are increasingly associated with human pathologies, including neurodegenerative and developmental disorders [4,5,6,7].
The COMMD (copper metabolism MURR1 domain)-containing subunit of the CCC complex includes several COMMD family proteins [1]. All members of this family share a unique C-terminal motif termed a COMM domain, which fosters homo- and hetero-dimerization of COMMD proteins and facilitates interactions with CCDC22 and CCDC93. On the other hand, the N-terminal region is unique in each COMMD protein, suggesting their diverse functions [8]. The first identified member of this family, COMMD1, was discovered to be mutated in Bedlington terriers with copper toxicosis [9]. Subsequently, COMMD1 was demonstrated to regulate the endosomal sorting of the copper transporter ATP7A [2]. COMMD1 also participates in the downregulation of nuclear factor kappa B (NF-κB)-dependent transcription [10,11].
The analysis of Commd10 conditional knockout mice with targeted deficiency to myeloid cells and macrophages demonstrated its direct role in propagating phagolysosomal maturation and clearing of monocyte-driven inflammation [12] and infection [13]. However, Commd10 is ubiquitously expressed, suggesting its role in other tissues [14,15]. Here, we examine the role of COMMD10 in the embryonic development of mice with a disrupted Commd10 gene.
2. Materials and Methods
2.1. Mice
Commd10Het mice were bought from the Jackson Laboratory (B6.Cg-Commd10Tg(Vav1-icre)A2Kio/J, Stock # 008610) [16]. Wild-type (WT) and Commd10Null embryos were generated by interbreeding of Commd10Het littermates. Animals were housed and bred in a specific pathogen-free animal facility and fed a standard diet. All mouse breeding and procedures were carried out according to the laboratory animal protocol approved by the IACUC. Animal genotyping was based on the detection of the intact Commd10 allele and iCre by real-time PCR using a DuPlex PCR approach with the following TaqMan assays:
Commd10-Fwd: CGGGTCTTCCCATCTCATTT
Commd10-Rev: TCAACTGGTTAGTCGGGATTG
Commd10 Probe: CAGACACACCCAGAGGCTCATTCATT
iCre-Fwd: TGGGCATTGCCTACAACA
iCre-Rev: ATCAGCATTCTCCCACCATC
iCre Probe: CGCATTGCCGAAATTGCCAGAATCA
2.2. Embryological Analysis
In order to harvest embryos at specified embryologic stages, timed pregnancies were set up by breeding Commd10Het mice. The embryos were considered 0.5 days post coitus (dpc) at noon on the day of detection of the vaginal plug. At embryonic days 8.5 (E8.5), E9.5, and E10.5, females were euthanized and embryos extracted. Embryonic genotyping was performed on genomic DNA purified from yolk sacs. Whole embryo images were obtained at total magnifications of 15× and 45× (combination of magnifications of 1.5× and 4.5× objective lens with 10× ocular lens) using an AmScope microscope with a MU1003 digital camera and AmScope software (AmScope).
2.3. Western Blot Analysis
Whole embryos were lysed in 1× Laemmli Sample Buffer (BIO-RAD, Hercules, CA, USA, 1610747) with 50 mM DTT, mixed with glass beads, and shaken in an Eppendorf shaker at 2000 RPM at 85 °C for 10 min. Samples were run on a 4–15% Mini-PROTEAN® TGX™ Protein Gel (BIO-RAD, 4561083); transferred to nitrocellulose membranes, which were blocked with Blotting-Grade Blocker (BIO-RAD, 1706404); and probed with anti-COMMD10 (Fisher Scientific, Waltham, MA, USA, PIPA531868; RRID: AB_2549341), anti-COMMD1 (Fisher Scientific, PIPA598616; RRID: AB_2813229), or anti-Sp1 antibody (Sigma-Aldrich, St. Louis, MO, USA, 07-645). Goat anti-rabbit IgG, HPR-linked (Cell Signaling Technologies, Danvers, MA, USA, 7074) was used as a secondary antibody. Sp1 levels were measured as loading controls.
2.4. RNA Extraction
WT and Commd10Null embryos at days E8.5, E9.5, and E10.5 were extracted from yolk sacs and immediately placed in Invitrogen™ RNAlater™ Stabilization Solution (Fisher Scientific, AM7023). They were kept at 4 °C for 24 h and transferred to −80 °C for long-term storage before RNA extraction. Total RNA was extracted using the RNeasy Plus Micro Kit (QIAGEN, Hilden, Germany, 74034) and QIAshredder (QIAGEN, 79656) according to the manufacturer’s instructions.
2.5. RNA-seq and Differential Expression (DE) Analysis
Total RNA purified from WT and Commd10Null embryos at E8.5, E9.5, and E10.5 was subjected to full transcriptome sequencing. At least three biological repeats were carried out for each condition. 3′-end RNA libraries were made using the Lexogen QuantSeq 3′ mRNA-seq Library Prep Kit FWD for Illumina. Sequencing was performed from single-end 75bp on an Illumina NextSeq High Output.
Post-sequence reads were quality-filtered for length and contaminants and were trimmed for Illumina adapters using BBDuk [17]. The resulting reads were pseudo-aligned to coding regions of the mouse reference genome (mm10) using STAR [18]. Gene annotation was performed via the R package biomaRt [19]. Differential expression was calculated using the Wald test implemented in the R package DESeq2 [20]. Significantly differentially expressed genes were defined as those that had both an absolute log2Fold change ≥ 1 and a false discovery rate (FDR) adjusted p-value ≤ 0.05 for each comparison independently.
2.6. Quantitative PCR (RT-qPCR)
Whole embryo total RNA was used to measure gene mRNA levels by real-time qPCR. Reverse transcription and cDNA amplification were performed in one tube using qScript™ XLT One-Step RT-qPCR ToughMix®, Low ROX™ (VWR Quanta Biosciences™, Beverly, MA, USA, 95134) on an Applied Biosystems 7500 Fast Real-Time PCR System (Fisher Scientific). Sample reactions were run in 3–6 replicates. Each mRNA analysis was run in a DuPlex PCR reaction with Gapdh as an internal control. Standard curves for each gene were run to verify the linear range of amplification. Input RNA was kept under 200 ng per reaction to stay within the linear range for Gapdh levels.
All data were analyzed in Microsoft Excel with the built-in analysis methods. TaqMan assays used for RT-qPCR are as follows (m–mouse assays):
mGapdh-Fwd: CCTGTTGCTGTAGCCGTATT
mGapdh-Rev: AACAGCAACTCCCACTCTTC
mGapdh Probe: TTGTCATTGAGAGCAATGCCAGCC
mSox10-Fwd: GCTATTCAGGCTCACTACAAGA
mSox10-Rev: GGACTGCAGCTCTGTCTTT
mSox10 Probe: ATGTCAGATGGGAACCCAGAGCAC
3. Results and Discussion
To examine the role of COMMD10 in embryonic development, we used B6.Cg-Commd10Tg(Vav1-icre)A2Kio/J mice (Jackson Laboratory; stock #008610). In these mice, the Vav1-iCre transgene is integrated into the intron between exons 5 and 6 of the Commd10 gene on chromosome 18 (Figure 1a) [16]. The insertion resulted in a functional knockout of Commd10 in homozygous (Commd10Null) mice [21]. Crossbreeding of Commd10 heterozygous (Commd10Het) littermates produced no Commd10Null newborn mice, while WT and heterozygous genotypes were born at the expected Mendelian ratio (Figure 1b). These results are consistent with those of a viability primary screen phenotypic assay performed on another Commd10 mutant mouse strain (Commd10tm1a(EUCOMM)Wtsi) from the EUCOMM consortium (strain #EPD065) at https://www.mousephenotype.org/data/genes/MGI:1916706 (accessed on 17 July 2022). However, the phenotype of these mice has not been reported in the literature. Thus, the essential role of COMMD10 in embryonic development was confirmed by using two different mouse strains with deficient COMMD10 expression.
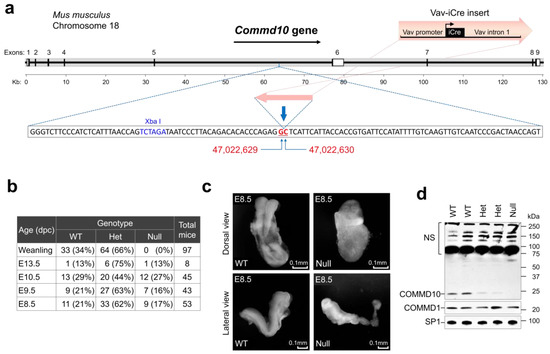
Figure 1.
COMMD10 deficiency results in embryonic lethality. (a) Schematic drawing (up-to-scale) of the Commd10 gene on mouse chromosome 18 shown as a thick grey line. Its direction of transcription is indicated by the black arrow above. Coding exons are represented as thin black boxes. Noncoding 5′- and 3′-untranslated regions are shown as open boxes. The Vav-iCre cassette sketch is shown above the track. The sequence around the Vav-iCre cassette insertion site is shown below the gene scheme in an inset window. Flanking the cassette, GC nucleotides are marked by red bold underlined font and indicated by blue arrows. Their exact positions in the genome are designated by numbers from Reference GRCm39 C57BL/6J below the sequence window. (b) Genotyping analysis of offspring of heterozygous Commd10Het mice (Het) mating. Commd10Null (Null) mice had never been born but embryo genotypes show the expected Mendelian distribution. (dpc): days post-coitus. (c) Morphological analysis of WT and Commd10Null (Null) embryos at E8.5 in dorsal (top panels) and lateral (bottom panels) views. (d) Western blot analysis of whole embryo lysates and anti-COMMD10 or anti-COMMD1 antibodies, as indicated. Anti-SP1 antibody was used as the loading control. NS: non-specific bands.
E8.5 Commd10Null embryos were visually abnormal and displayed abnormal neural plate morphology and growth retardation, but still remained comparable in size and yielded a comparable amount of RNA for analysis (Figure 1c). E9.5 and E10.5 mutant embryos showed progressive degradation and signs of tissue resorption (Figure S1a). Western blot analysis of E8.5 embryo lysates demonstrated lower levels of COMMD10 protein in Commd10Het embryos and its complete absence in Commd10Null embryos compared with WT embryos (Figure 1d).
To examine the root cause of the developmental failure of Commd10Null embryos, we carried out comparative transcriptome analyses of mutant and WT embryos (Figure S1b). Figure 2a shows the gene expression principal component analysis (PCA) plot. The cluster of WT samples on E8.5 appears stretched compared with other clusters, indicating some variability among WT samples on that day. The rest of the clusters are tight without any overlap. Importantly, the direction of embryonic development from E8.5 through E10.5 is reflected in the WT cluster distribution on the PCA plot (WT arrow). Interestingly, Commd10Null E8.5 and E9.5 clusters are located on opposite sides of the WT E8.5 samples. Importantly, both of these clusters are far from each other and from E10.5 samples (Figure 2a, C10_Null arrow). This segregation pattern suggests that the divergence point between WT and Commd10Null embryos took place not long before day E8.5. Thus, the Commd10Null E8.5 transcriptome represents an inflection point in embryogenesis from development to tissue resorption.
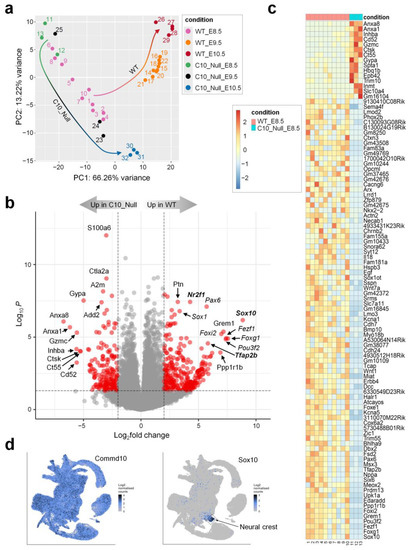
Figure 2.
Commd10Null embryos fail to develop beyond E8.5 due to impaired neural plate and neural crest development. (a) PCA plot of RNA-seq analysis in WT and Commd10Null (C10_Null) embryos at E8.5, E9.5, and E10.5. Sample clusters are shown in different colors. Colored arrows show direction of cluster shifts through E8.5 to E10.5 developmental timeframe for both genotypes. Changing arrow colors correlate with the corresponding sample cluster in a timeframe. (b) Volcano plot of RNA-seq analysis visualizing significant DEGs in WT vs. Commd10Null (C10_Null) E8.5 embryos: magnitude of change (x-axis) vs. statistically significant p-values (y-axis). Points that have a fold change less than 2 (log2 = 1) are shown in grey. Genes that are transcription factors are marked in italic font. Genes that are expressed in neural crest more highly than in any other cell type are shown in Bold font. (c) Heatmap of mRNA expression levels for top 100 significant DEGs in WT vs. Commd10Null E8.5 embryos by RNA-seq. (d) Distribution of Commd10 and Sox10 mRNA expression in WT embryos during early embryogenesis in a single-cell molecular map [22]. Presented plots were generated on a single-cell molecular map of mouse gastrulation and early organogenesis at https://marionilab.cruk.cam.ac.uk/MouseGastrulation2018/ (accessed on 2 September 2022). The full legend annotating cell clusters by different colors and the schematic map are shown in Figure S1c.
Figure 2b shows a volcano plot visualizing differentially expressed genes (DEGs) in WT vs. Commd10Null embryos at E8.5 and displaying wide areas of scattered genes on both sides of the y-axis. We sorted all significant DEGs by the absolute value of log2FoldChange and chose the top 100 DEGs to plot on a heatmap (Figure 2c). Among these top 100 DEGs, only 15 were upregulated in Commd10Null embryos, and the 85 remaining genes were downregulated in contrast to those in WT embryos. Interestingly, the 85 DEGs that are downregulated in mutant embryos include 20 transcription factors, at least 11 cytokines/growth factors/cell surface receptors, and 30 genes with unknown function. The rest of these DEGs encode structural proteins, modifying enzymes, and proteins involved in ion channel function, cell adhesion, and other metabolic cellular processes.
To find the specific embryonic lineage where each of these DEGs is expressed, we searched a single-cell molecular map of mouse gastrulation and early organogenesis at https://marionilab.cruk.cam.ac.uk/MouseGastrulation2018/ (accessed on 2 September 2022) [22]. This interactive atlas demonstrates specific mRNA expression profiles during mouse embryonic development between E6.5 and E8.5. As shown in Figure 2d, Commd10 is broadly expressed in all lineages during embryogenesis. The top most significantly (461-fold) downregulated gene in Commd10Null embryos at E8.5 is Sox10, a transcription factor with a central role in neural crest development and maturation of glia [23]. We have also validated Sox10 mRNA expression in WT and Commd10Null embryos at E8.5, E9.5, and E10.5 by RT-qPCR and found the highest Sox10 expression and the most drastic difference between the two genotypes at E8.5 (Figure S1d). In normal developing mouse embryos, Sox10 expression emerges after E8.0 almost exclusively in the neural crest (Figure 2d). The table in Figure 3a lists the top ten neural crest-specific markers according to the interactive atlas. Interestingly, six of those markers were differentially expressed in WT versus Commd10Null embryos, suggesting that there is a defect in neural crest development in Commd10Null embryos (Figure 2b–d and Figure 3b). Moreover, a list of significant DEGs, which define the trajectory of neurogenesis, includes numerous transcription factors critical for neural plate development, starting from rostral neuroectoderm at E6.5 and subsequent development of caudal neuroectoderm, spinal cord, forebrain/midbrain/hindbrain, and neural crest by E8.5.
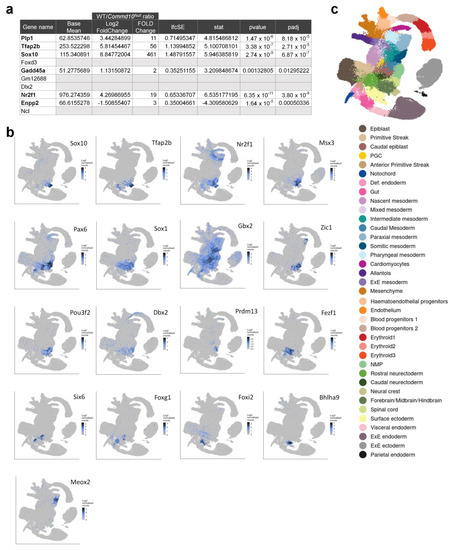
Figure 3.
Six of the top ten neural crest-specific markers are differentially expressed in WT versus Commd10Null embryos. (a) Table listing the top ten neural crest-specific markers, genes that are expressed in the neural crest more highly than in any other cell type. Six genes with differential expression in WT and Commd10Null embryos are shown in bold font. (b) Tissue distribution of mRNA expression of different transcription factors in WT embryos during early embryogenesis in the molecular map of whole dataset, as described in (c). (c) Legend for (b) annotating cell clusters by different colors, and a single-cell molecular map of mouse gastrulation and early organogenesis [22] up to day E8.5 of embryogenesis. All presented plots were generated on a single-cell molecular map of mouse gastrulation and early organogenesis at https://marionilab.cruk.cam.ac.uk/MouseGastrulation2018/ (accessed on 2 September 2022).
Besides Sox10, Commd10Null embryos exhibit significantly lower expression of transcription factors Tfap2b [24,25], Nr2f1 [26], Msx3 [27], Dbx2 with Pax6 [28,29], Sox1 [30], Gbx2 [31], Zic1 [32], Pou3f2 [33,34,35], Prdm13 [36], Fezf1 [37], Six6 [38,39], Foxg1 [40], and Foxi2 [41] (Figure 3). They all participate in the early stages of central nervous system development. Also significantly downregulated in Commd10Null embryos are genes encoding cytokines/growth factors involved in early embryonic neurogenesis, such as Ptn [42,43], Mdk [43,44], and Grem1 [45] (Figure 2b). In addition, transcription factors such as Meox2 [46], expressed in paraxial and somatic mesoderm, and Bhlha9 [47,48], expressed in surface ectoderm, are important for the expression of genes involved in signaling pathways essential for the formation and morphogenesis of somites and limbs in developing embryos (Figure 3). Taken together, these data are in agreement with the observation that WT embryos at E8.5 undergo continuous embryogenesis by means of cell proliferation, migration, and differentiation, particularly in the process of primary neurulation. This highly orchestrated process is defined by the expression of a number of transcription and growth factors that are coordinated in place and time. Significantly lower levels of these molecules in Commd10Null embryos may result in the termination of embryonic development.
On the other hand, there are no transcription factors or cytokine/growth factors among the top 15 DEGs upregulated in Commd10Null embryos at E8.5 as compared with their WT littermates (Figure 2c). While some of these genes, such as Anxa8 and Anxa1 [49], are modestly expressed in notochord, caudal neuroectoderm, and neural crest of the WT embryos, most are not expressed in developing neural tissue (Figure 4 and Figure 5). Instead, the majority of those genes are expressed in blood progenitors and erythroid tissue in particular (Gypa, Hbq1b, Epb42, Trim10 [50,51], Spta1). Interestingly, some of the upregulated DEGs in Commd10Null embryos may be involved in tissue remodeling and regression. Granzyme C (Gzmc) is increased 48-fold in Commd10Null embryos compared with WT, while Inhibin beta A chain (Inhba), a member of the inhibins/activins network of proteins, is increased 49-fold. Thus, embryonic cell death leading to tissue regression in E8.5 Commd10Null embryos may be caused by two main events. The first event is a failure of the neural plate and neural crest processes due to a substantial deficiency of transcription factor Sox10, together with lower expression of other transcription factors and cytokines/growth factors involved in early embryonic neurogenesis. The second event is based on the increased expression of proteins with potential embryo resorption abilities.
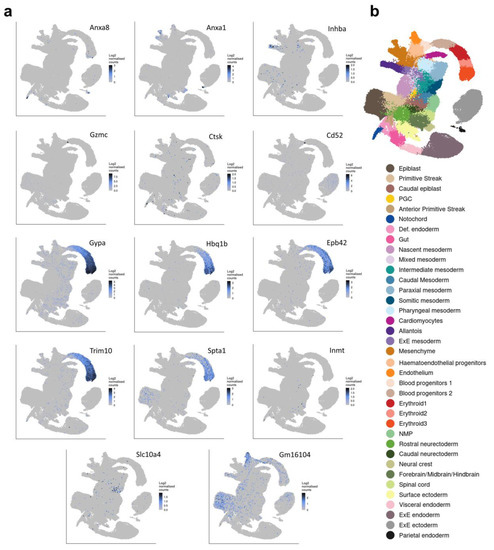
Figure 4.
Tissue distribution of 14 genes significantly upregulated in Commd10Null embryos on E8.5. (a) Single-cell molecular maps of mRNA expression in WT embryos for the top 14 genes significantly upregulated in Commd10Null embryos on E8.5. (b) Legend for (a) annotating cell clusters by different colors, and a single-cell molecular map of mouse gastrulation and early organogenesis [22] up to day E8.5 of embryogenesis. All presented plots were generated on a single-cell molecular map of mouse gastrulation and early organogenesis. at https://marionilab.cruk.cam.ac.uk/MouseGastrulation2018/ (accessed on 2 September 2022) website.
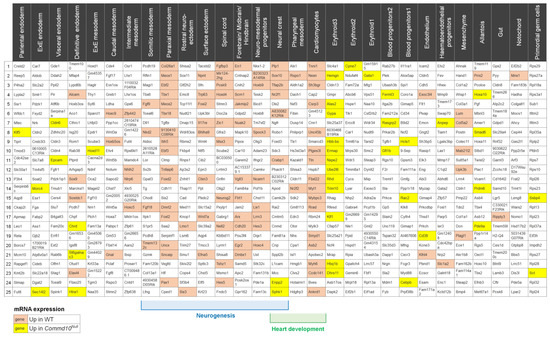
Figure 5.
Tissue distribution of differentially expressed genes in Commd10Null embryos on E8.5. The top row of table lists 29 cell lineages/tissues present in normal mouse embryos at the E8.5 stage of embryogenesis. The columns list the top 25 lineage-specific gene markers for each tissue. All lists were found on a single-cell molecular map of mouse gastrulation and early organogenesis. at https://marionilab.cruk.cam.ac.uk/MouseGastrulation2018/ (accessed on 22 October 2022) website. Genes that are significantly expressed at lower levels in Commd10Null embryos when compared with WT are shaded in red. Genes with higher expression in Commd10Null are shaded in yellow. Blue and green brackets below the table mark cell lineages/tissues involved in neurogenesis and heart development, respectively.
To verify our conclusions further, we examined the expression of statistically significant DEGs with the top 25 gene markers representing each embryonic cell type present in the mouse embryo at E8.5 (Figure 5). A single-cell molecular map of mouse gastrulation and early organogenesis [22] lists 29 different cell/tissue types for the E8.5 mouse embryo. The gene analysis revealed that the majority of genes with low expression in Commd10Null embryos are found in cells involved in early neural and heart development (Figure 5). Since recent studies demonstrated that neural crest cells develop into cardiomyocytes and contribute to heart development [52,53], gene expression deficiency in cardiomyocytes may be due to failed neural crest differentiation and/or cell migration.
We also performed gene functional enrichment analysis for the top 15–20 upregulated or downregulated DEGs using ToppGene Suite (https://toppgene.cchmc.org (accessed on 16 February 2023) [54]. We analyzed the top 20 genes downregulated in Commd10Null embryos and came up with a “GO: Biological Process” list of positive regulation of RNA biosynthetic process, epithelium development, animal organ morphogenesis, and brain and head development. We also analyzed the top 15 genes upregulated in Commd10Null embryos and selected the two top biological processes with the highest number of genes from the list: hemopoiesis and immune system development (Supplementary Tables).
Mice deficient in other members of the COMMD family, COMMD1 or COMMD9, were shown to be embryonically lethal. Commd1−/− embryos died between E9.5 and E10.5 due to defects in placenta vascularization [55]. Using genome-wide gene expression microarray analysis of embryonic RNA, the authors identified transcriptional upregulation of hypoxia-inducible factor 1 (HIF1) target genes in Commd1−/− embryos compared with their WT counterparts. Moreover, they demonstrated that COMMD1 may inhibit HIF1A stability and HIF1 activation by the physical association between the two proteins. Despite similarities in the timing of embryonic development failure between Commd1−/− and Commd10Null embryos, there were no similarities in gene expression patterns in the present study. Only Pfkp, one of eighteen hypoxia-associated DEGs upregulated in Commd1−/− versus WT embryos, was slightly upregulated in Commd10Null E8.5 embryos. Thus, the failure of Commd10Null embryos to thrive appears to have different underlying reasons compared to Commd1−/− embryos.
In contrast to Commd10Null embryos, Commd9−/− embryos die by E13.5 [56]. The authors found low levels of Hey1, Hey2, and Hes1 mRNA in the hearts of Commd9−/− embryos and concluded that the embryonic lethality of these mice was due to complex cardiovascular changes with signs of Notch deficiency. There were no differences in the mRNA expression of Notch or the genes listed above in Commd10Null embryos compared with WT. Taken together, these data indicate that COMMD1-, COMMD9-, and COMMD10-deficient mice display different underlying reasons for failed embryonic development and suggest that COMMD proteins play different critical roles during embryogenesis.
No direct connection between COMMD10 and Sox10 has been described in the scientific literature. We can only speculate as to how the absence of COMMD10 may lead to lower expression of Sox10 and, sequentially, other genes during embryogenesis. During normal embryogenesis, Sox10 mRNA appears in late gastrulating embryos (mouse E7.5) in the neural crest-forming region, and its gene expression depends on Wnt signaling [57,58]. Sox10 protein was also found to directly interact with β-catenin [59], which is activated in the canonical Wnt signaling pathway (reviewed in [60]). Wnt protein ligands bind to Frizzled family receptors (cell surface Fzd proteins and co-receptor Lrp5/6). Commd10Null embryos show significantly lower expression of Fzd3 and Fzd9 suggesting lower Wnt signaling potency. In addition, several Wnt ligands themselves were also dysregulated. There were higher levels of Wnt3 and Wnt9b while there were significantly lower levels of Wnt1, Wnt7a, and Wnt8b, suggesting dysregulation of Wnt signaling pathways in Commd10Null embryos. Wnt1-deficient mice exhibit a range of phenotypes, from early embryonic lethality to survival with severe ataxia [61]. Wnt7a signaling also controls multiple steps of neurogenesis [62]. It is plausible that by being part of the endosomal trafficking process inside the cell, COMMD10 may be involved in Wnt signaling regulation through as yet unknown mechanisms of Fzd receptor recycling or Wnt ligand secretion.
4. Limitations of the Study
The results described here characterize the timing of embryonic lethality of Commd10Null mice and also begin to demonstrate that neural plate developmental delay is the most likely cause of Commd10Null failed embryogenesis. The differential gene expression profile of Commd10Null as compared to normally developing WT embryos after E8.5 does not necessarily imply direct associations with COMMD10 deficiency. They rather verify the timing of embryonic failure by E8.5. Broader approaches and detailed analyses of earlier embryos are needed to pinpoint the exact role of COMMD10 in mouse embryogenesis, which are subjects of continued study and outside the scope of the present study.
5. Conclusions
Our study demonstrated that COMMD10 deficiency leads to embryonic lethality by day E8.5, most likely due to impaired neural plate and neural crest development processes resulting from the decreased expression of transcription factor Sox10 and several other genes. The molecular mechanism by which COMMD10 upregulates Sox10 expression remains unknown and merits further investigation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jdb11010013/s1, Figure S1: Complementary information for main figures; Supplementary Tables: GO:Biological Process of the top 15 genes upregulated in Commd10Null embryos and the GO:Biological Process of the top 20 genes downregulated in Commd10Null embryos; and a document with Supplementary figure titles and legends.
Author Contributions
Conceptualization, E.N.T. and I.F.D.; methodology, K.P.P., P.P. and G.W.; formal analysis, C.L. and M.B.; investigation, K.P.P., E.N.T., P.P. and A.V.T.; resources, I.F.D.; data curation, M.B.; writing—original draft preparation, A.V.T.; writing—review and editing, E.N.T., P.P., K.P.P. and I.F.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was carried out with help by the Oklahoma Medical Research Foundation Quantitative Analysis Core and supported by COBRE grant 1 P30 GM110766-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Institutional Review Board Statement
All housing and experimental use of mice were carried out at AAALAC-accredited facility in accordance with United States federal, state, local, and institutional regulations and guidelines governing the use of animals and were approved by OUHSC Institutional Animal Care and Use Committee (animal study protocol No. 20-028, approval date 8 May 2020).
Informed Consent Statement
Not applicable.
Data Availability Statement
Most data generated or analyzed during this study are included in this published article and its Supplementary Materials. Unprocessed RNA-seq raw data files and processed data files have been deposited on NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216492 (accessed on 13 March 2023). Further information and requests for materials should be directed to and will be fulfilled by the lead contact, Ian F. Dunn (ian-dunn@ouhsc.edu).
Acknowledgments
The authors thank Kathy J. Kyler for editing the manuscript, Hannah B. Homburg for assistance with mice, and Meng-Ling Wu for technical help with embryonic dissection techniques.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, K.E.; Healy, M.D.; Collins, B.M. Towards a molecular understanding of endosomal trafficking by Retromer and Retriever. Traffic 2019, 20, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Phillips-Krawczak, C.A.; Singla, A.; Starokadomskyy, P.; Deng, Z.; Osborne, D.G.; Li, H.; Dick, C.J.; Gomez, T.S.; Koenecke, M.; Zhang, J.S.; et al. COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Mol. Biol. Cell 2015, 26, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Bartuzi, P.; Billadeau, D.D.; Favier, R.; Rong, S.; Dekker, D.; Fedoseienko, A.; Fieten, H.; Wijers, M.; Levels, J.H.; Huijkman, N.; et al. CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. Nat. Commun. 2016, 7, 10961. [Google Scholar] [CrossRef]
- Qureshi, Y.H.; Baez, P.; Reitz, C. Endosomal Trafficking in Alzheimer's Disease, Parkinson's Disease, and Neuronal Ceroid Lipofuscinosis. Mol. Cell. Biol. 2020, 40, e00262-20. [Google Scholar] [CrossRef]
- Mallam, A.L.; Marcotte, E.M. Systems-wide Studies Uncover Commander, a Multiprotein Complex Essential to Human Development. Cell Syst. 2017, 4, 483–494. [Google Scholar] [CrossRef]
- Buckley, C.M.; Gopaldass, N.; Bosmani, C.; Johnston, S.A.; Soldati, T.; Insall, R.H.; King, J.S. WASH drives early recycling from macropinosomes and phagosomes to maintain surface phagocytic receptors. Proc. Natl. Acad. Sci. USA 2016, 113, E5906–E5915. [Google Scholar] [CrossRef]
- King, J.S.; Gueho, A.; Hagedorn, M.; Gopaldass, N.; Leuba, F.; Soldati, T.; Insall, R.H. WASH is required for lysosomal recycling and efficient autophagic and phagocytic digestion. Mol. Biol. Cell 2013, 24, 2714–2726. [Google Scholar] [CrossRef] [PubMed]
- Maine, G.N.; Burstein, E. COMMD proteins: COMMing to the scene. Cell. Mol. Life Sci. 2007, 64, 1997–2005. [Google Scholar] [CrossRef]
- van De Sluis, B.; Rothuizen, J.; Pearson, P.L.; van Oost, B.A.; Wijmenga, C. Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Hum. Mol. Genet. 2002, 11, 165–173. [Google Scholar] [CrossRef]
- Muller, P.A.; van de Sluis, B.; Groot, A.J.; Verbeek, D.; Vonk, W.I.; Maine, G.N.; Burstein, E.; Wijmenga, C.; Vooijs, M.; Reits, E.; et al. Nuclear-cytosolic transport of COMMD1 regulates NF-kappaB and HIF-1 activity. Traffic 2009, 10, 514–527. [Google Scholar] [CrossRef]
- Maine, G.N.; Mao, X.; Komarck, C.M.; Burstein, E. COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. EMBO J. 2007, 26, 436–447. [Google Scholar] [CrossRef]
- Mouhadeb, O.; Ben Shlomo, S.; Cohen, K.; Farkash, I.; Gruber, S.; Maharshak, N.; Halpern, Z.; Burstein, E.; Gluck, N.; Varol, C. Impaired COMMD10-Mediated Regulation of Ly6C(hi) Monocyte-Driven Inflammation Disrupts Gut Barrier Function. Front. Immunol. 2018, 9, 2623. [Google Scholar] [CrossRef] [PubMed]
- Ben Shlomo, S.; Mouhadeb, O.; Cohen, K.; Varol, C.; Gluck, N. COMMD10-Guided Phagolysosomal Maturation Promotes Clearance of Staphylococcus aureus in Macrophages. iScience 2019, 14, 147–163. [Google Scholar] [CrossRef] [PubMed]
- Burstein, E.; Hoberg, J.E.; Wilkinson, A.S.; Rumble, J.M.; Csomos, R.A.; Komarck, C.M.; Maine, G.N.; Wilkinson, J.C.; Mayo, M.W.; Duckett, C.S. COMMD proteins, a novel family of structural and functional homologs of MURR1. J. Biol. Chem. 2005, 280, 22222–22232. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, L.; Sun, Y.; Yang, M.; Wang, X.; Wu, X.; Huang, W.; Chen, L.; Pan, S.; Guan, J. Expression profile and bioinformatics analysis of COMMD10 in BALB/C mice and human. Cancer Gene Ther. 2020, 27, 216–225. [Google Scholar] [CrossRef]
- Goodwin, L.O.; Splinter, E.; Davis, T.L.; Urban, R.; He, H.; Braun, R.E.; Chesler, E.J.; Kumar, V.; van Min, M.; Ndukum, J.; et al. Large-scale discovery of mouse transgenic integration sites reveals frequent structural variation and insertional mutagenesis. Genome Res. 2019, 29, 494–505. [Google Scholar] [CrossRef]
- Bushnell, B. BBtools. Available online: http://sourceforge.net/projects/bbmap/ (accessed on 2 February 2022).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- de Boer, J.; Williams, A.; Skavdis, G.; Harker, N.; Coles, M.; Tolaini, M.; Norton, T.; Williams, K.; Roderick, K.; Potocnik, A.J.; et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003, 33, 314–325. [Google Scholar] [CrossRef]
- Pijuan-Sala, B.; Griffiths, J.A.; Guibentif, C.; Hiscock, T.W.; Jawaid, W.; Calero-Nieto, F.J.; Mulas, C.; Ibarra-Soria, X.; Tyser, R.C.V.; Ho, D.L.L.; et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 2019, 566, 490–495. [Google Scholar] [CrossRef]
- Pingault, V.; Zerad, L.; Bertani-Torres, W.; Bondurand, N. SOX10: 20 years of phenotypic plurality and current understanding of its developmental function. J. Med. Genet. 2022, 59, 105–114. [Google Scholar] [CrossRef]
- Zhao, F.; Satoda, M.; Licht, J.D.; Hayashizaki, Y.; Gelb, B.D. Cloning and characterization of a novel mouse AP-2 transcription factor, AP-2delta, with unique DNA binding and transactivation properties. J. Biol. Chem. 2001, 276, 40755–40760. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Bosserhoff, A.K.; Buettner, R.; Moser, M. A heart-hand syndrome gene: Tfap2b plays a critical role in the development and remodeling of mouse ductus arteriosus and limb patterning. PLoS ONE 2011, 6, e22908. [Google Scholar] [CrossRef]
- Zhou, C.; Tsai, S.Y.; Tsai, M.J. COUP-TFI: An intrinsic factor for early regionalization of the neocortex. Genes Dev. 2001, 15, 2054–2059. [Google Scholar] [CrossRef] [PubMed]
- Mehra-Chaudhary, R.; Matsui, H.; Raghow, R. Msx3 protein recruits histone deacetylase to down-regulate the Msx1 promoter. Biochem. J. 2001, 353, 13–22. [Google Scholar] [CrossRef]
- Zuber, M.E.; Gestri, G.; Viczian, A.S.; Barsacchi, G.; Harris, W.A. Specification of the vertebrate eye by a network of eye field transcription factors. Development 2003, 130, 5155–5167. [Google Scholar] [CrossRef]
- Takahashi, M.; Osumi, N. Pax6 regulates specification of ventral neurone subtypes in the hindbrain by establishing progenitor domains. Development 2002, 129, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Matsuo-Takasaki, M.; Tsuboi, I.; Kimura, K.; Salazar, G.T.; Yamashita, T.; Ohneda, O. Dual functions of hypoxia-inducible factor 1 alpha for the commitment of mouse embryonic stem cells toward a neural lineage. Stem Cells Dev. 2014, 23, 2143–2155. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Hu, J.; Zhang, J.; Zhou, X.; Li, X.; Gu, C.; Liu, T.; Xie, Y.; Liu, J.; Gu, M.; et al. Genome-wide association study identifies multiple susceptibility loci for craniofacial microsomia. Nat. Commun. 2016, 7, 10605. [Google Scholar] [CrossRef] [PubMed]
- Aruga, J.; Tohmonda, T.; Homma, S.; Mikoshiba, K. Zic1 promotes the expansion of dorsal neural progenitors in spinal cord by inhibiting neuronal differentiation. Dev. Biol. 2002, 244, 329–341. [Google Scholar] [CrossRef]
- Wapinski, O.L.; Vierbuchen, T.; Qu, K.; Lee, Q.Y.; Chanda, S.; Fuentes, D.R.; Giresi, P.G.; Ng, Y.H.; Marro, S.; Neff, N.F.; et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 2013, 155, 621–635. [Google Scholar] [CrossRef]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef]
- Treutlein, B.; Lee, Q.Y.; Camp, J.G.; Mall, M.; Koh, W.; Shariati, S.A.; Sim, S.; Neff, N.F.; Skotheim, J.M.; Wernig, M.; et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 2016, 534, 391–395. [Google Scholar] [CrossRef]
- Whittaker, D.E.; Oleari, R.; Gregory, L.C.; Le Quesne-Stabej, P.; Williams, H.J.; Torpiano, J.G.; Formosa, N.; Cachia, M.J.; Field, D.; Lettieri, A.; et al. A recessive PRDM13 mutation results in congenital hypogonadotropic hypogonadism and cerebellar hypoplasia. J. Clin. Investig. 2021, 131, e141587. [Google Scholar] [CrossRef]
- Hirata, T.; Nakazawa, M.; Yoshihara, S.; Miyachi, H.; Kitamura, K.; Yoshihara, Y.; Hibi, M. Zinc-finger gene Fez in the olfactory sensory neurons regulates development of the olfactory bulb non-cell-autonomously. Development 2006, 133, 1433–1443. [Google Scholar] [CrossRef]
- Jean, D.; Bernier, G.; Gruss, P. Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech. Dev. 1999, 84, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Toy, J.; Yang, J.M.; Leppert, G.S.; Sundin, O.H. The optx2 homeobox gene is expressed in early precursors of the eye and activates retina-specific genes. Proc. Natl. Acad. Sci. USA 1998, 95, 10643–10648. [Google Scholar] [CrossRef]
- Marçal, N.; Patel, H.; Dong, Z.; Belanger-Jasmin, S.; Hoffman, B.; Helgason, C.D.; Dang, J.; Stifani, S. Antagonistic effects of Grg6 and Groucho/TLE on the transcription repression activity of brain factor 1/FoxG1 and cortical neuron differentiation. Mol. Cell. Biol. 2005, 25, 10916–10929. [Google Scholar] [CrossRef] [PubMed]
- Wijchers, P.J.; Hoekman, M.F.; Burbach, J.P.; Smidt, M.P. Cloning and analysis of the murine Foxi2 transcription factor. Biochim. Biophys. Acta 2005, 1731, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Hienola, A.; Pekkanen, M.; Raulo, E.; Vanttola, P.; Rauvala, H. HB-GAM inhibits proliferation and enhances differentiation of neural stem cells. Mol. Cell. Neurosci. 2004, 26, 75–88. [Google Scholar] [CrossRef]
- Zou, P.; Muramatsu, H.; Sone, M.; Hayashi, H.; Nakashima, T.; Muramatsu, T. Mice doubly deficient in the midkine and pleiotrophin genes exhibit deficits in the expression of beta-tectorin gene and in auditory response. Lab. Investig. 2006, 86, 645–653. [Google Scholar] [CrossRef]
- Nakamura, E.; Kadomatsu, K.; Yuasa, S.; Muramatsu, H.; Mamiya, T.; Nabeshima, T.; Fan, Q.W.; Ishiguro, K.; Igakura, T.; Matsubara, S.; et al. Disruption of the midkine gene (Mdk) resulted in altered expression of a calcium binding protein in the hippocampus of infant mice and their abnormal behaviour. Genes Cells 1998, 3, 811–822. [Google Scholar] [CrossRef]
- Khokha, M.K.; Hsu, D.; Brunet, L.J.; Dionne, M.S.; Harland, R.M. Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat. Genet. 2003, 34, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Mankoo, B.S.; Skuntz, S.; Harrigan, I.; Grigorieva, E.; Candia, A.; Wright, C.V.; Arnheiter, H.; Pachnis, V. The concerted action of Meox homeobox genes is required upstream of genetic pathways essential for the formation, patterning and differentiation of somites. Development 2003, 130, 4655–4664. [Google Scholar] [CrossRef] [PubMed]
- Paththinige, C.S.; Sirisena, N.D.; Escande, F.; Manouvrier, S.; Petit, F.; Dissanayake, V.H.W. Split hand/foot malformation with long bone deficiency associated with BHLHA9 gene duplication: A case report and review of literature. BMC Med Genet. 2019, 20, 108. [Google Scholar] [CrossRef] [PubMed]
- Klopocki, E.; Lohan, S.; Doelken, S.C.; Stricker, S.; Ockeloen, C.W.; Soares Thiele de Aguiar, R.; Lezirovitz, K.; Mingroni Netto, R.C.; Jamsheer, A.; Shah, H.; et al. Duplications of BHLHA9 are associated with ectrodactyly and tibia hemimelia inherited in non-Mendelian fashion. J. Med Genet. 2012, 49, 119–125. [Google Scholar] [CrossRef]
- Patel, D.M.; Ahmad, S.F.; Weiss, D.G.; Gerke, V.; Kuznetsov, S.A. Annexin A1 is a new functional linker between actin filaments and phagosomes during phagocytosis. J. Cell Sci. 2011, 124, 578–588. [Google Scholar] [CrossRef]
- Blaybel, R.; Théoleyre, O.; Douablin, A.; Baklouti, F. Downregulation of the Spi-1/PU.1 oncogene induces the expression of TRIM10/HERF1, a key factor required for terminal erythroid cell differentiation and survival. Cell Res. 2008, 18, 834–845. [Google Scholar] [CrossRef]
- Yang, H.; Wang, X.X.; Zhou, C.Y.; Xiao, X.; Tian, C.; Li, H.H.; Yin, C.L.; Wang, H.X. Tripartite motif 10 regulates cardiac hypertrophy by targeting the PTEN/AKT pathway. J. Cell. Mol. Med. 2020, 24, 6233–6241. [Google Scholar] [CrossRef]
- Tang, W.; Martik, M.L.; Li, Y.; Bronner, M.E. Cardiac neural crest contributes to cardiomyocytes in amniotes and heart regeneration in zebrafish. eLife 2019, 8, e47929. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Wajid, S.; Demarest, B.L.; Yost, H.J. Loss of embryonic neural crest derived cardiomyocytes causes adult onset hypertrophic cardiomyopathy in zebrafish. Nat. Commun. 2018, 9, 4603. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef]
- van de Sluis, B.; Muller, P.; Duran, K.; Chen, A.; Groot, A.J.; Klomp, L.W.; Liu, P.P.; Wijmenga, C. Increased activity of hypoxia-inducible factor 1 is associated with early embryonic lethality in Commd1 null mice. Mol. Cell. Biol. 2007, 27, 4142–4156. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Koo, Y.; Mao, X.; Sifuentes-Dominguez, L.; Morris, L.L.; Jia, D.; Miyata, N.; Faulkner, R.A.; van Deursen, J.M.; Vooijs, M.; et al. Endosomal sorting of Notch receptors through COMMD9-dependent pathways modulates Notch signaling. J. Cell Biol. 2015, 211, 605–617. [Google Scholar] [CrossRef]
- Bhattarai, C.; Poudel, P.P.; Ghosh, A.; Kalthur, S.G. Comparative role of SOX10 gene in the gliogenesis of central, peripheral, and enteric nervous systems. Differentiation 2022, 128, 13–25. [Google Scholar] [CrossRef]
- Sutton, G.; Kelsh, R.N.; Scholpp, S. Review: The Role of Wnt/β-Catenin Signalling in Neural Crest Development in Zebrafish. Front. Cell Dev. Biol. 2021, 9, 782445. [Google Scholar] [CrossRef]
- Uka, R.; Britschgi, C.; Krättli, A.; Matter, C.; Mihic, D.; Okoniewski, M.J.; Gualandi, M.; Stupp, R.; Cinelli, P.; Dummer, R.; et al. Temporal activation of WNT/β-catenin signaling is sufficient to inhibit SOX10 expression and block melanoma growth. Oncogene 2020, 39, 4132–4154. [Google Scholar] [CrossRef]
- Zhong, Z.A.; Michalski, M.N.; Stevens, P.D.; Sall, E.A.; Williams, B.O. Regulation of Wnt receptor activity: Implications for therapeutic development in colon cancer. J. Biol. Chem. 2021, 296, 100782. [Google Scholar] [CrossRef]
- Thomas, K.R.; Capecchi, M.R. Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature 1990, 346, 847–850. [Google Scholar] [CrossRef]
- Qu, Q.; Sun, G.; Murai, K.; Ye, P.; Li, W.; Asuelime, G.; Cheung, Y.T.; Shi, Y. Wnt7a regulates multiple steps of neurogenesis. Mol. Cell. Biol. 2013, 33, 2551–2559. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).