Viruses as Living Systems—A Metacybernetic View
Abstract
1. Introduction
“The evolutionary history of viruses represents a fascinating, albeit murky, topic for virologists and cell biologists. Because of the great diversity among viruses, biologists have struggled with how to classify these entities, and how to relate them to the conventional tree of life. They may represent genetic elements that gained the ability to move between cells. They may represent previously free-living organisms that became parasites. They may be the precursors of life as we know it”.
“Viruses have several common characteristics: they are small, have DNA or RNA genomes, and are obligate intracellular parasites [requiring a host to reproduce]. The virus capsid functions to protect the nucleic acid from the environment, and some viruses surround their capsid with a membrane envelope”.([33]: p. 19).
2. Some Fundamentals
2.1. The Aristotelian Paradigm and Beyond
- the metabolic—which considers principles of materials exchange between the organism and its environment occurring in such a way that their general properties are not altered;
- the physiological—which is concerned with physiological functions like breathing, moving, digesting, a biochemical definition that identifies living systems through an ability to store hereditary information in nucleic acid molecules;
- the genetic—which is connected with the process of evolution and concerns how information is coded;
- the thermodynamic—which explains an ability to maintain low levels of entropy that explain order;
- the physics-based—which sees life as being composed of an ensemble of entities that share information coded in a physical substrate, this able to keep its entropy significantly lower than the maximal entropy of the ensemble;
- the physical—in which components of life are contained within distinct boundaries (like those of cells) resulting in locally increased order;
- the autopoietic—where organisms are self-governing, maintain their own identity, have information closure, self-relatedness, self-relational, and are adaptive through autopoietic (self-producing) processes.
- The chemical paradigm (also known as the neo-Aristotelian/metabolic paradigm) posits that life is an extremely complex form of chemistry and is completely described (in principle) by physical quantities. The paradigm has been represented by five basic biological processes that define the form of living: metabolism, temperature regulation, information processing, embryo development, and inheritance [52].
- The information paradigm reflects chemistry through information. It argues that information is not so much a “real observable” but rather a “fundamental observable”. While real observables may simply be pragmatic descriptions of a reality being observed, fundamental observables provide fundamental properties [28] that critically characterise it.
- The code paradigm embraces the idea that meaning is the basis of code, and that both information and meaning exist in every living system. This is because they are the inevitable results of the processes of copying and coding from which genes and proteins are produced, and this can be traced to meaning as a fundamental observable. Molecular coding is also an expression of something essential to all observable systems: their levels of complexity.
2.2. From Controversy to Fragmentation in Biology
2.3. From Functionality to Organisation
2.4. From Autonomy to Autopoietic Theory
2.5. Homeostasis
“Fifteen years ago, scientists celebrated the first draft of the sequenced human genome. At the time, they predicted that humans had between 25,000 and 40,000 genes that code for proteins. That estimate has continued to fall. Humans actually seem to have as few as 19,000 such genes—a mere 1–2% of the genome. The key to our complexity lies in how these genes are regulated by the remaining 99% of our DNA, known as the genome’s ‘dark matter’”.
“…as much as 98 percent—of our DNA do not code for proteins. Much of this ‘dark matter genome’ is thought to be nonfunctional evolutionary leftovers that are just along for the ride. However, hidden among this noncoding DNA are many crucial regulatory elements that control the activity of thousands of genes. What is more, these elements play a major role in diseases such as cancer, heart disease, and autism, and they could hold the key to possible cures”.
3. Understanding Viruses
“The term ‘adaptive’ mutation was used by Delbrück to indicate mutations formed in response to an environment in which the mutations were selected. The term does not imply that non-adaptive (unselected) mutations would not also be induced, or that the useful mutations would be induced preferentially (this latter idea being called ‘directed’ mutation). ‘Adaptive mutation’ was adopted subsequently by Tlsty in her examination of gene amplification in rat cells. She distinguished mutations that pre-exist at the time a cell is exposed to a selective environment versus (adaptive) mutations formed after exposure to the environment”.
3.1. The Nature of Viruses
3.2. Viruses as Parasites
“living cells, whether human, animal, plant, or bacterial, have double-stranded DNA (dsDNA) as their genetic material. Viruses, on the other hand, have genomes, or genetic material, that can be composed of DNA or RNA (but not both). Genomes are not necessarily double-stranded, either; different virus types can also have single-stranded DNA (ssDNA) genomes, and viruses with RNA genomes can be single-stranded or double-stranded. Any particular virus will only have one type of nucleic acid genome, however, and so viruses are not encountered that have both ssDNA and ssRNA genomes, for example”.
- (i)
- the directed arms race—where the host develops resistance and the virus follows by increasing its infectivity, the outcome being a directional evolution of increasing resistance and increasing infectivity. As an illustration of this, Brüssow and Brüssow [132] refer to the Russian flu in 1889, and Sharma [133] to the Spanish Flu of 1918 when the 3rd wave was mild and signalled the end of the pandemic, and suggests this might be the way of COVID-19 with the new Omicron variant;
- (ii)
- fluctuating selection dynamics—where parasites and hosts can experience oscillatory cycles, and where the densities of these interacting species dynamically fluctuate through time, resulting in non-directional evolution [134] where, for example, in marine virus–bacterial systems, viruses might infect different bacterial populations at different times, preventing resistance, this resulting in very high levels of genetic diversity in the host.
3.3. Viruses as Learners
3.4. Viruses as Autopoietic Organisms
- (i)
- to make or manufacture or create or form something from components or raw materials;
- (ii)
- to cause a particular result or situation to happen or exist.
3.5. The Evolution of Populations of Viruses
“An autopoiesis system organises the production of its own components, so that these components are continuously re-generated and the system can therefore maintain the very network process that produces them. Living beings are characterized by their continuous self-production, so they are an autopoietic organization. Even though the theory of autopoiesis is based on cellular life, viruses can fit in this definition [through their own processes of]…organisation”.
4. A Metacybernetic View of Autopoiesis
4.1. The Nature of Metacybernetics
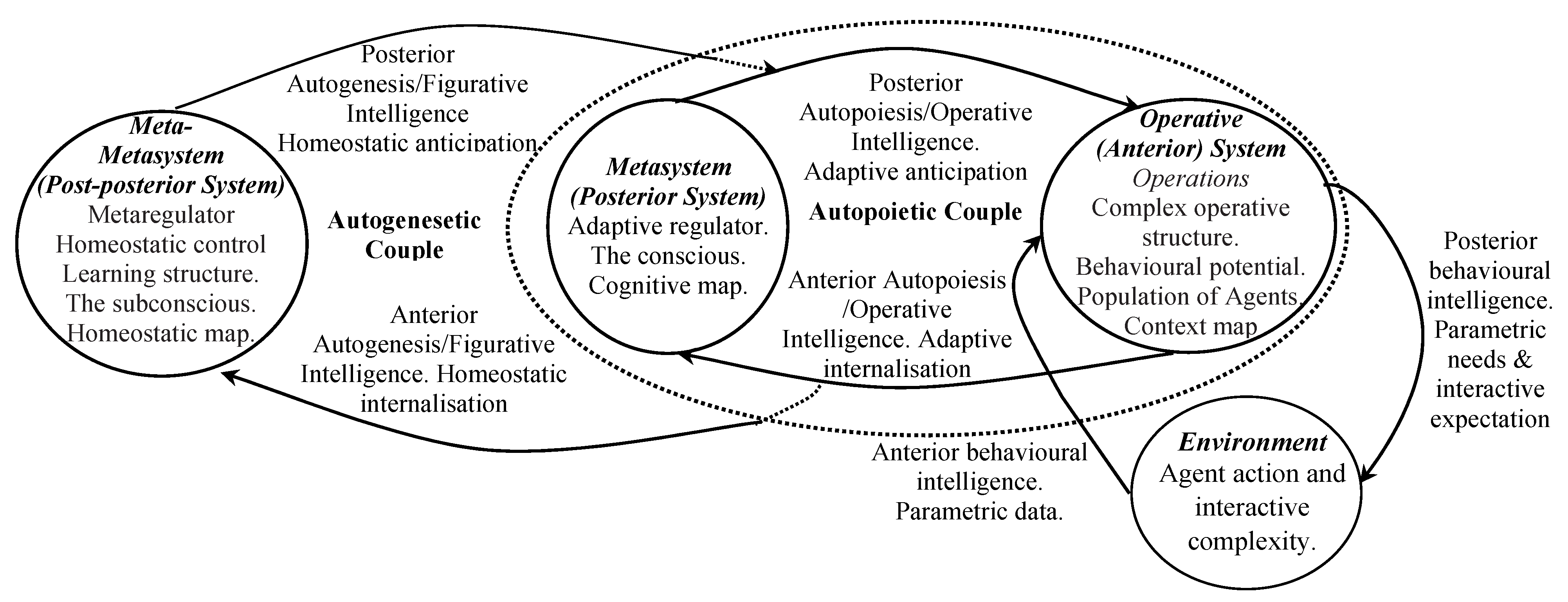
4.2. General Third Order Agency Model
4.3. Modelling Biological Living Systems
“an approach in biomedical research to understanding the larger picture—be it at the level of the organism, tissue, or cell—by putting its pieces together. It’s in stark contrast to decades of reductionist biology, which involves taking the pieces apart”([199]: p. 10).
“Few scientists will voluntarily characterise their work as reductionistic. Yet, reductionism is at the philosophical heart of the molecular biology revolution. Holistic science, the opposite of reductionistic science, has also acquired a bad name, perhaps due to an unfortunate association of the word “holistic” with new age pseudoscience…A fundamental tenet of systems biology is that cellular and organismal constituents are interconnected, so that their structure and dynamics must be examined in intact cells and organisms rather than as isolated parts…[the approach is] “holistic” because [it relies] on the “fundamental interconnectedness of all things…”([200]: p. 1401).
“Traditionally, science has taken a reductionist approach, dissecting biological systems into their constituent parts and studying them in isolation. Entire scientific careers have been devoted to studying only one gene or protein in order to understand its function. Although scientists have made progress using this method, this reductionist approach limits biological insights into the human body. As a result, efforts to treat many complex diseases have also faced limited success. Reductionism, by its nature, cannot comprehend the complexity of biological systems, the properties of which cannot be explained or predicted by studying their individual components”([201]: p. 1).
- DNA is a long stable molecule that contains a unique genetic code for a living system.
- RNA is a long (less stable) molecule that processes protein, carrying genetic information of many viruses from the cell to the cytoplasm (material outside the cell nucleus); it has various forms that include mRNA, rRNA, rRNA, tRNA and crRNA [211].
- Messenger RNA (mRNA) carries and transcribes the genetic code of the genome into a form that can be read and used to make proteins, and carries genetic information from the nucleus to the cytoplasm of a cell, and is multifunctional (cf. [212]).
- Ribosomal RNA (rRNA) is located in the cytoplasm of a cell, where ribosomes are found, and directs the translation of mRNA into proteins.
- Transfer RNA (tRNA) which, like rRNA, is located in the cellular cytoplasm and is involved in protein synthesis. Transfer RNA brings or transfers amino acids (protein building blocks) to the ribosome that corresponds to each three-nucleotide codon of rRNA. The amino acids then can be joined together and processed to make polypeptides and proteins.
- CRISPR array RNA (crRNA) is normally discussed within the context of gene editing, but which is also a feature of viruses [213], and that constitutes a model of the environment that has been autopoietically assimilated into a virus, and then accommodated into its genome.
- LncRNA (long noncoding RNA) regulate target gene expression through the interactions between their higher-order structures and major partner proteins in higher order structure connected with the dark/regulatory genome [214]. It is therefore an agency directly connected with dark/regulatory genome metaregulation (cf. [135]).
- The genome of a virus is an arrangement of genes, and regulates the capsid. Genome expression uses coded information that regulates capsid expression. Bacteria and cells also have genomes stored in their chromones (encoded genetic material in DNA molecules). In cells the chromosome is stored within the nucleus, in bacteria in the nucleoid.
- The capsid is the viruses operative shell. Capsid expression selectively produces certain proteins, which are biological compounds like enzymes, hormones, and antibodies.
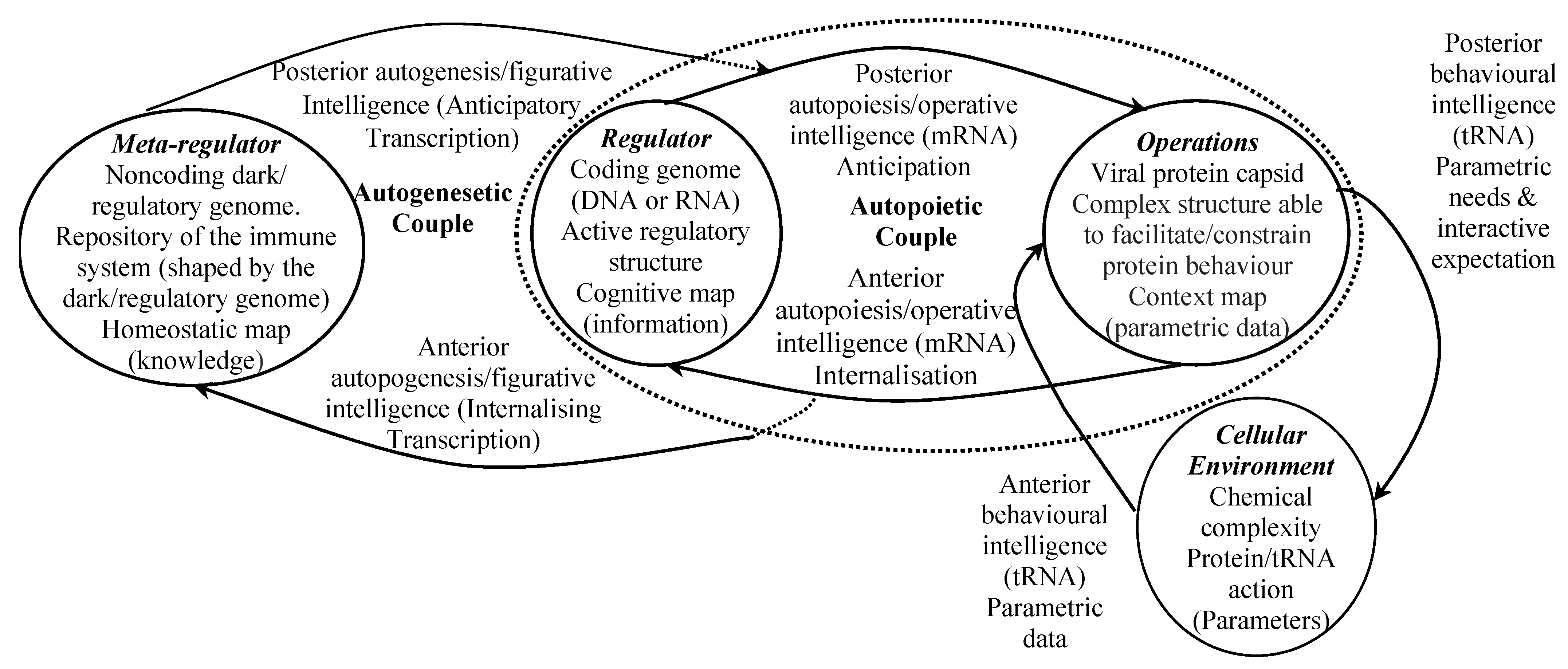
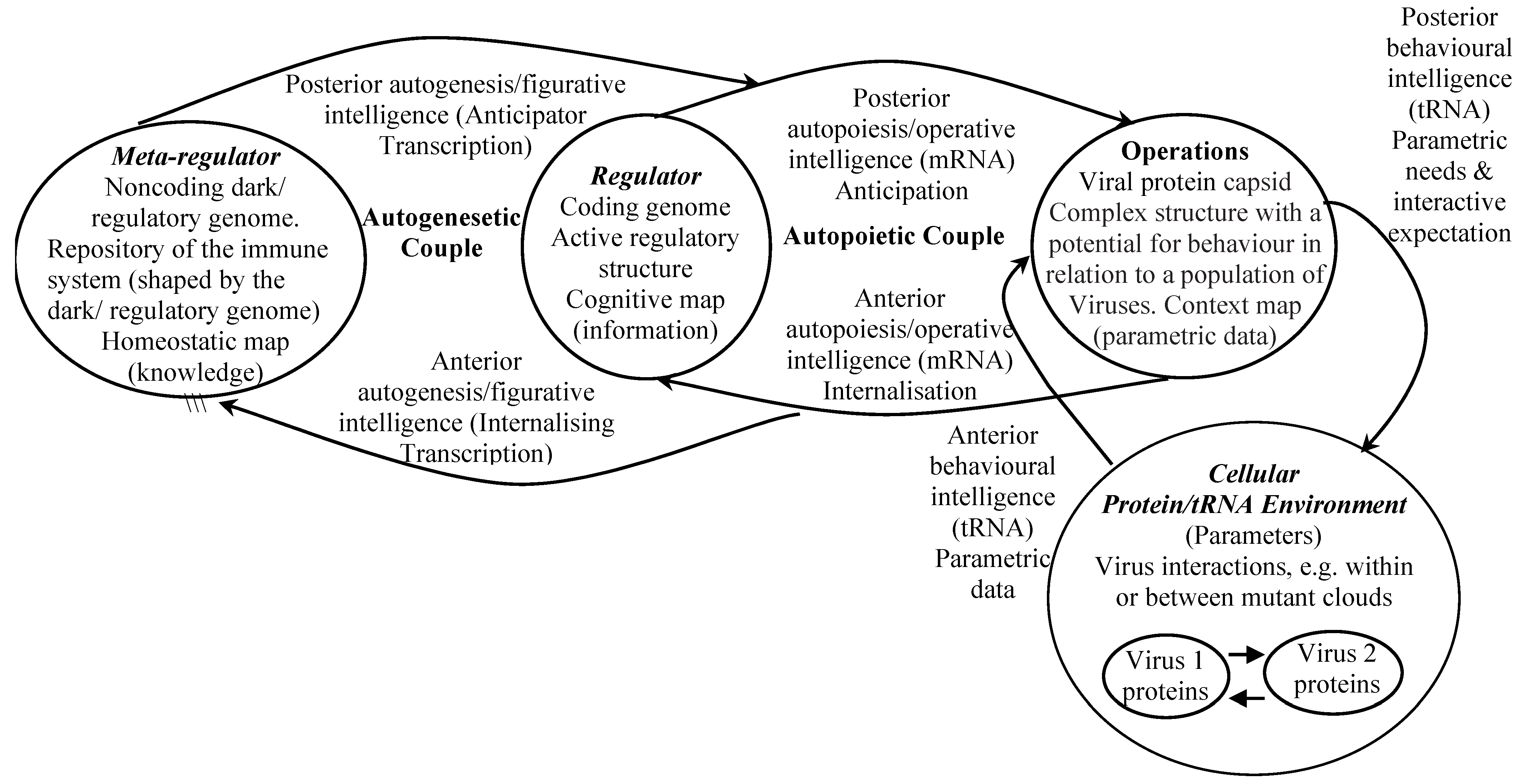
- The dark/regulatory genome is part of the nucleic acid that exists outside the known genes, and operates as a virus metaregulator. Dark/regulatory genome expression targets genes in the genome which it regulates, and influences the viruses immune system where it has one.
- Epigenetics refers to the relationship between the expression of the dark/regulatory genome, the genome and the capsid, and the environment.
- Protein synthesis occurs through translation, and during transcription an element of the genome is transcribed (copied similarly but not identically to the source) into mRNA which is then translated to produce a protein; during translation, mRNA along with tRNA and ribosomes (RNA and proteins responsible for assembling the proteins of the cell) work together to produce proteins. We note that in a virus the source element may be either DNA or RNA.
- Proteins are biological molecules in cells used for functions that vary from cellular support to cell signalling and cellular locomotion; illustrations are antibodies, enzymes and some hormones. Some proteins are enzymes capable of creating some substances and decomposing others; viral enzymes catalyse the integration of virally derived DNA into the DNA of a host cell in the nucleus; this forms a provirus that can be activated to produce viral proteins.
- Cytoplasm consists of all the contents outside of the nucleus (a structure containing a cell’s hereditary information which controls its growth and reproduction) and enclosed within the cell membrane of a cell and has various functions like protein synthesis and hormone and cellular waste removal.
4.4. Evolutionary Processes
4.5. Causal Agency Efficacy: The Intelligences as Information Channels
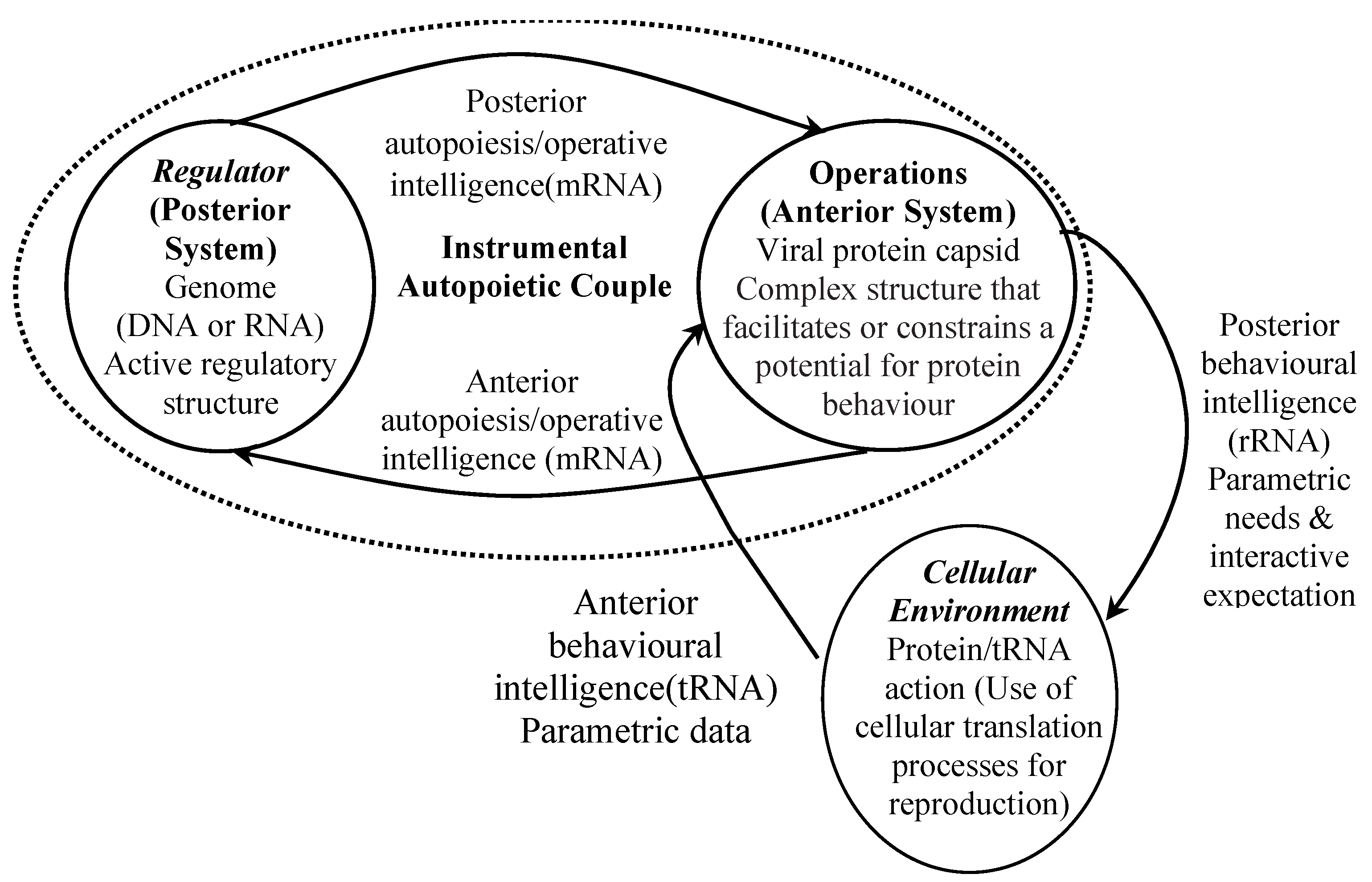
4.6. Virus Autopoietic Efficacy
- (i)
- show how a virus infection arises naturally, out of Equation (9), and
- (ii)
- show what biological parameters affect the value R and therefore what values of them tend to maximize R, thereby weakening the strength of the effect.
5. Case Study
5.1. SARS-CoV-2 Capsid Expression and Cybernetic Interactive Processes
- ACE as a ‘bad’ enzyme (τbad) causes activity leading to vasoconstriction (the narrowing of blood vessels by small muscles in their walls), oxidative stress (a phenomenon caused by imbalance between production and the accumulation of oxygen reactive species), inflammation and apoptosis (programmed cell death);
- ACE2, the ‘good’ enzyme (τgood) counters the preceding activities of ACE by altering ratios of hormones and amino acids (the two types of protein that form a basis for life).
5.2. Capsid Expression and the EPI Principle
5.3. SARS-CoV-2 as a Learning Virus
6. Discussion and Conclusions
“attempt to validate a view of that reality. The nature of facts, however, very much depends upon the context and framework from which one views them. Stafford Beer has called facts ‘fantasies that you can trust.’ Now, trust is ‘a firm belief in the honesty, veracity, justice, strength, etc., of a person or thing.’ Since trust occurs through belief, it should be realised that it can vary from individual to individual, from group to group, or from time to time. Beliefs are also culture based”([163]: p. 42).
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mallapapaty, S. Where Did Omicron Come from? Three Key Theories, Nature. Available online: https://www.nature.com/articles/d41586-022-00215-2?utm_source=Nature+Briefing&utm_campaign=b124caafb6-briefing-dy-2022028&utm_medium=email&utm_term=0_c9dfd39373-b124caafb6-46902170 (accessed on 20 March 2022).
- Hart, A. Son of Omicron? Cosmos Magazine. Available online: https://cosmosmagazine.com/health/COVID/new-omicron-sub-variant/ (accessed on 20 February 2022).
- Iacobucci, G. Covid-19: Unravelling the conundrum of omicron and deaths. Br. Med. J. 2022, 376, o254. [Google Scholar] [CrossRef] [PubMed]
- Wessner, D. The Origins of Viruses. Nat. Educ. 2010, 3, 37. [Google Scholar]
- Bormashenko, E.; Fedorets, A.A.; Dombrovsky, L.A.; Nosonovsky, M. Survival of Virus Particles in Water Droplets: Hydrophobic Forces and Landauer’s Principle. Entropy 2021, 23, 181. [Google Scholar] [CrossRef]
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479–480, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, G. General Characteristics of Viruses. 2021. Available online: https://bio.libretexts.org/@go/page/3232 (accessed on 10 February 2022).
- Brown, N.; Bhella, D. Are Viruses Alive? Microbiology Today Magazine. 10 May 2016, pp. 58–61. Available online: https://microbiologysociety.org/asset/405D51A4-5D04-4BFF-A407F9CE2F27CC28/ (accessed on 10 February 2022).
- Stadtländer, C.K.-H. Systems biology: Mathematical modeling and mod-el analysis. J. Biol. Dyn. 2018, 12, 1751–3758. [Google Scholar] [CrossRef][Green Version]
- Stegmaier, W. Subjects as Temporal Clues to Orientation: Nietzsche and Luhmann on Subjectivity. In Nietzsche and the Problem of Subjectivity; Constâncio, J., Mayer Branco, M.J., Ryan, B., Eds.; De Gruyter: Berlin, Germany, 2015; pp. 487–511. [Google Scholar]
- Seibt, J. “Process Philosophy,” in Stanford Encyclopaedia of Philosophy. 2017. Available online: https://plato.stanford.edu/entries/process-philosophy/ (accessed on 15 February 2022).
- Stegmaier, W. What Is Orientation? A Philosophical Investigation; Müller, R.G., Translator; De Gruyter: Berlin, Germany, 2019. [Google Scholar] [CrossRef]
- Sallie, R. Replicative homeostasis: A fundamental mechanism mediating selective viral replication and escape mutation. Virol. J. 2005, 2, 10. [Google Scholar] [CrossRef]
- Hoffmann, B. The Strange Story of the Quantum; Dover Publications: New York, NY, USA, 1959; Available online: https://www.google.co.uk/books/edition/The_Strange_Story_of_the_Quantum/ye0hDgAAQBAJ?hl=en&gbpv=1 (accessed on 20 January 2022).
- Farnsworth, K. An organisational systems-biology view of viruses explains why they are not alive. Biosystems 2020, 200, 104324. [Google Scholar] [CrossRef]
- Racaniello, V. Are Viruses Alive? Available online: www.virology.ws/are-viruses-alive (accessed on 10 January 2022).
- Cleland, C. The Quest for a Universal Theory of Life: Searching for Life as We Don’t Know It; Cambridge University Press: Cambridge, UK, 2019. [Google Scholar]
- Sutton, C. Unified Field Theory in Physics. Encyclopaedia Britannica. 1988. Available online: https://www.britannica.com/science/unified-field-theory (accessed on 20 February 2022).
- Aubry, S. New Theory for Elementary Chemical Reactions: A Key for Understanding Biochemistry? 2014. Available online: https://www.academia.edu/29316234/APS_123-QED_New_theory_for_elementary_chemical_reactions_a_key_for_understanding_biochemistry (accessed on 7 January 2022).
- Grandpierre, A.; Chopra, D.; Kafatos, M. The Universal Principle of Biology: Determinism, Quantum Physics and Spontaneity. NeuroQuantology 2014, 12, 364–373. [Google Scholar] [CrossRef]
- Bauer, E. Theoretical Biology; Akadémiai Kiadó: Budapest, Hungary, 1935; (originally In Russian, reprinted 1967: In Hungarian). [Google Scholar]
- Moggach, D. Bruno Bauer. The Stanford Encyclopedia of Philosophy. Zal-ta, E.N., Ed.; Spring 2022 Edition. 2022. Available online: https://plato.stanford.edu/archives/spr2022/entries/bauer/ (accessed on 20 January 2022).
- Elek, G.M.M. The living matter according to Ervin Bauer (1890–1938), (on the 75th anniversary of his tragic death) (History). Acta Physiol. Hung. 2013, 100, 124–132. [Google Scholar] [CrossRef]
- Grandpierre, A. The Biological Principle of Natural Sciences and the Logos of Life of Natural Philosophy: A Comparison and the Perspectives of Unifying the Science and Philosophy of Life. Analecta Husserl. 2011, 110, 711–727. [Google Scholar]
- Haken, H. Information and Self-Organisation: A Macroscopic Approach to Complex Systems; Springer: Heidelberg, Germany, 2006. [Google Scholar]
- Bonchev, D.G. Information Theoretic Complexity Measures. In Encyclopedia of Complexity and Systems Science; Meyers, R., Ed.; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Haken, H.; Portugali, J. Information and Self-Organization. Entropy 2017, 19, 18. [Google Scholar] [CrossRef]
- Meijer, D. Information: What Do You Mean? On the Formative Element of Our Universe. ResearchGate. 2013. Available online: https://www.researchgate.net/publication/275017053 (accessed on 12 March 2022).
- Collier, J. Intrinsic Information. In Information, Language and Cognition: Vancouver Studies in Cognitive Science; Hanson, P.P., Ed.; Oxford University Press (originally University of British Columbia Press): Oxford, UK, 1990; Volume 1, pp. 390–409. [Google Scholar]
- Yolles, M.; Frieden, B.R. Autopoiesis and Its Efficacy—A Metacybernetic View. Systems 2021, 9, 75. [Google Scholar] [CrossRef]
- Date, S. An Intuitive Look at Fisher Information. Towards Data Science. 7 August 2021. Available online: https://towardsdatascience.com/an-intuitive-look-at-fisher-information-2720c40867d8 (accessed on 1 February 2022).
- Frieden, R. Physics from Fisher Information: A Unification; Cambridge University Press: Cambridge, UK, 1998. [Google Scholar]
- Louten, J. Virus Structure and Classification. Essent. Hum. Virol. 2016, 19–29. [Google Scholar] [CrossRef]
- Mauron, A. Genomic Metaphysics. J. Mol. Biol. 2002, 319, 957–962. [Google Scholar] [CrossRef]
- Briones, C.D.E. Minority report: Hidden memory genomes in HIV-1 quasispecies and possible clinical implications. AIDS Rev. 2008, 10, 93–109. [Google Scholar]
- Alonso, M.E.; Pernaute, B.; Crespo, M.; Gomez-Skarmeta, J.L.; Manzanares, M. Understanding the regulatory genome. Int. J. Dev. Biol. 2009, 53, 1367–1378. [Google Scholar] [CrossRef]
- Wanke, K.A.; Devanna, P.; Vernes, S.C. Understanding Neurodevelopmental Disorders: The Promise of Regulatory Variation in the 3′UTRome. Biol. Psychiatry 2018, 83, 548–557. [Google Scholar] [CrossRef]
- Konopka, A.; Lindemann, S.; Fredrickson, J. Dynamics in microbial communities: Unraveling mechanisms to identify principles. ISME J. 2015, 9, 1488–1495. [Google Scholar] [CrossRef]
- von Foerster, H. Cybernetics; Circular Causal and Feedback Mechanisms in Biological and Social Systems; Josiah Macy, Jr. Foundation: New York, NY, USA, 1952. [Google Scholar]
- Von Foerster, H. Cybernetics of Cybernetics. In Understanding Understanding; Springer: New York, NY, USA, 2003; (a republication from 1979). [Google Scholar] [CrossRef]
- Yolles, M. Metacybernetics: Towards a General Theory of Higher Order Cybernetics. Systems 2021, 9, 34. [Google Scholar] [CrossRef]
- Schwarz, E. Autogenesis. SSRN. 2021. Available online: https://ssrn.com/abstract=3826203 (accessed on 7 March 2022).
- Yolles, M.; Fink, G. A Configuration Approach to Mindset Agency Theory—A Formative Trait Psychology with Affect Cognition & Behaviour; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2021. [Google Scholar]
- BioEd. Structure and Function of Living Things. BioEd Online. 2022. Available online: https://www.bioedonline.org/online-courses/educator-certification/generalist-4-8/structure-and-function-of-living-things/ (accessed on 20 March 2022).
- Boylan, M. Aristotle’s Biology. Internet Encyclopaedia of Philosophy. 2004. Available online: https://iep.utm.edu/aris-bio/ (accessed on 20 February 2022).
- Heylighen, F. Principles of Systems and Cybernetics: An evolutionary perspective. In Cybernetics and Systems ’92; Trappl, C., Ed.; World Science: Singapore, 1992; pp. 3–10. [Google Scholar]
- Trewavas, A. A brief history of systems biology. Plant Cell 2006, 18, 2420–2430. [Google Scholar] [CrossRef]
- Margulis, L.; Sagan, D. What Is Life? University of California Press: Berkeley, CA, USA, 2000. [Google Scholar]
- Álvarez-Vázquez, J. Animated Machines, Organic Souls: Maturana and Aristotle on the Nature of Life. Int. J. Nov. Res. Humanit. Soc. Sci. 2016, 3, 67–78. [Google Scholar]
- Maturana, H.R.; Varela, F.J. De Máquinas y Seres Vivos; Editorial Universitaria: Santiago, Chile, 1973. [Google Scholar]
- Barbieri, M. The Paradigms of Biology. Biosemiotics 2013, 6, 33–59. [Google Scholar] [CrossRef]
- Niklas, K.J.; Cobb, E.D. Size-dependent variation in plant form. Curr. Biol. 2017, 27, R900–R905. [Google Scholar] [CrossRef] [PubMed]
- Frieden, B. Fisher Information. University of Arizona. Arizona. 2009. Available online: https://wp.optics.arizona.edu/rfrieden/fisher-information/ (accessed on 15 February 2022).
- Mahoney, J. Revisiting General Theory in Historical Sociology. Soc. Forces 2004, 83, 459–489. [Google Scholar] [CrossRef]
- Dawkins, R. The Selfish Gene; Oxford University Press: Oxford, UK, 1976. [Google Scholar]
- Wilkins, J.S.; Bourrat, P. Replication and Reproduction. Stanford Encyclopedia of Philosophy. 25 September 2018. (Revised from 2001 Version). Available online: https://plato.stanford.edu/entries/replication/ (accessed on 10 February 2022).
- Fisher, R. The Genetical Theory of Natural Selection; Clarendon Press: Oxford, UK, 1930. [Google Scholar]
- Chaitanya, K.V. Structure and Organization of Virus Genomes. In Genome and Genomics: From Archaea to Eukaryotes; Springer: Singapore, 2019; pp. 1–30. [Google Scholar] [CrossRef]
- Koonin, E.S.P. Are viruses alive? The replicator paradigm sheds decisive light on an old but misguided question. Stud. Hist. Philos. Sci. Part C Stud. Hist. Philos. Biol. Biomed. 2016, 59, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.; Welch, J. A formal theory of the selfish gene. J. Evol. Biol. 2011, 24, 1801–1813. [Google Scholar] [CrossRef]
- Godfrey-Smith, P. The Replicator in Retrospect. Biol. Philos. 2000, 15, 403–423. [Google Scholar] [CrossRef]
- Griffiths, P.E.; Gray, R.D.; Journal of Philosophy Inc. Developmental Systems and Evolutionary Explanation. J. Philos. 1994, 91, 277–304. [Google Scholar] [CrossRef]
- Wilson, B. Developmental Genetics, EDTECH. 2018. Available online: www.google.co.uk/books/edition/Developmental_Genetics/LePEDwAAQBAJ?hl=en&gbpv=1&dq=Wilson.+B.+(2018).+Developmental+Genetics,+EDTECH,&pg=PR13&printsec=frontcover (accessed on 20 February 2022).
- Bhattacharyya, S. Ability as ‘performance’: Analyzing the ableness of ‘life’ through a critical study of The Shawshank Redemption and The Dark Knight. Rupkatha J. Interdiscip. Stud. Humanit. 2021, 13, 1–9. [Google Scholar] [CrossRef]
- Razeto-Barry, P. Autopoiesis 40 years Later. A Review and a Reformulation, Origins of Life and Evolution of Biospheres. PubMed 2012, 42, 543–567. [Google Scholar] [CrossRef]
- David, V.; Robert, L.; Serge, T.; Tom, Z. Embodied cognition and circular causality: On the role of constitutive autonomy in the reciprocal coupling of perception and action. Front. Psychol. 2015, 6, 1660. [Google Scholar] [CrossRef]
- Maturana, H.R.; Varela, F.J. The Tree of Knowledge: The Biological Roots of Human Understanding. Access: New Science Library/Shambhala Publications. 1987. Available online: https://uranos.ch/research/references/Maturana1988/maturana-h-1987-tree-of-knowledge-bkmrk.pdf (accessed on 20 February 2022).
- Herrera Pérez, C.; Ziemke, T. Aristotle, autonomy and the explanation of behaviour. Pragmat. Cogn. 2007, 15, 547–571. [Google Scholar] [CrossRef]
- Heath, M. Cognition in Aristotle’s Poetics. Mnemosyne 2009, 62, 51–75. [Google Scholar] [CrossRef]
- Colombetti, G. Enaction, Sense-making and Emotion. In Enaction: Towards a New Paradigm for Cognitive Science; Stewart, J., Gapenne, O., Di Paolo, E., Eds.; MIT Press: Cambridge MA, USA, 2010; pp. 145–164. [Google Scholar]
- Rosen, R. Anticipatory Systems; Pergamon Press: New York, NY, USA, 1985. [Google Scholar]
- Yolles, M.; Dubois, D. Anticipatory Viable Systems. Int. J. Comput. Anticip. Syst. 2001, 9, 3–20. [Google Scholar]
- Prigogine, I. Introduction to Thermodynamics of Irreversible Processes, 2nd ed.; Interscience: New York, NY, USA, 1961. [Google Scholar]
- Balietti, S.; Mäs, M.; Helbing, D. On Disciplinary Fragmentation and Scientific Progress. PLoS ONE 2015, 10, e0118747. [Google Scholar] [CrossRef]
- Wilson, D.S.; Sober, E. Reviving the Superorganism. J. Theor. Biol. 1989, 136, 337–356. [Google Scholar] [CrossRef]
- Herrero-Uribe, L. Viruses, definitions and reality. Rev. Biol. Trop. 2011, 59, 993–998. [Google Scholar] [CrossRef][Green Version]
- Tetz, V.V.; Tetz, G.V. A new biological definition of life. Biomol. Concepts 2019, 11, 1–6. [Google Scholar] [CrossRef]
- Carter, S.; Scott, M. Biology for Majors II, Luman Learning. 2021. Available online: https://courses.lumenlearning.com/wm-biology2/chapter/properties-of-life/ (accessed on 9 February 2022).
- Raybould, R.; Sims, R. Searching the Dark Genome for Alzheimer’s Disease Risk Variants. Brain Sci. 2021, 11, 332. [Google Scholar] [CrossRef]
- Gericke, N.M.; Hagberg, M.; dos Santos, V.C.; Joaquim, L.M.; El-Hani, C.N.; Mariane, J.L. Conceptual Variation or Incoherence? Textbook Discourse on Genes in Six Countries. Sci. Educ. 2012, 23, 381–416. [Google Scholar] [CrossRef]
- CDC. What Is Epigenetics? Centre for Desease Control and Prevention, August 2020. Available online: https://www.cdc.gov/genomics/disease/epigenetics.htm#:~:text=Epigenetics%20is%20the%20study%20of,body%20reads%20a%20DNA%20sequence (accessed on 10 February 2022).
- Sarkar, S. Evolutionary Theory in the 1920s: The Nature of the “Synthesis”; Mitchell, S.D., Ed.; The Philosophy of Science: Pittsburgh, PA, USA, 2004; Volume 71, pp. 1215–1226. [Google Scholar]
- Haldane, J.B.S. A Mathematical Theory of Natural and Artificial Selection, Part I; Transactions of the Cambridge Philosophical Society: Cambridge, UK, 1924; Volume 23, pp. 19–41. [Google Scholar]
- Yewdall, N.A.; Mason, A.F.; van Hest, J.C.M. The hallmarks of living systems: Towards creating artificial cells. R. Society 2018, 8, 20180023. [Google Scholar] [CrossRef]
- Bich, L.; Arnellos, A. Autopoiesis, Autonomy, and Organizational Biology: Critical Remarks on “Life After Ashby”. Cybern. Hum. Knowing 2014, 19, 75–103. [Google Scholar]
- Collier, J. Autonomy and Process Closure as the Basis for Functionality. 2020. Available online: https://www.researchgate.net/publication/227891444_Autonomy_and_Process_Closure_as_the_Basis_for_Functionality (accessed on 9 February 2022).
- Mossio, M.; Saborido, C. Functions, Organization and Etiology: A Reply to Artiga and Martinez. Acta Biotheor. 2016, 64, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Niño EL-Hani, C.; Nunes-Neto, N.F. Function in Biology: Etiological and Organizational Perspectives. Acta Biol. Colomb. 2009, 14, 111–132. [Google Scholar]
- Collier, J. Interactively Open Autonomy Unifies Two Approaches to Function. In Computing Anticipatory Systems: CASY’03—Sixth International Conference; Dubois, D.M., Ed.; American Institute of Physics: Melville, NY, USA, 2004; pp. 228–235. [Google Scholar]
- Bich, L.; Etxeberria, A. Systems, Autopoietic. In Encyclopedia of Systems Biology; Dubitzsky, W., Wolkenhauer, D., Cho, K.H., Yokota, H., Eds.; Spinger: New York, NY, USA, 2013; pp. 2110–2113. [Google Scholar]
- de Carvalho Fidelis Braga, J. The Complex Nature of Autonomy. Doc. De Estud. Em Lingüística Teórica E Apl. 2008, 24, 441–468. [Google Scholar]
- Arnellos, A.; Spyrou, T.; Darzentas, J. Cybernetic embodiment and the role of autonomy in the design process. Kybernetes 2007, 36, 1207–1224. [Google Scholar] [CrossRef][Green Version]
- Bertschinger, N.; Olbrich, E.; Ay, N.; Jost, J. Autonomy: An information theoretic perspective. Biosystems 2008, 91, 331–345. [Google Scholar] [CrossRef]
- Bitbol, M.; Luisi, P.L. Autopoiesis with or without cognition: Defining life at its edge. J. R. Soc. Interface 2004, 1, 99–107. [Google Scholar] [CrossRef]
- Luisi, P. Autopoiesis: A review and a reappraisal. Naturwissenschaften 2003, 90, 49–59. [Google Scholar] [CrossRef]
- Claverie, J.M.; Abergel, C. Giant viruses: The difficult breaking of multiple epistemological barriers. Stud. Hist. Philos. Sci. Part C Stud. Hist. Philos. Biol. Biomed. Sci. 2016, 59, 89–99. [Google Scholar] [CrossRef]
- Boden, M. Autopoiesis and Life. Cogn. Sci. Q. 2000, 1, 117–145. [Google Scholar]
- Billman, G.E. Homeostasis: The Underappreciated and Far Too Often Ignored Central Organizing Principle of Physiology. Front. Physiol. 2020, 11, 200. [Google Scholar] [CrossRef] [PubMed]
- Turner, S. Homeostasis as a Fundamental Principle for a Coherent Theory of Brains, The Royal Society. 2019. Available online: https://royalsocietypublishing.org/doi/10.1098/rstb.2018.0373 (accessed on 25 February 2022).
- Williams, H. Homeostatic Adaptive Networks. Doctoral Thesis, University of Leeds, Leeds, UK, 2006. Available online: www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&cad=rja&uact=8&ved=2ahUKEwidjLuRrs31AhWPFMAKHQ0LCFcQFnoECAQQAQ&url=https%3A%2F%2Feth (accessed on 20 February 2022).
- Diéguez, A. Life as a Homeostatic Property Cluster. Biol. Theory 2013, 7, 180–186. [Google Scholar] [CrossRef]
- Chiller, J.M.; Louis, J.A.; Skidmore, B.J.; Weigle, W.O. Cellular Parameters of The Tolerant State Induced to Human Γ Globulin in Mice and of its Modulation by Bacterial Lipopolysaccharides. In Immunological Tolerance; Katz, H., Benacerraf, B., Eds.; Academic Press: Cambridge, MA, USA, 1974; pp. 373–390. [Google Scholar]
- Marques, R.E.; Marques, P.E.; Guabiraba, R.; Teixeira, M.M. Exploring the Homeostatic and Sensory Roles of the Immune System. Front. Immunol. 2016, 7, 125. [Google Scholar] [CrossRef]
- Ashby, W. Introduction to Cybernetics; Chapman & Hall: London, UK, 1956. [Google Scholar]
- Russell, S. What’s with the Spikes? Those Structures that Give Coronavirus Its Mame might be SARS-CoV-2’s Weak Point. Hutch News Service. 3 April 2020. Available online: https://www.fredhutch.org/en/news/center-news/2020/04/covid19-virus-spike-structure.html (accessed on 9 February 2022).
- Ferron, F.; Subissi, L.; Silveira De Morais, A.T.; Le, N.T.T.; Sevajol, M.; Gluais, L.; Decroly, E.; Vonrhein, C.; Bricogne, G.; Canard, B.; et al. Structural and molecular basis of mismatch correction and ribavirin excision from corona-virus. PNAS 2017, 115, E162–E171. [Google Scholar] [CrossRef]
- Chi, K.R. The dark side of the human genome. Nature 2016, 538, 275–277. [Google Scholar] [CrossRef]
- Bai, N.; Smith, D. The Mysterious 98%: Scientists Look to Shine Light on Our Dark Genome. University of California San Francisco Research. 2017. Available online: https://www.ucsf.edu/news/2017/02/405686/mysterious-98-scientists-look-shine-light-our-dark-genome (accessed on 20 February 2022).
- Wood, L. Uncovering How ‘Dark Matter’ Regions of the Genome Affect Inflammatory Diseases. Babaham Institute Report. 13 May 2020. Available online: https://www.babraham.ac.uk/news/2020/05/uncovering-how-dark-matter-regions-genome-affect-inflammatory-diseases (accessed on 10 February 2022).
- Nasrallah, R.; Imianowski, C.J.; Bossini-Castillo, L.; Grant, F.M.; Dogan, M.; Placek, L.; Kozhaya, L.; Kuo, P.; Sadiyah, F.; Whiteside, S.K.; et al. A distal enhancer at risk locus 11q13.5 promotes suppression of colitis by Treg cells. Nature 2020, 583, 447–452. [Google Scholar] [CrossRef]
- Ma, L.-G.; Zhao, H.-G.; Deng, X.W. Analysis of the mutational effects of the COP/DET/FUS loci on genome expres-sion profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development 2003, 130, 969–981. [Google Scholar] [CrossRef]
- Rogers, P. Clues to Schizophrenia and Bipolar Disorder Hidden in the Dark Ge-Nome. The Big Think. 7 March 2022. Available online: https://bigthink.com/health/dark-genome-schizophrenia-biopolar/ (accessed on 20 March 2022).
- Forterre, P.; Prangishvili, D. The Great Billion-year War between Ribosome- and Capsid-encoding Organisms (Cells and Viruses) as the Major Source of Evolutionary Novelties. Ann. N. Y. Acad. Sci. 2009, 1178, 65–77. [Google Scholar] [CrossRef]
- Lwoff, A. The concept of virus. J. Gen. Microbiol. 1957, 17, 239–253. [Google Scholar] [CrossRef]
- Luria, S. Viruses as infective genetic materials. In Immunity and Virus Infection; Najjar, V., Ed.; Wiley: New York, NY, USA, 1959; pp. 188–195. [Google Scholar]
- Morowitz, H.J. Energy Flow in Biology; Academic Press: New York, NY, USA, 1968. [Google Scholar]
- Harold, F. The Vital Force: A Study of Bioenergetics; W.H. Freemann: New York, NY, USA, 1986. [Google Scholar]
- Villarreal, L. Are viruses alive? Sci. Am. 2004, 29, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.; Coleman, M.; Weigele, P.; Rohwer, F.; Chisholm, S. Three Prochlorococcus cyanophage genomes: Signature features and ecological interpretation. PLoS Biol. 2005, 3, 791–806. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S. Evolving Responsively: Adaptive Mutation. Nat. Rev. Genet. 2001, 2, 504–515. [Google Scholar] [CrossRef]
- Moelling, K.; Broecker, F. Viroids and the Origin of Life. Int. J. Mol. Sci. 2021, 22, 3476. [Google Scholar] [CrossRef] [PubMed]
- Luong, L. Genetics, Connexions; Rice University: Houston, TX, USA, 2009. [Google Scholar]
- ScienceDaily. Viruses Don’t Have a Metabolism; but Some Have the Building Blocks for One. Science Daily. 6 April 2020. Available online: www.sciencedaily.com/releases/2020/04/200406092839.htm (accessed on 9 February 2022).
- Pradeu, T.; Kostyrka, G.; Dupré, J. Understanding Viruses: Philosophical Investigations (Editprial). Stud. Hist. Philos. Biol. Biomed. Sci. 2016. Available online: www.sciencedirect.com/science/article/pii/S1369848616300024 (accessed on 7 January 2022).
- Roossinck, M.J.; Bazán, E.R. Symbiosis: Viruses as Intimate Partners. Annu. Rev. Virol. 2017, 4, 123–139. [Google Scholar] [CrossRef]
- Moore, P.S.; Chang, Y. Common Commensal Cancer Viruses. PLoS Pathog. 2017, 13, e1006078. [Google Scholar] [CrossRef]
- Pradeu, T. Mutualistic viruses and the heteronomy of life. Stud. Hist. Philos. Sci. Part C Stud. Hist. Philos. Biol. Biomed. Sci. 2016, 59, 80–88. [Google Scholar] [CrossRef]
- Roossinck, M.J. Move over, Bacteria! Viruses Make Their Mark as Mutualistic Microbial Symbionts. J. Virol. 2015, 89, 6532–6535. [Google Scholar] [CrossRef]
- Roossinck, M. The good viruses: Viral mutualistic symbioses. Nat. Rev. Genet. 2011, 9, 99–108. [Google Scholar] [CrossRef]
- Grasis, J. The Intra-Dependence of Viruses and the Holobiont. Front. Immunol. 2017, 8, 1501. [Google Scholar] [CrossRef]
- Martiny, J.B.H.; Marston, M.F.; Riemann, L.; Middelboe, M. Antagonistic Coevolution of Marine Planktonic Viruses and Their Hosts. Annu. Rev. Mar. Sci. 2014, 6, 393–414. [Google Scholar] [CrossRef]
- Brüssow, H.; Brüssow, L. Clinical evidence that the pandemic from 1889 to 1891 commonly called the Russian flu might have been an earlier coronavirus pandemic. Microb. Biotechnol. 2021, 14, 1860–1870. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J. Will Omicron End COVID-19 Pandemic Like a Mild Variant Did to Spanish Flu 100 Years Back? Outlook. Available online: https://www.outlookindia.com/website/story/india-news-will-omicron-end-covid-pandemic-like-a-mild-variant-did-to-spanish-flu-100-years-back/407090 (accessed on 20 January 2022).
- Williams, E.S.C.P.; Morales, N.M.; Wasik, B.R.; Brusic, V.; Whelan, S.P.J.; Turner, P.E. Repeatable Population Dynamics among Vesicular Stomatitis Virus Lineages Evolved under High Co-infection. Front. Microbiol. 2016, 7, 370. [Google Scholar] [CrossRef]
- Case, L.K.; Wall, E.H.; Dragon, J.A.; Saligrama, N.; Krementsov, D.N.; Moussawi, M.; Zachary, J.F.; Huber, S.A.; Blankenhorn, E.P.; Teuscher, C. The Y chromosome as a regulatory element shaping immune cell transcriptomes and susceptibility to autoimmune disease. Genome Res. 2013, 23, 1474–1485. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, J. Memory in the innate and adaptive immune systems. Microbes Infect. 2004, 6, 1410–1417. [Google Scholar] [CrossRef]
- Maital, S.; Barzani, E. We Will Bounce Back: A Real-Time Coronavirus Diary; Samuel Neaman Institute: Haifa, Israel, 2020. [Google Scholar]
- Vossen, M.T.; Westerhout, E.M.; Söderberg-Nauclér, C.; Wiertz, E.J. Viral immune evasion: A masterpiece of evolution. Immunogenetics 2002, 54, 527–542. [Google Scholar] [CrossRef] [PubMed]
- Broecker, F.; Moelling, K. Evolution of Immune Systems from Viruses and Transposable Elements. Front. Microbiol. 2019, 10, 51. [Google Scholar] [CrossRef]
- Wessner, D.R. Discovery of the Giant Mimivirus. Nat. Educ. 2010, 3, 61. [Google Scholar]
- Klein, J.S.; Bjorkman, P.J. Few and Far between: How HIV May Be Evading Antibody Avidity. PLoS Pathog. 2010, 6, e1000908. [Google Scholar] [CrossRef]
- Naughtie, A. HIV ‘Invisibility Cloak’ Allows Virus to Evade Immune System. The Conversation. 6 November 2013. Available online: https://theconversation.com/hiv-invisibility-cloak-allows-virus-to-evade-immune-system-19918 (accessed on 20 February 2022).
- Nadin, E. A Viral Cloaking Device: Caltech Biologists Show How Human Cytomegalovirus Hides from the Immune System. Caltech. 18 July 2008. Available online: https://www.caltech.edu/about/news/viral-cloaking-device-caltech-biologists-show-how-human-cytomegalovirus-hides-immune-system (accessed on 10 February 2022).
- Gallagher, S. New Study Shows Viruses Can Have Immune Systems. Tufts Now. 9 February 2013. Available online: https://now.tufts.edu/news-releases/new-study-shows-viruses-can-have-immune-systems (accessed on 20 February 2022).
- Bondy-Denomy, J.; Pawluk, A.; Maxwell, K.; Davidson, A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 2012, 493, 429–432. [Google Scholar] [CrossRef]
- Zimmer, C. The Virus That Learns. Natl. Geogr. 2013. Available online: www.nationalgeographic.com/science/article/the-virus-that-learns (accessed on 10 January 2022).
- Holmes, R.K.; Jobling, M.G. Genetics. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Gal-veston: Austin, TX, USA, 1996. [Google Scholar]
- Ye, Z.; Zhang, Q. Characterization of CRISPR RNA transcription by exploiting stranded metatranscriptomic data. RNA 2016, 22, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Villalobos, M.W.D. Living Systems: Autonomy, Autopoiesis and Enaction. Philos. Technol. 2014, 28, 225–239. [Google Scholar] [CrossRef]
- Maturana, H.R.; Varela, F.J. Autopoiesis and Cognition, the Realization of the Living; Reidel: Dordrecht, The Netherlands, 1980. [Google Scholar]
- Turabian, J. Biopsychosocial Causality in General Medicine: Knot, Ball, and Tangle. Epidemiol. Int. J. 2019, 3, 1–9. [Google Scholar] [CrossRef]
- Steward, H. Actions as Processes. Philosophical Perspectives. Philos. Mind 2012, 26, 373–388. [Google Scholar]
- Meincke, A. Autopoiesis, biological autonomy and the process view of life. Eur. J. Philos. Sci. 2018, 9, 5. [Google Scholar] [CrossRef]
- Maturana, H. The organization of the living: A theory of the living organization. Int. J. Man-Mach. Stud. 1975, 7, 313–332. [Google Scholar] [CrossRef]
- Varela, F. Principles of Biological Autonomy; Elsevier: New York, NY, USA, 1979. [Google Scholar]
- Varela, F. Organism: A meshwork of selfless selves. In Organism and the Origin of Self, Dordrecht; Tauber, A., Ed.; Kluwer Academic Publishers: Norwell, MA, USA, 1991; pp. 79–107. [Google Scholar]
- Di Paolo, E.; Thompson, E. The enactive approach. In The Routledge Handbook of Embodied Cognition; Routledge: Abingdon-on-Thames, UK, 2014; pp. 86–96. [Google Scholar]
- Bensaude-Vincent, B. Self-Assembly, Self-Organization: A Philosophical Perspective on a Major Challenge of Nanotechnology. 2006. Available online: https://halshs.archives-ouvertes.fr/halshs-00350831/ (accessed on 21 February 2022).
- Amoroso, R.L.; Amoroso, P.J. The Fundamental Limit and Origin of Complexity in Biological Systems: A New Model for the Origin of Life. In Proceedings of the CP718, Computing Anticipatory Systems: CASYS03—Sixth International Conference, Liège, Belgium, 11–16 August 2003; Dubois, D.M., Ed.; American Institute of Physics: College Park, MD, USA, 2004. [Google Scholar]
- Drãgãnescu, M. On the structural phenomenological theories of consciousness. Noetic J. 1997, 1, 28–33. [Google Scholar]
- Miller, W.B., Jr.; Enguita, F.J.; Leitão, A.L. Non-Random Genome Editing and Natural Cellular Engineering in Cognition-Based Evolution. Cells 2021, 10, 1125. [Google Scholar] [CrossRef]
- Yolles, M. Management Systems: A Viable Approach; Financial Times Pitman: London, UK, 1999. [Google Scholar]
- Foster, P. Mechanisms of Stationary Phase Mutation: A Decade of Adaptive Mutation. Annu. Rev. Genet. 1999, 33, 57–88. [Google Scholar] [CrossRef]
- Foster, P.L. Adaptive mutation: Implications for evolution. BioEssays News Rev. Mol. Cell. Dev. Biol. 2000, 22, 1067–1074. [Google Scholar] [CrossRef]
- Chen, J.; Swofford, R.; Johnson, J.; Cummings, B.B.; Rogel, N.; Lindblad-Toh, K.; Haerty, W.; di Palma, F.; Regev, A. A quantitative framework for characterizing the evolutionary history of mammalian gene expression. Genome Res. 2018, 29, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Darren, P.; Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Ren, W.; Lan, J.; Ju, X.; Gong, M.; Long, Q.; Zhu, Z.; Yu, Y.; Wu, J.; Zhong, J.; Zhang, R.; et al. Mutation Y453F in the spike protein of SARSCoV-2-CoV-2 enhances interaction with the mink ACE2 receptor for host adaption. PLoS Pathog. 2021, 17, e1010053. [Google Scholar] [CrossRef] [PubMed]
- Munkevics, M. How Do Viruses Evolve and Why it Happens so Quickly. Monkey Gene. 2020. Available online: https://www.monkeygene.com/how-viruses-evolve/?gclid=CjwKCAiA866PBhAYEiwANkIneKkZ2tbVc0HyxUwVx0xBYNmRW8p3JQWhB60VB2Ly40JZnb8yIA7r6xoCpAIQAvD_BwE (accessed on 20 February 2022).
- QBI. Harnessing Viruses to Repair Damaged Nerve Cells. Queensland Brain Institute, University of Queensland, 2020. Available online: https://qbi.uq.edu.au/brain/nature-discovery/harnessing-viruses-repair-damaged-nerve-cells (accessed on 20 February 2022).
- Jacobsen, R. Artificial Proteins Never Seen in the Natural World Are Becoming New COVID Vaccines and Medicines. Scientific American. 1 July 2021. Available online: https://www.scientificamerican.com/article/artificial-proteins-never-seen-in-the-natural-world-are-becoming-new-covid-vaccines-and-medicines/ (accessed on 20 January 2022).
- Griffin, S.; Masood, M.I.; Nasim, M.J.; Sarfraz, M.; Ebokaiwe, A.P.; Schäfer, K.-H.; Keck, C.M.; Jacob, C. Natural Nanoparticles: A Particular Matter Inspired by Nature. Antioxidants 2017, 7, 3. [Google Scholar] [CrossRef]
- Stern, A.; Andino, R. Viral Evolution: It Is All about Mutations. In Viral Pathogenesis, 3rd ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 233–240. [Google Scholar] [CrossRef]
- Domingo, E.; Escarmís, C.A.; Sevilla, N.; Moya, A.; Elena, S.F.; Quer, J.; Novella, I.S.; Holland, J.J. Basic concepts in RNA virus evolution. FASEB J. 1996, 10, 859–864. [Google Scholar] [CrossRef]
- Simmonds, P.; Aiewsakun, P.; Katzourakis, A. Prisoners of war—Host adaptation and its constraints on virus evolution. Nat. Rev. Microbiol. 2019, 17, 321–328. [Google Scholar] [CrossRef]
- Lawton, G. Viruses Have Busy Social Lives That We Could Manipulate to Defeat Them. New Scientist. 21 October 2020. Available online: https://www.newscientist.com/article/mg24833052-900-viruses-have-busy-social-lives-that-we-could-manipulate-to-defeat-them/ (accessed on 20 February 2022).
- West, S.A.; Griffin, A.S.; Gardner, A.; Diggle, S.P. Social Evolution Theory for Microorganisms. Nat. Rev. Genet. 2006, 4, 597–607. [Google Scholar] [CrossRef]
- Guttinger, S. A Virus Is Not a Thing, Part 1: The Case for a Process View of Viruses; London School of Economics, Dept. Philosophy: London, UK, 2020. [Google Scholar]
- Arnold, C. How Mutant Viral Swarms Spread Disease. Quantum Magazine. 25 August 2015. Available online: https://www.quantamagazine.org/how-mutant-viral-swarms-spread-disease-20150825 (accessed on 21 February 2022).
- Yolles, M. The Socio-Cultural Dynamics of Development: Part 3 Agency Theory. Kybernetes 2019, 49, 1871–1898. [Google Scholar] [CrossRef]
- Whitley, R. Business Systems; Manchester Business School: Manchester, UK, 1994. [Google Scholar]
- Dopfer, K.; Foster, J.; Potts, J. Micro–meso–macro. J. Evol. Econ. 2004, 14, 263–279. [Google Scholar] [CrossRef]
- Dopfer, K. The Origins of Meso Economics: Schumpeter’s Legacy; Max Lank Institute: Munich, Germany, 2006. [Google Scholar]
- Dopfer, K. Mesoeconomics: A unified approach to systems complexity and evolution. In Handbook on the Economic Complexity of Technological Change; Antonelli, C., Ed.; Edward Elgar Publishing: Cheltenham, MA, USA, 2011; pp. 341–346. [Google Scholar]
- Schwarz, E. Can Real Life Complex Systems Be Interpreted with the Usual Dualist Physicalist Epistemology—Or Is a Holistic Approach Necessary? ResearchGate. 2002. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.542.1330&rep=rep1&type=pdf (accessed on 20 February 2022).
- Casti, J.; Karlkvist, A. Towards a Theory of Models for Living Systems; Birkhauser: Boston, UK, 1989. [Google Scholar]
- Frieden, B.G.R. Information Dynamics in Living Systems: Prokaryotes, Eukaryotes, and Cancer. PLoS ONE 2011, 6, e22085. [Google Scholar] [CrossRef] [PubMed]
- Gare, A. Systems Theory and Complexity: Introduction. Democr. Nat. 2000, 6, 327–339. [Google Scholar] [CrossRef]
- Cohen, J.; Stewart, I. The Collapse of Chaos; Viking, Penguin Books: London, UK, 1995. [Google Scholar]
- Halbe, J.; Adamowski, J.; Bennett, E.M.; Pahl-Wostl, C.; Farahbakhsh, K. Functional organization analysis for the design of sustainable engineering systems. Ecol. Eng. 2014, 73, 80–91. [Google Scholar] [CrossRef]
- Mossio, M.; Saborido, C.; Moreno, A. An Organizational Account of Biological Functions. Br. J. Philos. Sci. 2009, 60, 813–841. [Google Scholar] [CrossRef]
- Luhmann, N. The Autopoiesis of Social Systems. In Sociocybernetic Paradoxes: Observation, Control and Evolution of Self-Steering Systems; Geyer, F., Zeuwen, J., Eds.; Sage: London, UK, 1986; pp. 172–192. [Google Scholar]
- Seidl, D. Luhmann’s Theory of Autopoietic Social Systems; Ludwig-Maximilians-Universität München-Munich School of Management: Munich, Germany, 2004. [Google Scholar]
- Schwarz, E. A Generic Model for the Emergence and Evolution of Natural Systems toward Complexity and Autonomy. In Proceedings of the 36th Annual Meeting of the ISSS, Denver, CO, USA, 18–21 May 2016; Volume II, p. 766. [Google Scholar]
- Piaget, J. The Psychology of Intelligence; Routledge: London, UK, 1950. [Google Scholar]
- Golledge, R. Cognitive maps. In Encyclopedia of Social Measurement; Elsevier: Amsterdam, The Netherlands, 2005; pp. 329–339. [Google Scholar] [CrossRef]
- Mitchell, A.; Lim, W. Cellular perception and misperception: Internal models for decision-making shaped by evolutionary experience. BioEssays 2016, 38, 845–849. [Google Scholar] [CrossRef]
- Emerson, E. Researcher Teases Out Secrets from Surprisingly ‘Intelligent’ Viruses. University of California News. 19 October 1998. Available online: https://news.usc.edu/9791/researcher-teases-out-secrets-from-surprisingly-intelligent-viruses/ (accessed on 20 February 2022).
- Wanjek, C. Systems Biology as Defined by NIH: An Intellectual Resource for Integrative Biology. Natl. Inst. Health Catal. 2011, 19, 6. [Google Scholar]
- Fang, F.C.; Casadevall, A. Reductionistic and Holistic Science. Infect. Immun. 2011, 79, 1401–1404. [Google Scholar] [CrossRef]
- Galitski, T. Reductionism Gives Way to Systems Biology. Genet. Eng. Biotechnol. News 2012, 32, 6. [Google Scholar] [CrossRef]
- Trafton, A. Controlling RNA in Living Cells. MIT News. 25 April 2016. Available online: https://news.mit.edu/2016/controlling-rna-living-cells-0425 (accessed on 21 February 2022).
- Helen A Foster, H.A.; Bridger, J.M. The genome and the nucleus: A marriage made by evolution. Chromosoma 2005, 114, 212–229. [Google Scholar] [CrossRef]
- Augustyn, A. Membrane. In Encyclopedia Britannica; Encyclopedia Britannica: Chicago, IL, USA, 2022. [Google Scholar]
- Yeates, T.O.; Thompson, M.C.; Bobik, T.A. The protein shells of bacterial microcompartment organelles. Curr. Opin. Struct. Biol. 2011, 21, 223–231. [Google Scholar] [CrossRef]
- Walsh, D.; Mathews, M.B.; Mohr, I. Tinkering with Translation: Protein Synthesis in Virus-Infected Cells. Cold Spring Harb. Perspect. Biol. 2012, 5, a012351. [Google Scholar] [CrossRef] [PubMed]
- Graham, F. Largest Bacterium ever Discovered Is 2 cm Long. Nature Briefing. 24 February 2022. Available online: https://www.nature.com/articles/d41586-022-00579-5 (accessed on 2 March 2022).
- Mokobi, F. 7 Types of RNA with Structure and Functions. Top Form. 2020. Available online: https://microbenotes.com/types-of-rna/ (accessed on 20 February 2022).
- Cheriyedath, S. Types of RNA mRNA, rRNA and tRNA. 2020. Available online: https://www.news-medical.net/life-sciences/-Types-of-RNA-mRNA-rRNA-and-tRNA.aspx (accessed on 22 January 2022).
- Wang, D.; Farhana, A. Biochemistry, RNA Structure. 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK558999/ (accessed on 14 January 2022).
- Ying, S.Y.; Chang, D.C.; Lin, S.L. The MicroRNA (miRNA): Overview of the RNA Genes that Modulate Gene Function. Mol. Biotechnol. 2007, 38, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Munir, A.; Mehmood, A.; Azam, S. Structural and Function Prediction of Musa acuminata subsp. Malaccensis Protein. Int. J. Bioautomotion 2016, 20, 19–30. [Google Scholar]
- LePage, M. Giant Viruses Have Weaponised CRISPR against Their Bacterial Hosts. New Scientist. 30 March 2019. Available online: https://www.newscientist.com/article/2197422-giant-viruses-have-weaponised-crispr-against-their-bacterial-hosts/ (accessed on 2 January 2022).
- Martin, L.; Chang, H.Y. Uncovering the role of genomic “dark matter” in human disease. J. Clin. Investig. 2012, 122, 1589–1595. [Google Scholar] [CrossRef]
- Lawton, J.A.; Estes, M.K.; Prasad, B.V.V. Mechanism of genome transcription in segmented dsRNA viruses. Adv. Virus Res. 2000, 55, 185–229. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hong, T.; Parameswaran, S.; Ernst, K.; Marazzi, I.; Weirauch, M.T.; Bass, J.I.F. Human Virus Transcriptional Regulators. Cell 2020, 182, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Cortes, M.G.; Trinh, J.T.; Guan, J.; Balázsi, G.; Zeng, L. Coupling of DNA Replication and Negative Feedback Controls Gene Expression for Cell-Fate Decisions. iScience 2018, 6, 1–12. [Google Scholar] [CrossRef]
- Frederick, E. RNA Molecules Are Masters of Their Own Destiny. Harvard MIT Health Science and Technology News. 21 December 2020. Available online: https://hst.mit.edu/news-events/rna-molecules-are-masters-their-own-destiny (accessed on 21 February 2022).
- Istrail, S.; Peter, I.S. How Does the Regulatory Genome Work? J. Comput. Biol. 2019, 26, 685–695. [Google Scholar] [CrossRef]
- Hensel, M.U.; Menges, A.; Weinstock, M. Morphogenesis and Emergence. In The Digital Turn in Architecture; Carpo, M., Ed.; Wiley: Chichester, UK, 2012; pp. 1992–2012. [Google Scholar]
- Frieden, B. Science from Fisher Information. A Unification; Cambridge Universty Press: Cambridge, UK, 2004. [Google Scholar] [CrossRef]
- Frieden, B.R.; Gatenby, R.A. Principle of maximum Fisher information from Hardy’s axioms applied to statistical systems. Phys. Rev. E 2013, 88, 042144. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Frieden, B.R. Application of information theory and extreme physical information to carcinogenesis. Cancer Res. 2002, 62, 3675–3684. [Google Scholar]
- McLaughlin-Drubin, M.E.; Munger, K. Viruses associated with human cancer. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2008, 1782, 127–150. [Google Scholar] [CrossRef]
- Würdinger, T.; Verheije, M.H.; Raaben, M.; Bosch, B.J.; Haan, C.A.M.D.; Van Beusechem, V.W.; Rottier, P.J.M.; Gerritsen, W.R. Targeting non-human coronaviruses to human cancer cells using a bispecific single-chain antibody. Gene Ther. 2005, 12, 1394–1404. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Goshua, A. A Man in Italy Got COVID-19. Then His Cancer Went into Remission. Medical Examiner. 29 April 2021. Available online: http://slate.com/technology/2021/04/cancer-COVID-remission-treatment-viruses.html (accessed on 2 January 2022).
- Würdinger, T.; Verheije, M.H.; van Beusechem, V.W.; de Haan, C.A.M.; Rottier, P.J.M.; Gerritsen, W.R. Coronaviruses Are Able to Efficiently Eradicate Human Tumor Cells If Provided with the Appropriate Virus Receptor. Mol. Ther. 2003, 7, S182. [Google Scholar]
- Howells, A.; Marelli, G.; Lemoine, N.R.; Wang, Y. Oncolytic Viruses—Interaction of Virus and Tumor Cells in the Battle to Eliminate Cancer. Front. Oncol. 2017, 7, 195. [Google Scholar] [CrossRef] [PubMed]
- van Trees, H. Detection, Estimation and Modulation Theory, Part I; Wiley: New York, NY, USA, 1968. [Google Scholar]
- Awazu, A.; Tanabe, T.; Kamitani, M.; Tezuka, A.; Nagano, A.J. Broad distribution spectrum from Gaussian to power law appears in stochastic variations in RNA-seq data. Sci. Rep. 2018, 8, 8339. [Google Scholar] [CrossRef] [PubMed]
- Stern, D. Evolution of Gene Expression. 2021. Available online: https://www.oxfordbibliographies.com/view/document/obo-9780199941728/obo-9780199941728-0085.xml (accessed on 22 January 2022).
- Gumpper, R.H.; Li, W.; Luo, M. Constraints of Viral RNA Synthesis on Codon Usage of Negative-Strand RNA Virus. J. Virol. 2019, 93, e01775-18. [Google Scholar] [CrossRef] [PubMed]
- Chaitin, G.J. Toward a mathematical definition of ‘life’. In Aximum Entropy Principle; Levine, R.D., Tribus, M., Eds.; MIT Press: Cambridge, MA, USA, 1979. [Google Scholar]
- Wu, J.-P.; Liao, W.; Udenigwe, C.C. Revisiting the mechanisms of ACE inhibitory peptides from food proteins. Trends Food Sci. Technol. 2017, 69, 214–219. [Google Scholar] [CrossRef]
- Sriram, K.; Insel, P.; Loomba, R. What Is the ACE2 Receptor? Am. Soc. Biochem. Mol. Biol. 2020. Available online: https://www.asbmb.org/asbmb-today/science/051620/what-is-the-ace2-receptor (accessed on 16 May 2020).
- Sriram, K.; Insel, P.; Loomba, R. What Is the ACE2 Receptor, How Is It Connected to Coronavirus and Why might It Be Key to Treating COVID-19? The Experts Explain. The Conversation. 4 May 2020. Available online: https://theconversation.com/what-is-the-ace2-receptor-how-is-it-connected-to-coronavirus-and-why-might-it-be-key-to-treating-covid-19-the-experts-explain-136928 (accessed on 24 January 2022).
- Procko, E. The sequence of human ACE2 is suboptimal for binding the S spike protein of SARS coronavirus 2. bioRxiv 2020. bioRxiv: 2020.03.16.994236. [Google Scholar] [CrossRef]
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020, 126, 1456–1474. [Google Scholar] [CrossRef]
- Li, W.; Li, M.; Ou, G. COVID-19, cilia, and smell. FEBS J. 2020, 28, 3672–3676. [Google Scholar] [CrossRef]
- Pagliaro, P.; Penna, C. ACE/ACE2 Ratio: A Key Also in 2019 Corona-Virus Disease (COVID-19)? Opinion 2020, 7, 335. [Google Scholar] [CrossRef]
- Babtie, A.C.; Stumpf, M.P.H. How to deal with parameters for whole-cell modelling. J. R. Soc. Interface 2017, 14, 20170237. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Karki, C.B.; Du, D.; Li, H.; Wang, J.; Sobitan, A.; Teng, S.; Tang, Q.; Li, L. Spike Proteins of SARS-CoV and SARS-CoV-2 Utilize Different Mechanisms to Bind with Human ACE2. Front. Mol. Biosci. 2020, 7, 591873. [Google Scholar] [CrossRef] [PubMed]
- Frieden, B. Principle of minimum loss of Fisher information. In Handbook of Statistics: Information Geometry; North-Holland, Elsevier: Amsterdam, The Netherlands, 2021; Chapter 6. [Google Scholar]
- Ritchie, H.; Mathieu, E.; Rodés-Guirao, L.; Appel, C.; Giattino, C.; Ortiz-Ospina, E.; Hasell, J.; Macdonald, B.; Beltekian, D.; Roser, M. Coronavirus Pandemic (COVID-19). 2021. Available online: https://ourworldindata.org/coronavirus (accessed on 22 January 2022).
- Paulsson-Habegger, L.; Snabaitis, A.K.; Wren, S.P. Enzyme inhibition as a potential therapeutic strategy to treat COVID-19 infection. Bioorganic Med. Chem. 2021, 48, 116389. [Google Scholar] [CrossRef] [PubMed]
- Mousavizadeh, L.G.S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020, 54, 159–163. [Google Scholar] [CrossRef]
- Thomas, S. The Structure of the Membrane Protein of SARS-CoV-2 Resembles the Sugar Transporter SemiSWEET. Pathog Immun. 2020, 5, 342–363. [Google Scholar] [CrossRef]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef]
- Bhardwaj, K.K.; Verma, A.K. Corona Outbreak: Is It Nature’s Wake-Up Call? Int. J. Appl. Biol. Pharm. Technol. 2021, 12, 301–321. [Google Scholar]
- Sibley, C. Regulation of gene expression through production of unstable mRNA isoforms. Biochem. Soc. Trans. 2014, 42, 1196–1205. [Google Scholar] [CrossRef]
- Yolles, M. Organizations as Complex Systems: An introduction to Knowledge Cybernetics; Information Age Publishing, Inc.: Greenwich, CT, USA, 2006. [Google Scholar]
- Chodasewicz, K. Evolution, reproduction and definition of life. Theory Biosci. Theor. Den Biowiss. 2014, 133, 39–45. [Google Scholar] [CrossRef]
- Kelty-Stephen, D.G.; Dixon, J.A. When Physics Is Not “Just Physics”: Complexity Science Invites New Measurement Frames for Exploring the Physics of Cognitive and Biological Development. Crit. Rev. Biomed. Eng. 2012, 40, 471–483. [Google Scholar] [CrossRef] [PubMed]
- van Bakel, H.; Nislow, C.; Blencowe, B.J.; Hughes, T.R. Most “Dark Matter” Transcripts Are Associated with Known Genes. PLoS Biol. 2010, 8, e1000371. [Google Scholar] [CrossRef]
- GEP. Why mRNA Vaccines Aren’t Gene Therapies. Genomics Education Program of the UK National Health Service. 11 June 2021. Available online: https://www.genomicseducation.hee.nhs.uk/blog/why-mrna-vaccines-arent-gene-therapies/ (accessed on 9 January 2022).
- Editorial. Let’s talk about lipid nanoparticles. Nat Rev Mater. 2021, 9, 99. [Google Scholar] [CrossRef]
- Lee, S.; Ryu, J.-H. Influenza Viruses: Innate Immunity and mRNA Vaccines. Front. Immunol. 2021, 12, 106472. [Google Scholar] [CrossRef] [PubMed]
- Kollewe, J. COVID-19 Vaccines: The Contracts, Prices and Profits. The Guardian. 11 August 2021. Available online: https://www.theguardian.com/world/2021/aug/11/covid-19-vaccines-the-contracts-prices-and-profits (accessed on 24 January 2022).
- Kollewe, J. Pfizer Accused of Pandemic Profiteering as Profits Double. The Guardian. 8 February 2022. Available online: https://www.theguardian.com/business/2022/feb/08/pfizer-covid-vaccine-pill-profits-sales (accessed on 21 February 2022).
- Jemielniak, D.; Krempovych, Y. An analysis of AstraZeneca COVID-19 vaccine misinformation and fear mongering on Twitter. Public Health 2021, 200, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Unicef. Artists across Africa Help Spread the Word That Vaccines are Safe and Effective. UNICEF. 2021. Available online: https://www.unicef.org/niger/stories/artists-across-africa-help-spread-word-vaccines-are-safe-and-effective (accessed on 21 February 2022).
- Guildford, A. Why the AstraZeneca Vaccine is Linked with Rare Blood Clots: New Insights. Medical News Today. 17 March 2022. Available online: https://www.medicalnewstoday.com/articles/why-the-astrazeneca-vaccine-is-linked-with-rare-blood-clots-new-insights (accessed on 22 March 2022).
- Mauro, V.P.; Chappell, S.A. A critical analysis of codon optimization in human therapeutics. Trends Mol. Med. 2014, 20, 604–613. [Google Scholar] [CrossRef]
- Robertson, S. Research Suggests Pfizer-BioNTech COVID-19 Vaccine Reprograms Innate Immune Responses. News Medical. 12 May 2021. Available online: https://www.news-medical.net/news/20210510/Research-suggests-Pfizer-BioNTech-COVID-19-vaccine-reprograms-innate-immune-responses.aspx (accessed on 22 January 2022).
- Konstantin Föhse, F.K.; Geckin, B.; Overheul, G.J.; van de Maat, J.; Kilic, G.; Bulut, O.; Dijkstra, H.; Lemmers, H.; Sarlea, S.A.; Reijnders, M.; et al. The BNT162b2 mRNA vaccine against SARS-CoV-2 reprograms both adaptive and innate immune responses. MedRxiv 2021. [Google Scholar] [CrossRef]
- Oster, M.E.; Shay, D.K.; Su, J.R.; Gee, J.; Creech, C.B.; Broder, K.R.; Edwards, K.; Soslow, J.H.; Dendy, J.M.; Schlaudecker, E.; et al. Myocarditis Cases Reported After mRNA-Based COVID-19 Vaccination in the US From December 2020 to August 2021. JAMA 2021, 231, 331–340. [Google Scholar] [CrossRef]
- Diaz, G.A.; Parsons, G.T.; Gering, S.K.; Meier, A.R.; Hutchinson, I.V.; Robicsek, A. Myocarditis and Pericarditis After Vaccination for COVID-19. JAMA 2021, 326, 1210–1212. [Google Scholar] [CrossRef]
- Florins, A.; Gillet, N.; Asquith, B.; Boxus, M.; Burteau, C.; Twizere, J.C.; Urbain, P.; Vandermeers, F.; Debacq, C.; Sanchez-Alcaraz, M.T.; et al. Cell dynamics and immune response to BLV infection: A unifying model. Front. Biosci. 2007, 12, 1520–1531. [Google Scholar] [CrossRef]
- Wong, W.F.; Kohu, K.; Chiba, T.; Sato, T.; Satake, M. Interplay of transcription factors in T-cell differentiation and function: The role of Runx. Immunology 2011, 132, 157–164. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yolles, M.; Frieden, R. Viruses as Living Systems—A Metacybernetic View. Systems 2022, 10, 70. https://doi.org/10.3390/systems10030070
Yolles M, Frieden R. Viruses as Living Systems—A Metacybernetic View. Systems. 2022; 10(3):70. https://doi.org/10.3390/systems10030070
Chicago/Turabian StyleYolles, Maurice, and Roy Frieden. 2022. "Viruses as Living Systems—A Metacybernetic View" Systems 10, no. 3: 70. https://doi.org/10.3390/systems10030070
APA StyleYolles, M., & Frieden, R. (2022). Viruses as Living Systems—A Metacybernetic View. Systems, 10(3), 70. https://doi.org/10.3390/systems10030070






