Abstract
Although COVID-19 is not a pandemic anymore, the virus frequently mutates, resulting in new strains and presenting global public health challenges. The lack of oral antiviral drugs makes it difficult to treat him, which makes the creation of broadly acting antivirals necessary to fight current and next epidemics of viruses. Using the molecular docking approach, 118 compounds derived from marine organisms and 92 previously synthesized compounds were screened to assess their binding affinity for the main protease and papain-like protease enzymes of SARS-CoV-2. The best candidates from the xanthene, benzoxazole, and coumarin classes were identified. Marine-derived compounds showed slightly better potential as enzyme inhibitors, though the binding affinities of synthesized compounds were similar, with the best candidates displaying affinity values between 0.2 and 0.4 mM. Xanthenes, among both marine origin and synthesized compounds, emerged as the most promising scaffolds for further research as inhibitors. The papain-like protease was found to be more druggable than the main protease. Additionally, all top candidates met the criteria for various drug-likeness properties, indicating good oral bioavailability and low risk of adverse effects. This research provides valuable insights into the comparative affinities of marine origin and synthesized compounds from the xanthene, coumarin, and benzoxazole classes, highlighting promising candidates for further in vitro and in vivo studies.
1. Introduction
In late 2019, a highly contagious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was discovered in Wuhan, China. This virus quickly spread around the world, resulting in a disease known as COVID-19 (Coronavirus Disease 2019), which is responsible for severe acute respiratory syndrome and has affected millions of people worldwide, and the number is still increasing [1,2]. Officially, on 11 March 2020, the World Health Organization announced COVID-19 as a pandemic. The disease has spread to almost all countries on all continents. The number of reported cases of COVID-19 underestimates the true magnitude of the problem, given the unavailability of tests and the significant proportion of people who are asymptomatic or have such mild disease that they remain unrecognized/undiagnosed. According to data from the World Health Organization (WHO), by 5 December 2024, a total of 776,897,200 cases of SARS-CoV-2 infection were confirmed worldwide [3], including 7,076,329 deaths [4]. COVID-19 is shifting from pandemic to endemic, but the etiological agent, SARS-CoV-2, is still spreading and causing public health issues around the world. New variants, with multiple mutations in their spike proteins, are now able to evade neutralization by antibodies in vaccinated and convalescent individuals [5]. Consequently, there is a pressing need to develop broadly acting antivirals to address current and future virus outbreaks. Coronaviruses are enveloped viruses with single-stranded, positively oriented ribonucleic acid (RNA). Due to the presence of glycoproteins in the envelope, under an electron microscope, they have the appearance of a crown, which is how they got their name [6]. They belong to the Coronaviridae family, Orthocoronavirinae subfamily, and Nidovirales order. The Orthocoronavirinae subfamily is divided into four genera: Alphacoronavirus, Betacoronavirus, Deltacoronavirus, and Gammacoronavirus [7]. Members of this large family can cause respiratory, digestive, and neurological diseases in humans and various animal species, including camels, cats, and bats.
Coronaviruses can pass from one species to another and cause diseases of varying intensity in humans, from colds to severe infections, such as Middle East respiratory syndrome (MERS) or heavy acute respiratory syndrome (SARS) [8]. SARS-CoV-2 is a beta-coronavirus with a round or oval shape, varying in diameter from approximately 60 to 140 nm. The virus is sensitive to ultraviolet radiation and heat [9]. Like most coronaviruses, SARS-CoV-2 evolves rapidly.
Therefore, there are several types of viruses with different characteristics. Genetic evolution can occur over months or years and is often observable and measurable. Evolution was minimal at the beginning, and the D614G variant of the virus was dominant worldwide, which could be easily transmitted but did not cause severe illness [10]. However, due to frequent changes in the virus’s genome, many different types of the virus began to emerge [11,12]. Therefore, despite significant efforts by scientists and health communities to manage, treat, and develop vaccines for SARS-CoV-2 infections, the risk to human health remains high [13]. Treatment of conditions caused by SARS-CoV-2, RNA virus with high transmissible and mutation rate continues to be a global health challenge due to limited orally bioavailable antiviral drugs. The development of antivirals with alternative mechanisms of action has arisen [14,15,16]. Main protease (Mpro) and papain-like protease (PLpro) are proteins essential for viral replication and thus should be considered very important biochemical targets for drug development [17]. PLpro mediates the cleavage of the viral polyproteins pp1a and pp1ab at three sites to produce nonstructural proteins nsp1, nsp2, and nsp3 that are necessary for viral replication. It also antagonizes the host immune response by cleaving ubiquitin and the ubiquitin-like interferon-stimulated gene 15 (ISG15) protein from host protein conjugates [18]. Mpro is a cysteine protease that contains a catalytic dyad consisting of the nucleophilic cysteine 145 (C145) and histidine 41 (H41) [19], cleaves interleukin-1 receptor-associated kinase 1 (IRAK1), TAK1 binding protein (TAB1), and mRNA-decapping enzyme 1A (DCP1A) to disturb the production of pro-inflammatory cytokines and attenuate the immune defense [20]. Although Mpro and PLpro are both nucleophilic cysteine proteases, they differ in their structures, catalytic mechanisms, and substrate selectivities. Mpro is predominantly homodimeric and employs an active site Cys–His dyad to catalyze the hydrolysis of the SARS-CoV-2 polyproteins at 11 sites. On the contrary, PLpro is likely monomeric and employs an active site Cys–His–Asp triad to catalyze the hydrolysis of the polyprotein at the nsp1/2, nsp2/3, and nsp3/4 sites [21]. PLpro shares structural similarities with various human cysteine proteases, potentially leading to cross-reactivity issues with inhibitor development, whereas Mpro has no homologous human proteases, therefore offering specific inhibition of its proteolytic activity without significant impact on host cells, which makes Mpro a more favorable target for anti-SARS-CoV-2 drug development [22]. Inhibiting the catalytic activity of Mpro and PLpro in SARS-CoV-2, which constitutes a key aspect for the virus replication, offers promising strategies for mitigating COVID-19 by disrupting viral replication, reducing viral load, and enhancing host immune response, thus potentially ameliorating disease severity [23].
Marine natural products are important biomedical resources for the development of marine pharmaceuticals, bioactive compounds, and agents. They have bioactive properties such as antioxidant, anti-infection, anti-inflammatory, anticoagulant, anti-diabetic, cancer treatment, and immune improvement. Marine natural products were studied for their anti-infection properties against coronavirus, SARS-CoV-2 and its major variants (such as Delta and Omicron), tuberculosis, H. pylori, and HIV infections, as well as their potential as biomedical resources for infection-related cardiovascular disease (irCVD), diabetes, and cancer [24].
Prioritizing screening of compounds against SARS-CoV-2 in the search for innovative potential therapies to treat or prevent COVID-19 can be aided by a thorough evaluation of the antiviral properties linked to well-known marine natural products. Numerous compounds with antiviral properties, including those that suppress coronaviruses, have been found in both terrestrial and marine sources [25]. Flavonoids, phlorotannins, alkaloids, terpenoids, peptides, lectins, polysaccharides, lipids, and other marine-derived biologically active substances can affect coronaviruses during the stages of viral particle penetration and entry into the cell, viral nucleic acid replication, and virion release from the cell; they can also act on the host’s cellular targets. These natural chemicals could be a valuable tool in the fight against coronaviruses [26]. Many studies have found that natural compounds have activity against the coronavirus SARS-CoV-2, which causes SARS. Recent research indicates that natural compounds derived from the isoprenoid, peptide, polyketide, binaphthoquinone, and polyphenol structural groups have potent anti-SARS-CoV-2 activity in vitro. Natural products classified by molecular targets include SARS-CoV-2 chymotrypsin-like protease (3CLpro)/major protease (Mpro) are dieckol, tannic acid, savinin, naringenin, esculetin-4-carboxylic acid ethyl ester, and amentoflavone [27,28].
Recently, the repurposed drug plitidepsin (also known as dehydrodidemnin B, a depsipeptide from the marine ascidian Aplidium albicans) was shown to be more than 20 times more potent than remdesivir against SARS-CoV-2 [29].
Other natural compounds with antiviral activity reported in the literature are stachyflin, active against the influenza A virus [30]; topsentin and mycalamide, active against coronavirus A59 [31]; N-butyl harmine, which inhibits HIV viral replication [32]; tetrandrine, active against human coronavirus OC43 [33]; saikosaponin B2, active against human coronavirus 229E [34]; and epigallocatechin gallate, active against SARS-CoV-2 strain [35]. Three separate marine natural products were found to have antiviral properties: homofascaplysin A, (+)-aureole, and bromophycolide A. These compounds were shown to be able to prevent the replication of SARS-CoV-2 at quantities that do not harm human airway epithelial cells. These substances represented a good one for future investigation to find novel treatments for SARS-CoV-2 [25]. New diketopiperazines (aspamides) and indolyl diketopiperazines (aspamides) have been isolated from the solid culture of Aspergillus versicolor, an endophyte with the sea crab (Chiromantes haematocheir), together with 11 known diketopiperazines and intermediates. The coronavirus 3-chymotrypsin-like protease (Mpro) of the SARS-CoV-2 was used to virtually screen all isolated compounds. The compounds that were mentioned had the highest docking scores of all the screened molecules, suggesting that they could be useful in the fight against SARS-CoV-2 [36]. Numerous other studies were conducted in an attempt to screen for potential natural origin inhibitors against SARS-CoV-2 [37,38,39].
Marine natural items offer higher therapeutic benefits for COVID-19 treatment than other chemical drugs, which have been linked to negative cardiovascular consequences. Protecting this environment is crucial for the long-term development of the ocean economy. As vast, innovative, and promising medicinal resources for anti-infection against SARS-CoV-2 and irCVD, marine natural compounds’ novel potential processes may involve many targets and pathways controlling human immunity and reducing inflammation [40].
We have previously reported numerous syntheses, structures, and biological activity for various categories of compounds. For many years, we have been studying 4-hydroxycoumarin derivatives, aryl-substituted xanthenes, and aryl-substituted benzoxazoles. Docking, a highly advanced computer approach for predicting the location of drug binding to a critical protein, was used extensively in those studies. In numerous studies, we examined Lipinski’s rule of five, which refers to the probability of manufactured compounds and prospective medications passing per os [41,42,43]. Based on the above, we utilized the SeaSAR v12 software to analyze 118 compounds from marine organisms as well as 92 previously synthesized compounds to test their binding to enzymes important for the life of SARS-CoV-2: the main protease of SARS-CoV-2 and the papain-like protease. An important method that enables this is docking, a computational process that predicts the binding of a ligand (drug) to a receptor and its orientation at the binding site. Also, to test the possible use of compounds of this type per os, it is necessary to test Lipinski’s rule of five and other drug-likeness parameters, as well as their toxicity. The compounds that showed the best results in the preliminary tests were subjected to more detailed analyses.
2. Materials and Methods
SeeSAR v12.1 (SeeSAR version 12.1; BioSolveIT GmbH, Sankt Augustin, Germany, 2022, www.biosolveit.de) software was used for the molecular docking study. The 3D structures of two SARS-CoV-2 proteins were selected and used as targets: the main protease (PDB ID: 7ZQV) in complex with the antiviral compound AG7404 (rupintrivir-like molecule, AG7800), and the papain-like protease (PDB ID: 7TZJ) in complex with inhibitor 3k (piperidine structure inhibitor). Natural compounds of coumarin, xanthene, and benzoxazole structure (118 structures in total) that naturally occur in marine organisms, as well 92 compounds from our in-house library, previously synthesized (56 xanthenes, 23 coumarins, 11 benzoxazoles, thymoquinone, and amino thymoquinone) [42,43,44,45,46,47,48,49,50,51], were selected and prepared as ligands. Molecules used for benchmarking with our tested compounds were FDA-approved Mpro inhibitor nirmatrelvir [52] and experimental drug Jun12682 [53] as PLpro inhibitor. For the parameter settings for the docking analysis, the initial settings of the SeeSAR program were retained. The initial settings in SeeSAR are typically designed to accommodate a broad range of compounds, including marine origin natural compounds. Marine origin compounds often possess unique structural characteristics, such as high molecular flexibility, unusual ring systems, and complex stereochemistry, whereas our synthesized compounds are more rigid, often planar, and lack complex stereochemistry. The hydrogen dehydration scoring function Equation (1) (HYDE) was used in SeeSAR to test the docking affinity estimate [54,55]. The best candidate molecules were selected based on their estimated affinity.
In Equation (1), ΔGdehydration represents the change in dehydration energy, while ΔGH-bonds represents the change in hydrogen bond (H-bond) energy for each atom i in the protein–ligand complex. HYDE’s scoring function is determined in terms of desolvation and hydration, which are conventionally related to octanol–water partition coefficients. The ligand molecule is completely dehydrated during the binding process.
The FlexX docking function in SeeSAR places the ligand in the binding site. It is based on an incremental construction algorithm [56,57]. Ligands are divided into so-called fragments; the initial fragment (or combinations thereof) is placed at multiple locations in the binding pocket and is achieved using a simple but very fast pre-scoring scheme. From the set n solutions, the ligand is further built, fragment by fragment, and the temporary solutions are scored against each other. The best results are presented in a report at the end.
Optibrium—ADME Properties Prediction
Physicochemical properties are an important factor to consider when deciding whether a compound is a promising drug candidate. Early assessment of ADME parameters can predict toxicity issues and absorption challenges in the early stages of drug discovery, saving time and resources. SeeSAR uses the optional StarDrop module (Optibrium) to predict many parameters, such as rotatable bonds, molecular weight, topological polar surface area (TPSA), and ADME (absorption, distribution, metabolism, excretion) parameters such as affinity for CYP enzymes, blood–brain barrier passage (BBB), logS, and more. A detailed explanation, as well as references to the methods behind the calculation of the different Optibrium properties, can be found in the Optibrium Reference Guide, Optibrium Ltd., StarDrop, Waterbeach, UK, 2022 (www.optibrium.com).
3. Results
Molecular docking studies were used to screen and examine the affinity for 118 molecules that occur naturally in marine organisms, as well as for 92 synthetic compounds synthesized previously. As the reference compounds, for the docking analysis with the main protease, the FDA-approved Mpro inhibitor nirmatrelvir was used, while for the docking analysis with the papain-like protease, an experimental drug, jun12682, was used as a PLpro inhibitor. Detailed results for all compounds are available as Supplementary Material.
The results of the docking study of the best marine origin (Figure 1) and synthesized compounds (Figure 2) with the main protease of SARS-CoV-2 are shown in Table 1 and Table 2.
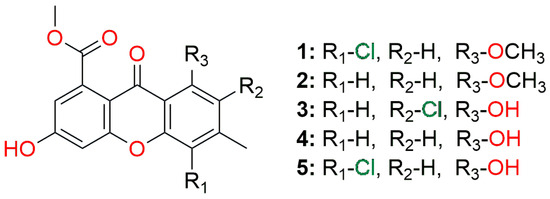
Figure 1.
The general structure of the best marine origin compounds (Table 1) screened against the main protease.
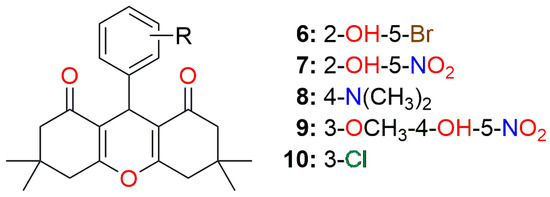
Figure 2.
The general structure of the best synthesized compounds (Table 2) screened against the main protease.

Table 1.
Results of the docking study and OPTIBRIUM parameters for the top five marine origin molecules with the main protease SARS-CoV-2 (PDB ID: 7ZQV).

Table 2.
Results of the docking study and OPTIBRIUM parameters for the top five synthesized molecules with the main protease of SARS-CoV-2 (PDB ID: 7ZQV).
According to the molecular docking analysis with the Mpro, the lower/upper boundary for the estimated affinity for nirmatrelvir (Figure 3) was estimated to be 2.448/243.213 mM (Supplementary Tables S1 and S2).
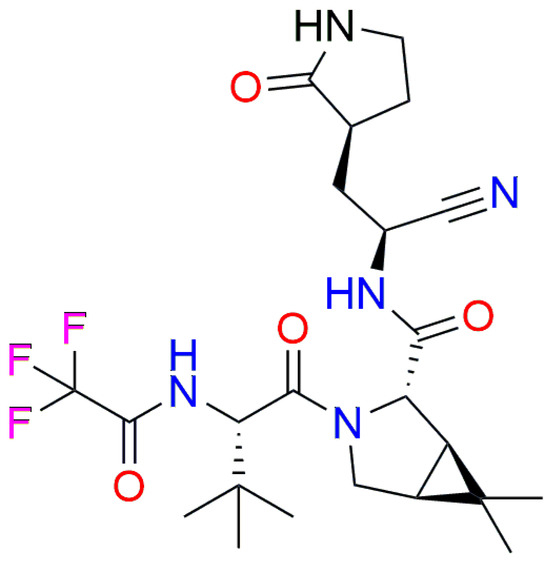
Figure 3.
Structure of nirmatrelvir.
Although nirmatrelvir is an FDA-approved Mpro inhibitor, surprisingly, its affinity showed to be almost 500-fold weaker in affinity toward Mpro compared to compound 6 and 1200-fold weaker compared to compound 1.
The results of the docking study of the best marine origin (Figure 4) and synthesized compounds (Figure 5) with the SARS-CoV-2 papain-like protease are shown in Table 3 and Table 4.
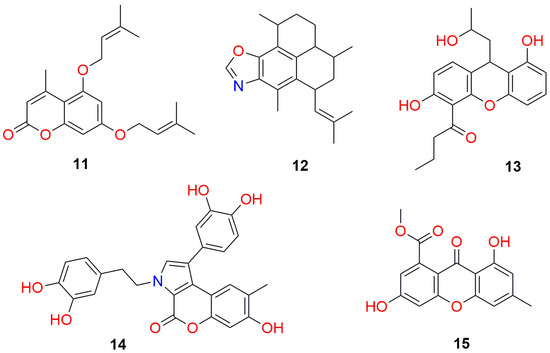
Figure 4.
Structures of the best marine origin compounds (Table 3) screened against papain-like protease.
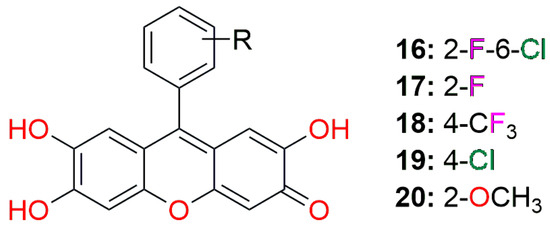
Figure 5.
The general structure of the best synthesized compounds (Table 4) screened against papain-like protease.

Table 3.
Results of the docking study and OPTIBRIUM parameters for the top five marine origin molecules with the papain-like protease SARS-CoV-2 (PDB ID: 7TZJ).

Table 4.
Results of the docking study and OPTIBRIUM parameters for the top five synthesized molecules with the papain-like protease SARS-CoV-2 (PDB ID: 7TZJ).
Since there are no FDA-approved drugs for targeting SARS-CoV-2 PLpro, we used the experimental drug Jun12682 (Figure 6) for benchmarking. According to the molecular docking analysis with the papain-like protease, the lower/upper boundary for the estimated affinity for Jun12682 was estimated to be 2.173/215.899 mM (Supplementary Tables S3 and S4). This inhibitor also came up as much weaker in affinity toward PLpro compared to tested marine and synthesized compounds.
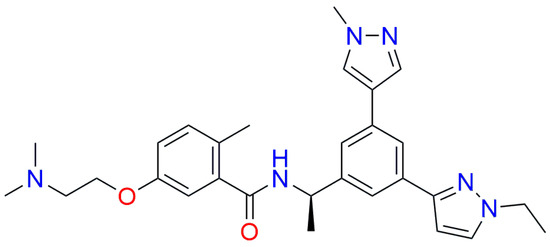
Figure 6.
Structure of Jun12682.
Due to a large pool of results, our discussion will cover the best five compounds for each target enzyme and group of ligands in terms of binding affinity values and drug-likeness properties.
A docking study on the main protease SARS-CoV-2 (PDB ID: 7ZQV) revealed interesting insight into ligand–enzyme interactions. All of the five best marine origin compounds are of xanthene structures. The basis of these compounds is 2-hydroxy-4-acetoxy-7-methylxanthen-9-one, which comes in several variations, as 5-hydroxy or 5-methoxy, and with 6- or 8-chloro substituents (Figure 1, Figure 7 and Figure 8).
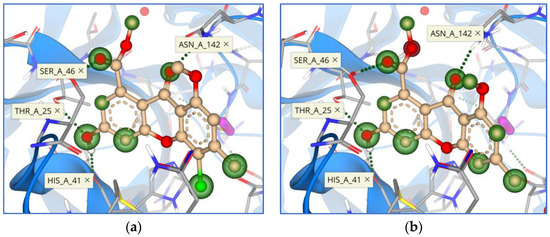
Figure 7.
Binding mode and contacting amino acid residues of the compounds 1 (a) and 2 (b) within the main protease active site shown with HYDE contributions of individual atoms to the total affinity (green circled atoms).
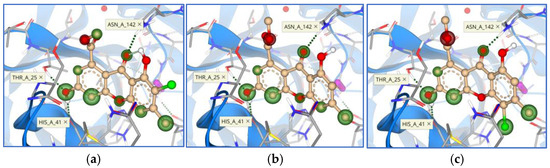
Figure 8.
Binding modes and contacting amino acid residues of the marine compounds 3 (a), 4 (b), and 5 (c) within the main protease active site, with HYDE contributions of individual atoms to the total affinity (green circled atoms).
The upper boundary for estimated affinity (mM) ranged from 0.224 and 0.442 for the first two compounds, while it drastically increased for the third compound and above (>3.5 mM) (Table 1). This 10-fold decrease in the affinity was surprising since the structures are quite similar; however, this took us to another conclusion that position 5, as methoxy substituted in the first two structures, was crucial for achieving higher affinity.
A chlorine atom is present in three of the five best candidates, which seems to also play an important role in the process of binding to the main protease of the SARS-CoV-2 virus.
Amino acid residues that were most frequently represented in interactions with ligands were Thr 25, His 41, Ser 46, and Asn 142 (Figure 7 and Figure 8). The most interesting interaction for all compounds is with the hydrogen bond between the 2-hydroxy group and His 41, which is part of the catalytic dyad of this enzyme.
Figures represent HYDE contributions of individual atoms to the total affinity (green circled atoms). The higher the contribution of the atom to total affinity, the bigger the green circle. Figures also show red circled atoms as the opposite contribution to green atoms.
In the top 20 derivatives (Supplementary Material Table S1), other core structures worth mentioning are coumarin derivatives and condensed coumarin systems with furan and pyrrole. Not a single derivative of the tested benzoxazoles is among the top 20 derivatives.
A docking study of synthesized derivatives of xanthene, coumarin, and benzoxazole moieties on main protease showed that predominantly the best candidates were 9-aryl-substituted derivatives of xanthene-1,8-dione (Figure 2, Figure 9 and Figure 10).
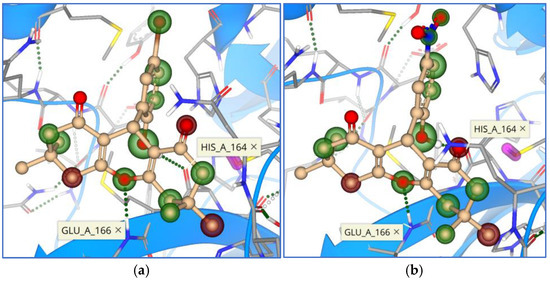
Figure 9.
Binding mode and contacting amino acid residues of the synthesized compounds 6 (a) and 7 (b) within the main protease active site shown with HYDE contributions of individual atoms to the total affinity (green circled atoms).
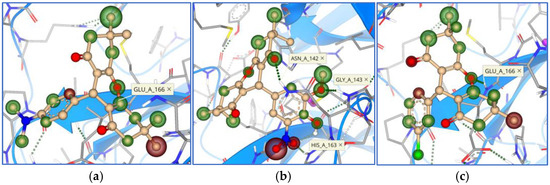
Figure 10.
Binding modes and contacting amino acid residues of the synthesized compounds 8 (a), 9 (b), and 10 (c) within the main protease active site, with HYDE contributions of individual atoms to the total affinity (green circled atoms).
Of the 20 most potent derivatives (Supplementary Material Table S2), 14 of them are derivatives of xanthene-1,8-dione, and the remaining 6 are derivatives of 3-cinnamoyl-4-hydroxycoumarin. Unlike xanthene-3-one derivatives, whose basic xanthene heterocycle is planar, xanthene-1,8-diones are not planar, and their geometry shows a certain curvature of the basic tricyclic system. It seems that this flexibility of the xanthene-1,8-diones is responsible for the better interaction with the main protease and the better binding affinity values.
On the other hand, in coumarin derivatives, the core is planar. However, the coumarin heterocycle is smaller than that of xanthene, and a 3-cinnamoyl substituent part of the molecule is more flexible compared to planar coumarin. We can conclude that these compounds can also occupy the appropriate position in the active site of the main protease.
Only the best among those synthesized, compound 9-(5-bromo-2-hydroxyphenyl)-3,3,6,6-tetramethylxanthene-1,8(2H)-dione, was comparable in binding affinity (0.473 mM) with the marine origin compounds, while the rest four were at least 2-fold weaker (>0.747 mM) (Table 2).
Unlike marine origin compounds, synthesized xanthene-1,8-diones did not interact with the His 41 residue. The most frequent amino acids involved in interaction were Asn 142, Gly 143, Ser 144, His 163, His 164, and Glu 166 (Figure 9 and Figure 10).
Synthesized xanthen-3-one derivatives and benzoxazole derivatives did not reach high places in terms of receptor binding affinity (available in Supplementary Material Table S2) compared to the previously discussed compounds.
A docking study on papain-like protease SARS-CoV-2 (PDB ID: 7TZJ) resulted in quite heterogeneous top five marine origin candidates (Table 3). Two out of the best five candidates are of coumarin core, two are xanthenes, and one is a benzoxazole derivative (Figure 4). For some of them, additional condensed cyclic structures are attached to the main core. Observed amino acid residue interactions with the ligands were with Asp 164, Arg 166, Tyr 264, Gly 266, and Tyr 268 (Figure 11 and Figure 12).
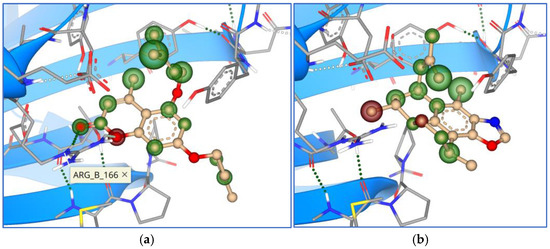
Figure 11.
Binding mode and contacting amino acid residues of the marine compounds 11 (a) and 12 (b) within papain-like protease active site shown with HYDE contributions of individual atoms to the total affinity (green circled atoms).
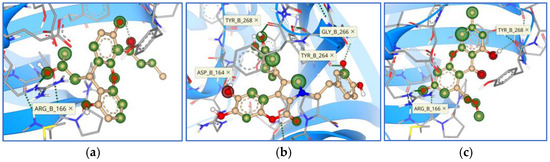
Figure 12.
Binding modes and contacting amino acid residues of the marine compounds 13 (a), 14 (b), and 15 (c) within papain-like protease active site, with HYDE contributions of individual atoms to the total affinity (green circled atoms).
From this study, we retrieved the best of all the candidates, 4-methyl-5,7-bis((3-methylbut-2-en-1-yl)oxy)coumarin (Figure 4, compound 11, Figure 11), with the upper boundary affinity value of only 0.01 mM, positioning it as 30-fold stronger inhibitor than the rest of the tested compounds.
The affinity values for the other four compounds were in the range from 0.299 to 0.483 mM, which indicates papain-like protease is a more easily and druggable target by our set of investigated compounds compared to the main protease. Another interesting finding was that compound 2,5-dihydroxy-4-acetoxy-7-methylxanthen-9-one (Figure 4, compound 15) from the five best marin origin compounds tested against papain-like protease (Table 3, compound 15), is also among the top five compounds from Table 1 (compound 4). Although quite different in the affinity values toward these two enzymes, this is the only candidate from the top tier that could be considered a dual target inhibitor.
Contrary to the docking study of synthesized compounds with the main protease (Table 2), for the inhibition of papain-like protease, the best candidates are derivatives of 9-aryl-substituted 2,6,7-trihydroxyxanthen-3-one with the planar main xanthene core. Different only by the substituents on the aromatic ring at position 9 (Figure 5, Figure 13 and Figure 14), four out of the five best candidates are rich in halogen substituents, and only the fifth one is the methoxy derivative.
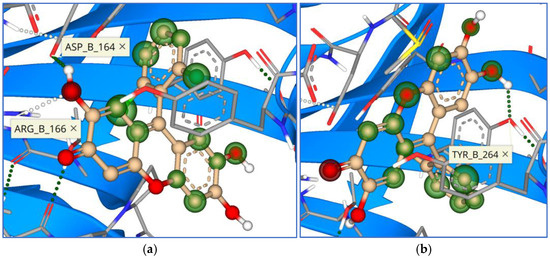
Figure 13.
Binding mode and contacting amino acid residues of the best synthesized compounds 16 (a) and 17 (b) within papain-like protease active site shown with HYDE contributions of individual atoms to the total affinity (green circled atoms).
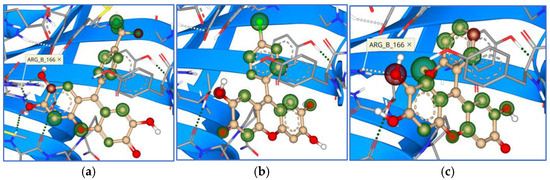
Figure 14.
Binding modes and contacting amino acid residues of the best synthesized compounds 18 (a), 19 (b), and 20 (c) within papain-like protease active site, with HYDE contributions of individual atoms to the total affinity (green circled atoms).
Calculated affinity values ranged from 0.171 to 0.286 mM (Table 4), indicating synthesized xanthene-3-one derivatives as overall best candidates against papain-like protease, confirming the previous conclusion that papain-like protease is an easily and more druggable target. Observed interactions were similar with marine compounds, mostly with amino acid residues Asp 164, Arg 166, and Tyr 264 (Figure 13 and Figure 14).
4. Discussion
The druggability of SARS-CoV-2 main protease (Mpro) and papain-like protease (PLpro) is shaped by key structural differences that affect how drugs interact with their active sites and their inhibition potential. Mpro has a well-defined, highly conserved active site with a catalytic dyad (Cys145 and His41), allowing for flexibility in substrate binding and making it a promising target for small molecules [58,59]. In contrast, PLpro features a more complex catalytic triad (Cys111, His272, and Asp286), and its larger, more flexible active site presents challenges for inhibitor design [53]. The higher binding affinity of tested compounds toward PLpro could be attributed to its larger and more flexible active site, as well as its dynamic conformational flexibility. These factors provide a broader surface area and more diverse interactions for potential ligands, improving binding affinity.
Docking studies offer valuable preliminary insights into potential ligand–protein interactions, but they come with significant limitations that must be considered. These include the potential for false positives and false negatives, limited ability to represent dynamic flexibility, and challenges in accurately modeling solvent effects and protein flexibility. To overcome these limitations, docking results are often supplemented with more advanced methods, such as molecular dynamics simulations, free energy calculations, or experimental validation.
Genetic variations have a profound impact on both the binding affinity and druggability of biomolecules. By altering protein structures, active sites, and interaction dynamics, genetic mutations can affect how well a drug binds and how effective it is in treating diseases. This understanding plays a crucial role in drug design, therapeutic development, and overcoming challenges like drug resistance.
Xanthene derivatives display different binding modes across various proteases, owing to variations in the active sites, catalytic mechanisms, and the flexibility of the target enzymes. Mechanistically, they typically engage with the protease’s active site through a mix of hydrogen bonding, hydrophobic interactions, and steric effects. These derivatives often form critical interactions with residues essential for catalytic activity. This is particularly evident in the interactions between marine origin xanthene compounds and Mpro, where they form hydrogen bonds with His41, a key amino acid in the catalytic dyad.
When exploring visually the mechanism and modes of interaction between tested compounds and the Mpro, only the top five marine compounds kept uniform orientation within the active site (Figure 15), while synthesized compounds, even if they are of similar structure, bound in different orientations to the Mpro (Figure 16).
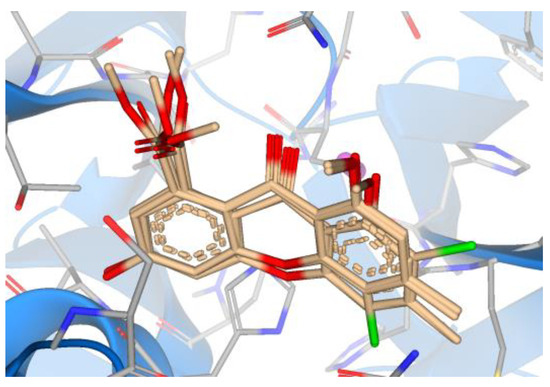
Figure 15.
Overlay of the top five marine compounds within the active site of Mpro.
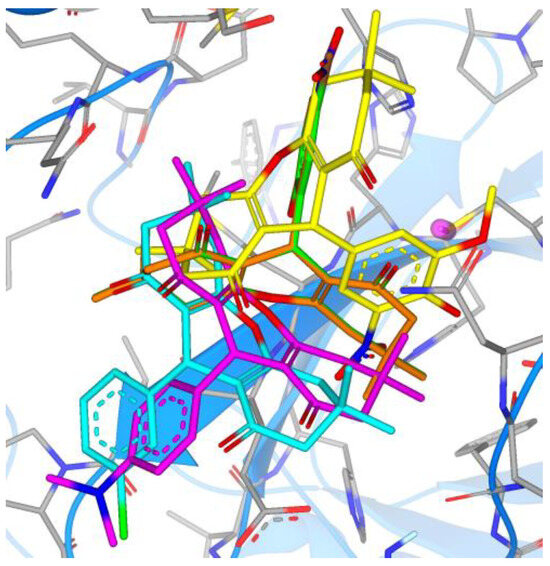
Figure 16.
Overlay of the top five synthesized compounds within the active site of Mpro (6—green, 7—orange, 8—magenta, 9—cyan, 10—yellow).
Compounds 6 and 7 overlayed perfectly in one binding mode, while compounds 8 and 9 overlayed in a different mode. Compound 10 (yellow in Figure 16) was bound in a totally different mode, with the 9-ary substituent away from the active site.
Comparing the binding modes of the tested marine compounds with PLpro, it is evident that molecules also bound in different modes, which was not unexpected due to their different structures (Figure 17). On the contrary, synthesized xanthene-3-ones showed a uniform mode of binding, where four out of five compounds overlayed well, and only compound 20 had the opposite orientation (Figure 18).
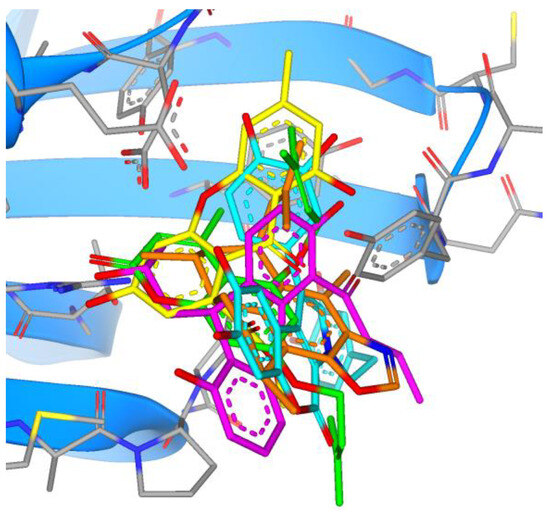
Figure 17.
Overlay of the top five marine compounds within the active site of PLpro (11—green, 12—orange, 13—magenta, 14—cyan, 15—yellow).
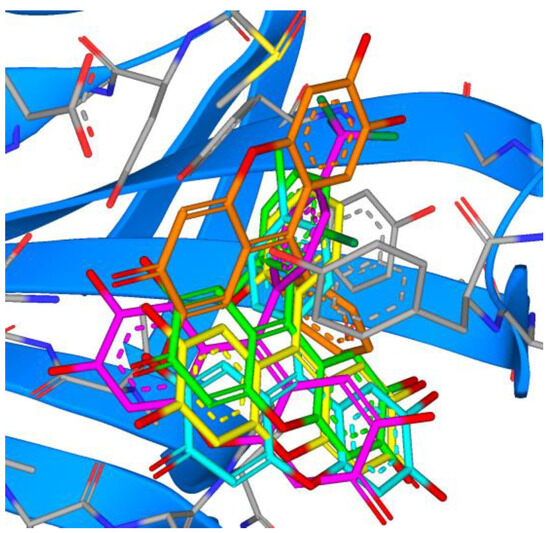
Figure 18.
Overlay of the top five synthesized compounds within the active site of PLpro (16—green, 17—orange, 18—magenta, 19—cyan, 20—yellow).
In the initial stages of testing biologically active compounds, a number of molecular parameters and pharmacokinetic properties are used to provide information about the potential of some compounds to become drugs. The ability of a drug to penetrate a membrane is one of the first parameters tested in the new drug modeling process. Lipinski’s rule of five is a well-known set of parameters that is useful in this process. According to this rule, a molecule will be able to passively pass through the intestinal membrane if its properties are within the following range: molecular weight less than 500 g/mol, log P value less than 5 representing its lipophilicity, no more than 5 hydrogen bond donors and no more than 10 hydrogen bond acceptors [60]. Further research extended this set by several more conditions, such as molar refraction (a measure of the total polarity of molecules) in the range of 40 to 130, topological polar surface area (TPSA) less than or equal to 140 Å2, and less than 10 rotatable bonds (RB) [61,62] that closely describe the permeability and flexibility of the drug. Therefore, if a compound follows these rules and parameters, it is expected to be orally bioavailable. If a molecule violates any two or more of the above requirements, it is assumed that it will not be able to penetrate by passive intestinal diffusion. In addition to absorption, other pharmacokinetic parameters of a compound can be predicted based on lipophilicity, including distribution, elimination, and toxicity [61]. Additional physical–chemical properties related to previous parameters are logD and logS. The logarithm of the n-octanol/water partition coefficient at the physiological pH 7.4 (logD) is used to describe the relationship between lipophilicity and hydrophilicity of an ionized compound. LogS @ pH 7.4 is the logarithm of intrinsic aqueous solubility at physiological pH 7.4 in μM for ionizable compounds. Human intestinal absorption (HIA) and plasma protein binding (PPB) are important for drug transport through the body [63]. Compounds that are predicted to be absorbed by greater than or equal to 30% are classified with a ‘+’, while compounds that are predicted to be absorbed by <30% are classified with a ‘−’. Drugs with high protein binding tend to have longer half-lives than those with lower values. Predictions are ‘low’ for compounds that are <90% bound and ‘high’ for compounds that are >90% bound. The more the drug is bound to plasma proteins, the smaller the fraction of free drug for therapeutic effect [64].
Permeability through the blood–brain barrier (BBB) is a prerequisite for compounds to show their potential effects on the central nervous system [65,66]. CYP2C9 pKi prediction represents affinity estimates of the compound to bind at the enzyme involved in several metabolic drug pathways. CYP2D6 classification represents the predicted compound’s value to be in one out of four categories: ‘low’ for compounds with a pKi < 5, ‘medium’ for compounds with a pKi between 5 and 6, ‘high’ for compounds with a pKi between 6 and 7, and very high for compounds with a pKi > 7. The involvement of P-glycoprotein 1 protein, also known as multidrug resistance protein 1 (MDR1) and ATP-binding cassette subfamily B member 1 (ABCB1)), is relevant for many unwanted effects. P-glycoprotein 1 transport classification predicts whether a compound is likely to be a substrate of P-gp or not. Prediction of pIC50 values for inhibition of human Ether-a-go-go Related Gene (hERG) potassium channels expressed in mammalian cells is highly relevant for drug effects on the heart. The top five marine origin compounds screened on the main protease were predicted to have good oral bioavailability. Log P values ranged from 2.93 to 3.86, and the rest of the parameters (Mw, number of hydrogen bond donors and acceptors, the number of rotatable bonds, MR and TPSA values) fit the rules for drug-likeness. As can be seen from Table 1, all compounds have positive intestinal absorption, while plasma protein binding values are high. The top five synthesized compounds screened on the main protease were also predicted to have good oral bioavailability. Log P values for these compounds were somewhat higher (3.88–5.00) but still within the recommended range of 4/15, as were the rest of the parameters. As can be seen from Table 2, all compounds have positive intestinal absorption. The top five marine origin compounds tested against papain-like protease were predicted to also have good oral bioavailability as their log P values ranged from 2.51 to 4.28 with one exception of compound 7, whose value was 6.46 (Table 3). The compounds also fit the rest of the rules, except for compound 9, which had six H-bond donors and a TPSA value of around 150 Å2. As previously stated, even if a compound violates one of the rules, it is still expected to have good oral bioavailability. As can be seen from Table 3, all compounds except 9 have positive intestinal absorption, while plasma protein binding values are mixed. The top five synthesized compounds tested against papain-like protease were predicted to have good oral bioavailability as well, with log P values ranging from 3.23 to 3.95. The rest of the parameters also fit within drug-likeness rules. As can be seen from Table 4, all compounds have positive intestinal absorption, while plasma protein binding values are high except for compound 20.
Although our findings of potent inhibitors for SARS-CoV-2’s Mpro and PLpro are applicable to other coronaviruses due to the conservation of these proteases, significant differences in their structure, function, and interactions with the host could influence their druggability. For instance, the high affinity of PLpro in SARS-CoV-2 may not be fully applicable to other viruses if there are structural variations in the protease or if the virus utilizes different immune modulation mechanisms. Thus, while these findings provide valuable guidance for the development of broad-spectrum antiviral agents, further research is necessary to better understand the specific characteristics of other coronaviruses and their proteases.
The detailed understanding of how ligands interact with the active sites of main protease and papain-like protease from docking studies can serve as the foundation for selecting lead compounds that show promise in vitro. Docking studies help identify the most potent inhibitors based on binding affinity and predicted pharmacodynamics. These lead compounds can then be tested for their ability to inhibit the proteases in laboratory settings.
The molecular insights gained from docking studies of SARS-CoV-2 proteases (Mpro and PLpro) are critical for guiding in vivo and clinical translation. These insights can help identify promising lead compounds, optimize them for better efficacy and safety profiles, and ensure their potential for clinical success. By using in silico predictions as a foundation, researchers can streamline the drug discovery process, ultimately accelerating the development of effective antiviral therapies for COVID-19 and potentially other coronavirus infections.
This study significantly contributes to the field of SARS-CoV-2 protease inhibitors by introducing novel chemical scaffolds (xanthene, coumarin, and benzoxazole derivatives) derived from marine sources and synthesized compounds. These compounds offer new binding modes and mechanisms of inhibition, particularly against both Mpro and PLpro. The study also advances the concept of broad-spectrum antiviral agents. Integrating both marine and synthetic chemistry opens up new avenues for drug discovery and provides contributions to enhance our understanding of protease inhibitors and set the stage for future therapeutic development.
5. Conclusions
Although COVID-19 is transitioning from a pandemic to an endemic, the etiological agent, SARS-CoV-2, continues to spread and pose public health challenges globally. The virus frequently mutates, leading to the emergence of various strains. Despite considerable efforts by scientists and healthcare communities to manage, treat, and develop vaccines against SARS-CoV-2, the risk to human health remains significant. Treating conditions caused by SARS-CoV-2 remains a global health challenge due to the limited availability of orally bioavailable antiviral drugs. There is an urgent need to develop broadly acting antivirals to address current and future virus outbreaks. By utilizing molecular docking for screening 118 compounds derived from marine organisms and 92 previously synthesized compounds to assess their binding affinity for the main protease and papain-like protease enzymes of SARS-CoV-2, the best marine origin and synthesized candidates have been identified from the xanthene, benzoxazole, and coumarin classes of compounds. Marine origin compounds emerged as slightly better potential inhibitors for these enzymes, although the binding affinity values for synthesized compounds were quite similar, with overall affinity values for the best candidates ranging from 0.2 to 0.4 mM. Among both marine origin and synthesized compounds, xanthenes have proven to be the most promising scaffolds for further research as inhibitors. Between the two investigated targets, the PLpro was found to be more druggable compared to the Mpro. All top candidates also met the criteria for the wide arsenal of different drug-likeness properties for good oral bioavailability and low probability of adverse effects. Tested compounds also appeared better in comparison to the FDA-approved drug nirmatrelvir as an Mpro inhibitor and experimental drug Jun12682 as a PLpro inhibitor. The molecular insights obtained from docking studies of SARS-CoV-2 proteases (Mpro and PLpro) are essential for informing in vivo studies and clinical applications. These insights can assist in identifying promising lead compounds, refining them to enhance efficacy and safety, and ensuring their potential for clinical success. By leveraging in silico predictions as a starting point, researchers can streamline the drug discovery process, speeding up the development of effective antiviral treatments for COVID-19 and potentially other coronavirus infections. This extended research provided us valuable insight into the comparative affinity of marine origin and synthesized compounds of xanthene, coumarin, and benzoxazole classes with top compounds as promising candidates to consider for further in vitro and in vivo studies.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/scipharm93010002/s1, Table S1: Marine compounds with Mpro (7ZQV); Table S2: Synthesized compounds with Mpro (7ZQV); Table S3: Marine compounds with PLpro (7TZJ); Table S4: Synthesized compounds with PLpro (7TZJ).
Author Contributions
Conceptualization, S.Š.-H. and A.O.; methodology, E.V. and A.O.; validation, M.M., E.V. and M.S.; formal analysis, A.O., M.M. and E.V.; investigation, A.O. and E.V.; resources, M.S., M.P., A.S. and L.H.; data curation, A.O. and L.H.; writing—original draft preparation, S.Š.-H. and M.S.; writing—review and editing, everyone; visualization, A.O., M.P. and A.S.; supervision, S.Š.-H.; project administration, S.Š.-H. and A.O.; funding acquisition, S.Š.-H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
Acknowledgments
This research was supported by funding through grant number 27-02-11-41250-15/21 provided by Canton Sarajevo Ministry of Science, Higher Education and Youth.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Fan, H.; Qin, S.; Cui, Y. Emergence and characterization of the SARS-CoV-2 JN. 1 variant: Global prevalence and implications for public health. Zoonoses 2024, 4, 994. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.; Lau, E.H.; Wong, J.Y.; et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Number of COVID-19 Cases Reported to WHO (Cumulative Total). Available online: https://data.who.int/dashboards/covid19/cases?n=o (accessed on 5 December 2024).
- Number of COVID-19 Deaths Reported to WHO (Cumulative Total). Available online: https://data.who.int/dashboards/covid19/deaths?n=o (accessed on 5 December 2024).
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; Peacock, S.J.; Barclay, W.S.; De Silva, T.I.; Towers, G.J.; Robertson, D.L. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Javed, S.; Farhan, B.A.; Shabbir, M.; Tahseen, A.; Ahmedah, H.T.; Moga, M. Morphology, Pathogenesis, Genome Organization, and Replication of Coronavirus (COVID-19). In The COVID-19 Pandemic, 1st ed.; Apple Academic Press: Palm Bay, FL, USA, 2022; pp. 3–43. [Google Scholar] [CrossRef]
- Zmasek, C.M.; Lefkowitz, E.J.; Niewiadomska, A.; Scheuermann, R.H. Genomic evolution of the coronaviridae family. Virology 2022, 570, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Hu, B.; Wang, L.F. Zoonotic Origin and Evolution of SARS Coronavirus. In Genetics and Evolution of Infectious Diseases, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2024; pp. 841–861. [Google Scholar] [CrossRef]
- Scheller, C.; Krebs, F.; Minkner, R.; Astner, I.; Gil-Moles, M.; Wätzig, H. Physicochemical properties of SARS-CoV-2 for drug targeting, virus inactivation and attenuation, vaccine formulation and quality control. Electrophoresis 2020, 41, 1137–1151. [Google Scholar] [CrossRef]
- Kumar, R.; Srivastava, Y.; Muthuramalingam, P.; Singh, S.K.; Verma, G.; Tiwari, S.; Tandel, N.; Beura, S.K.; Panigrahi, A.R.; Maji, S.; et al. Understanding mutations in human SARS-CoV-2 spike glycoprotein: A systematic review & meta-analysis. Viruses 2023, 15, 856. [Google Scholar] [CrossRef]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Singh, D.; Yi, S.V. On the origin and evolution of SARS-CoV-2. Exp. Mol. Med. 2021, 53, 537–547. [Google Scholar] [CrossRef]
- Chakraborty, C.; Bhattacharya, M.; Dhama, K. SARS-CoV-2 vaccines, vaccine development technologies, and significant efforts in vaccine development during the pandemic: The lessons learned might help to fight against the next pandemic. Vaccines 2023, 11, 682. [Google Scholar] [CrossRef]
- Jadhav, P.; Huang, B.; Osipiuk, J.; Zhang, X.; Tan, H.; Tesar, C.; Endres, M.; Jedrzejczak, R.; Tan, B.; Deng, X.; et al. Structure-based design of SARS-CoV-2 papain-like protease inhibitors. Eur. J. Med. Chem. 2024, 264, 116011. [Google Scholar] [CrossRef]
- Kakavandi, S.; Zare, I.; VaezJalali, M.; Dadashi, M.; Azarian, M.; Akbari, A.; Ramezani Farani, M.; Zalpoor, H.; Hajikhani, B. Structural and non-structural proteins in SARS-CoV-2: Potential aspects to COVID-19 treatment or prevention of progression of related diseases. Cell Commun. Signal. 2023, 21, 110. [Google Scholar] [CrossRef] [PubMed]
- Onyango, O.H. In Silico Models for Anti-COVID-19 Drug Discovery: A Systematic Review. Adv. Pharmacol. Pharm. Sci. 2023, 2023, 4562974. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Shahabi, D.; Imhoff, M.E.; Kovela, S.; Sharma, A.; Hattori, S.I.; Higashi-Kuwata, N.; Mitsuya, H.; Mesecar, A.D. SARS-CoV-2 papain-like protease (PLpro) inhibitory and antiviral activity of small molecule derivatives for drug leads. Bioorg. Med. Chem. Lett. 2023, 96, 129489. [Google Scholar] [CrossRef]
- Sanders, B.C.; Pokhrel, S.; Labbe, A.D.; Mathews, I.I.; Cooper, C.J.; Davidson, R.B.; Phillips, G.; Weiss, K.L.; Zhang, Q.; O’Neill, H.; et al. Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2. Nat. Commun. 2023, 14, 1733. [Google Scholar] [CrossRef]
- Funk, L.M.; Poschmann, G.; Rabe von Pappenheim, F.; Chari, A.; Stegmann, K.M.; Dickmanns, A.; Wensien, M.; Eulig, N.; Paknia, E.; Heyne, G.; et al. Multiple redox switches of the SARS-CoV-2 main protease in vitro provide opportunities for drug design. Nat. Commun. 2024, 15, 411. [Google Scholar] [CrossRef] [PubMed]
- Melano, I.; Lo, Y.; Su, W. Characterization of host substrates of SARS-CoV-2 main protease. Front. Microbiol. 2023, 14, 1251705. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.H.; Brewitz, L.; Lukacik, P.; Strain-Damerell, C.; Walsh, M.A.; Schofield, C.J.; Duarte, F. Studies on the selectivity of the SARS-CoV-2 papain-like protease reveal the importance of the P2′ proline of the viral polyprotein. RSC Chem. Biol. 2024, 5, 117–130. [Google Scholar] [CrossRef]
- Citarella, A.; Dimasi, A.; Moi, D.; Passarella, D.; Scala, A.; Piperno, A.; Micale, N. Recent advances in SARS-CoV-2 main protease inhibitors: From nirmatrelvir to future perspectives. Biomolecules 2023, 13, 1339. [Google Scholar] [CrossRef]
- Waqas, M.; Ullah, S.; Halim, S.A.; Rehman, N.U.; Ali, A.; Jan, A.; Muhsinah, A.B.; Khan, A.; Al-Harrasi, A. Targeting papain-like protease by natural products as novel therapeutic potential SARS-CoV-2. Int. J. Biol. Macromol. 2024, 258, 128812. [Google Scholar] [CrossRef]
- Banerjee, P.; Mandhare, A.; Bagalkote, V. Marine natural products as a source of new drugs: An updated patent review. Expert. Opin. Ther. Pat. 2021, 32, 317–363. [Google Scholar] [CrossRef]
- Chhetri, B.K.; Tedbury, P.R.; Sweeney-Jones, A.M.; Mani, L.; Soapi, K.; Manfredi, C.; Sorscher, E.; Sarafianos, S.G.; Kubanek, J. Marine natural products as leads against SARS-CoV-2 infection. J. Nat. Prod. 2022, 85, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Zaporozhets, T.S.; Besednova, N.N. Biologically active compounds from marine organisms in the strategies for combating coronaviruses. AIMS Microbiol. 2020, 6, 470. [Google Scholar] [CrossRef]
- Abdallah, H.M.; El-Halawany, A.M.; Sirwi, A.; El-Araby, A.M.; Mohamed, G.A.; Ibrahim, S.R.; Koshak, A.E.; Asfour, H.Z.; Awan, Z.A.; A. Elfaky, M. Repurposing of some natural product isolates as SARS-COV-2 main protease inhibitors via in vitro cell free and cell-based antiviral assessments and molecular modeling approaches. Pharmaceuticals 2021, 14, 213. [Google Scholar] [CrossRef]
- Wang, S.C.; Chen, Y.; Wang, Y.C.; Wang, W.J.; Yang, C.S.; Tsai, C.L.; Hou, M.H.; Chen, H.F.; Shen, Y.C.; Hung, M.C. Tannic acid suppresses SARS-CoV-2 as a dual inhibitor of the viral main protease and the cellular TMPRSS2 protease. Am. J. Cancer Res. 2020, 10, 4538. [Google Scholar] [CrossRef] [PubMed]
- White, K.M.; Rosales, R.; Yildiz, S.; Kehrer, T.; Miorin, L.; Moreno, E.; Jangra, S.; Uccellini, M.B.; Rathnasinghe, R.; Coughlan, L.; et al. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science 2021, 371, 926–931. [Google Scholar] [CrossRef]
- Minagawa, K.; Kouzuki, S.; Yoshimoto, J.U.N.; Kawamura, Y.; Tani, H.; Iwata, T.; Terui, Y.; Nakai, H.; Yagi, S.; Hattori, N.; et al. Stachyflin and acetylstachyflin, novel anti-influenza A virus substances, produced by Stachybotrys sp. RF-7260 I. Isolation, structure elucidation and biological activities. J. Antibiot. 2002, 55, 155–164. [Google Scholar] [CrossRef]
- Tsujii, S.; Rinehart, K.L.; Gunasekera, S.P.; Kashman, Y.; Cross, S.S.; Lui, M.S.; Pomponi, S.A.; Diaz, M.C. Topsentin, bromotopsentin, and dihydrodeoxybromotopsentin: Antiviral and antitumor bis(indolyl)imidazoles from Caribbean deep-sea sponges of the family Halichondriidae. Structural and synthetic studies. J. Org. Chem. 1988, 53, 5446–5453. [Google Scholar] [CrossRef]
- Ishida, J.; Wang, H.K.; Oyama, M.; Cosentino, M.L.; Hu, C.Q.; Lee, K.H. Anti-AIDS agents. 46. Anti-HIV activity of harman, an anti-HIV principle from Symplocos setchuensis, and its derivatives. J. Nat. Prod. 2001, 64, 958–960. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.E.; Min, J.S.; Jang, M.S.; Lee, J.Y.; Shin, Y.S.; Park, C.M.; Song, J.H.; Kim, H.R.; Kim, S.; Jin, Y.H.; et al. Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus OC43 infection of MRC-5 human lung cells. Biomolecules 2019, 9, 696. [Google Scholar] [CrossRef]
- Cheng, P.N.L.; Chiang, L.; Lin, C. Antiviral effects of saikosaponins on human coronavirus 229e in vitro. Clin. Exp. Pharmacol. Physiol. 2006, 33, 612–616. [Google Scholar] [CrossRef]
- Hong, S.; Seo, S.H.; Woo, S.J.; Kwon, Y.; Song, M.; Ha, N.C. Epigallocatechin gallate inhibits the uridylate-specific endoribonuclease Nsp15 and efficiently neutralizes the SARS-CoV-2 strain. J. Agric. Food Chem. 2021, 69, 5948–5954. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhu, X.; Hao, L.; Zhao, M.; Hua, Q.; An, F. Bioactive indolyl diketopiperazines from the marine derived endophytic Aspergillus versicolor DY180635. Mar. Drugs 2020, 18, 338. [Google Scholar] [CrossRef] [PubMed]
- Cheohen, C.F.; Esteves, M.E.; da Fonseca, T.S.; Leal, C.M.; Assis, F.D.; Campos, M.F.; Rebelo, R.S.; Allonso, D.; Leitão, G.G.; da Silva, M.L.; et al. In silico screening of phenylethanoid glycosides, a class of pharmacologically active compounds as natural inhibitors of SARS-CoV-2 proteases. Comput. Struct. Biotechnol. J. 2023, 21, 1461–1472. [Google Scholar] [CrossRef] [PubMed]
- Vegad, U.G.; Gajjar, N.D.; Nagar, P.R.; Chauhan, S.P.; Pandya, D.J.; Dhameliya, T.M. In silico screening, ADMET analysis and MD simulations of phytochemicals of Onosma bracteata Wall. as SARS CoV-2 inhibitors. 3 Biotech 2023, 13, 221. [Google Scholar] [CrossRef]
- Mohapatra, P.K.; Chopdar, K.S.; Dash, G.C.; Mohanty, A.K.; Raval, M.K. In silico screening and covalent binding of phytochemicals of Ocimum sanctum against SARS-CoV-2 (COVID 19) main protease. J. Biomol. Struct. Dyn. 2023, 41, 435–444. [Google Scholar] [CrossRef]
- Hu, C. Marine natural products and human immunity: Novel biomedical resources for anti-infection of SARS-CoV-2 and related cardiovascular disease. Nat. Prod. Bioprospecting 2024, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Salihović, M.; Osmanović, A.; Špirtović-Halilović, S.; Roca, S.; Meščić, A.; Vujisić, L.; Trifunović, S.; Završnik, D.; Sofić, E. Synthesis, structural, conformational and DFT studies of N-3 and O-4 alkylated regioisomers of 5-(hydroxypropyl) pyrimidine. J. Mol. Struct. 2015, 1091, 170–176. [Google Scholar] [CrossRef]
- Špirtović-Halilović, S.; Veljović, E.; Salihović, M.; Osmanović, A.; Šapčanin, A.; Softić, D.; Roca, S.; Trifunović, S.; Škrijelj, N.; Škrbo, S.; et al. Synthesis, Microbiological Activity and In Silico Investigation for Some Synthesized Coumarin Derivatives. Croat. Chem. Acta 2020, 93, 23–31. [Google Scholar] [CrossRef]
- Bilajac, E.; Osmanović, A.; Glamočlija, U.; Veljović, E.; Imamović, B.; Bečić, E.; Roca, S.; Salihović, M.; Završnik, D.; Špirtović-Halilović, S. Synthesis, In Silico Study and Antitumor Activity of Coumarin Compounds in Lymphoma Cells. Farmacia 2023, 71, 1263–1273. [Google Scholar] [CrossRef]
- Završnik, D.; Špirtović-Halilović, S.; Softić, D. Synthesis, structure and antibacterial activity of 3-substituted derivatives of 4-hydroxycoumarin. Period. Biol. 2011, 113, 93–97. [Google Scholar]
- Veljović, E.; Špirtović-Halilović, S.; Muratović, S.; Valek Žulj, L.; Roca, S.; Trifunović, S.; Osmanović, A.; Završnik, D. 9-Aryl substituted hydroxylated xanthen-3-ones: Synthesis, structure and antioxidant potency evaluation. Croat. Chem. Acta 2015, 88, 121–127. [Google Scholar] [CrossRef]
- Veljović, E.; Špirtović-Halilović, S.; Muratović, S.; Osmanović, A.; Badnjević, A.; Gurbeta, L.; Tatlić, B.; Zorlak, Z.; Imamović, S.; Husić, Đ.; et al. Artificial neural network and docking study in design and synthesis of xanthenes as antimicrobial agents. In CMBEBIH 2017, Proceedings of the International Conference on Medical and Biological Engineering, Sarajevo, Bosnia and Herzegovina, 16–18 March 2017; Badnjević, A., Ed.; Springer: Singapore, 2017; Volume 62, pp. 617–626. [Google Scholar] [CrossRef]
- Zukić, S.; Veljović, E.; Špirtović-Halilović, S.; Muratović, S.; Osmanović, A.; Trifunović, S.; Novaković, I.; Završnik, D. Antioxidant, Antimicrobial and Antiproliferative Activities of Synthesized 2,2,5,5-Tetramethyl-9-aryl-3,4,5,6,7,9-hexahydro-1H-xanthene-1,8(2H)-dione Derivatives. Croat. Chem. Acta 2018, 91, 1–9. [Google Scholar] [CrossRef]
- Veljović, E.; Špirtović-Halilović, S.; Muratović, S.; Osmanović, A.; Novaković, I.; Trifunović, S.; Završnik, D. Synthesis and biological evaluation of xanthen-1,8-dione derivatives. Bull. Chem. Technol. Bosnia Herzegovina 2018, 51, 13–18. [Google Scholar]
- Glamočlija, U.; Padhye, S.; Špirtović-Halilović, S.; Osmanović, A.; Veljović, E.; Roca, S.; Novaković, I.; Mandić, B.; Turel, I.; Kljun, J.; et al. Synthesis, biological evaluation and docking studies of benzoxazoles derived from thymoquinone. Molecules 2018, 23, 3297. [Google Scholar] [CrossRef]
- Bilajac, E.; Glamočlija, U.; Osmanović, A.; Mahmutović, L.; Sezer, A.; Roca, S.; Špirtović-Halilović, S.; Salihović, M.; Hromić-Jahjefendić, A.; Veljović, E. Analysis of Antitumor Potential of Xanthene Compounds in Lymphoma Cells. Croat. Chem. Acta 2023, 96, 59–68. [Google Scholar] [CrossRef]
- Spirtović-Halilović, S.; Salihović, M.; Osmanović, A.; Veljović, E.; Rahić, O.; Mahmutović, E.; Hadziabdić, J.; Novaković, I.; Roca, S.; Trifunović, S.; et al. In Silico Study of Microbiologically Active Benzoxazole Derivatives. Indian J. Pharm. Sci. 2023, 85, 767–777. [Google Scholar] [CrossRef]
- Owen, D.R.; Allerton, C.M.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- Tan, B.; Zhang, X.; Ansari, A.; Jadhav, P.; Tan, H.; Li, K.; Chopra, A.; Ford, A.; Chi, X.; Ruiz, F.X.; et al. Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model. Science 2024, 383, 1434–1440. [Google Scholar] [CrossRef] [PubMed]
- Schneider, N.; Lange, G.; Hindle, S.; Klein, R.; Rarey, M. A consistent description of HYdrogen bond and DEhydration energies in protein–ligand complexes: Methods behind the HYDE scoring function. J. Comput.-Aided Mol. Des. 2013, 27, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Reulecke, I.; Lange, G.; Albrecht, J.; Klein, R.; Rarey, M. Towards an Integrated Description of Hydrogen Bonding and Dehydration: Decreasing False Positives in Virtual Screening with the HYDE Scoring Function. ChemMedChem 2008, 3, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Rarey, M.; Kramer, B.; Lengauer, T.; Klebe, G.A. Fast Flexible Docking Method using an Incremental Construction Algorithm. J. Mol. Biol. 1996, 261, 470–489. [Google Scholar] [CrossRef]
- Gastreich, M.; Lilienthal, M.; Briem, H.; Claussen, H. Ultrafast de novo docking combining pharmacophores and combinatorics. J. Comput.-Aided Mol. Des. 2007, 20, 717–734. [Google Scholar] [CrossRef]
- Duan, Y.; Wang, H.; Yuan, Z.; Yang, H. Structural biology of SARS-CoV-2 Mpro and drug discovery. Curr. Opin. Struct. Biol. 2023, 82, 102667. [Google Scholar] [CrossRef]
- Zagórska, A.; Czopek, A.; Fryc, M.; Jończyk, J. Inhibitors of SARS-CoV-2 main protease (Mpro) as anti-coronavirus agents. Biomolecules 2024, 14, 797. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Congreve, M.; Carr, R.; Murray, C.; Jhoti, H. A ‘Rule of Three’ for fragment-based lead discovery? Drug Discov. Today 2003, 8, 876–877. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; Wang, Z.; Cai, Z. Prediction of human intestinal absorption by GA feature selection and support vector machine regression. Int. J. Mol. Sci. 2008, 9, 1961–1976. [Google Scholar] [CrossRef]
- Ghafourian, T.; Amin, Z. QSAR models for the prediction of plasma protein binding. BioImpacts 2013, 3, 21–27. [Google Scholar] [CrossRef]
- Daneman, R.; Prat, A. The blood–brain barrier. Cold Spring Harbor Perspect. Biol. 2015, 7, a020412. [Google Scholar] [CrossRef]
- Summerfield, S.G.; Read, K.; Begley, D.J.; Obradovic, T.; Hidalgo, I.J.; Coggon, S.; Lewis, A.V.; Porter, R.A.; Jeffrey, P. Central nervous system drug disposition: The relationship between in situ brain permeability and brain free fraction. J. Pharmacol. Exp. Ther. 2007, 322, 205–213. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Published by MDPI on behalf of the Österreichische Pharmazeutische Gesellschaft. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).