Interaction Network Provides Clues on the Role of BCAR1 in Cellular Response to Changes in Gravity
Abstract
1. Introduction
2. Results
2.1. Selection of Proteins
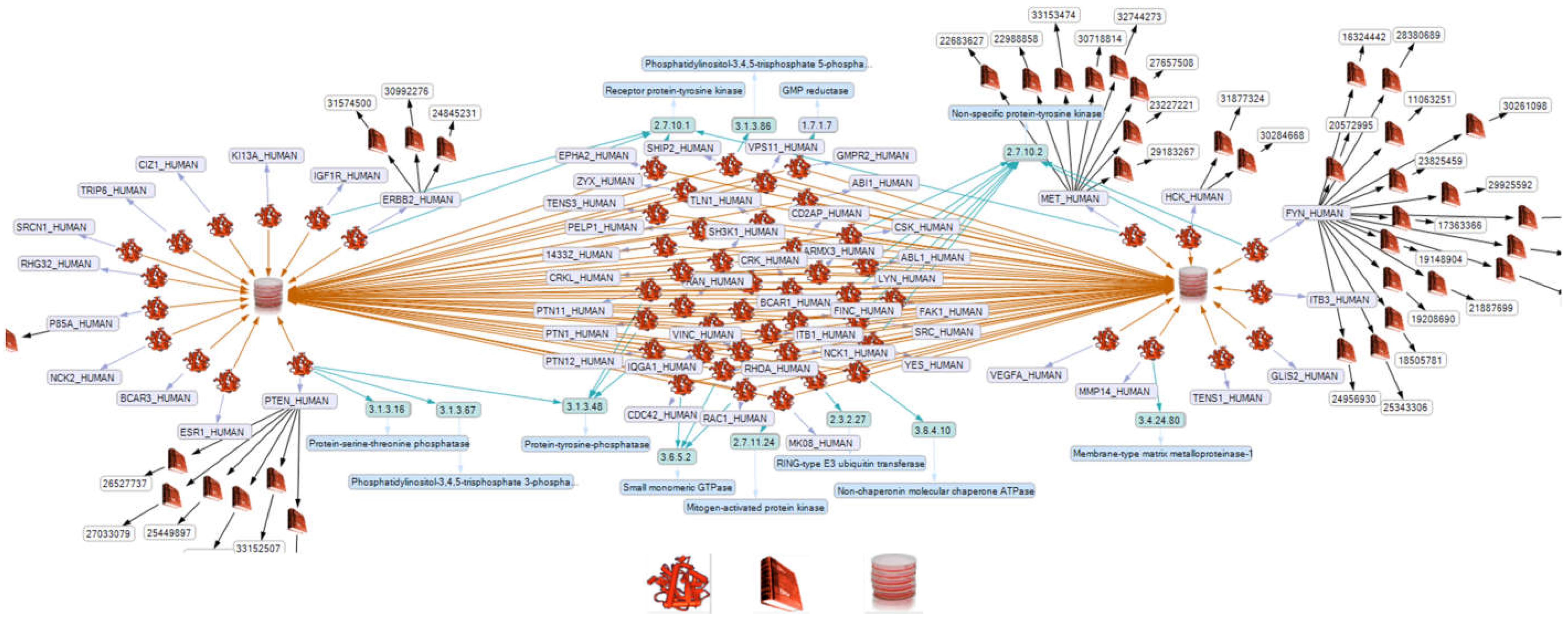
2.2. Network Formation of Selected Proteins
2.3. Influence of Selected Interaction on Cell Physiology
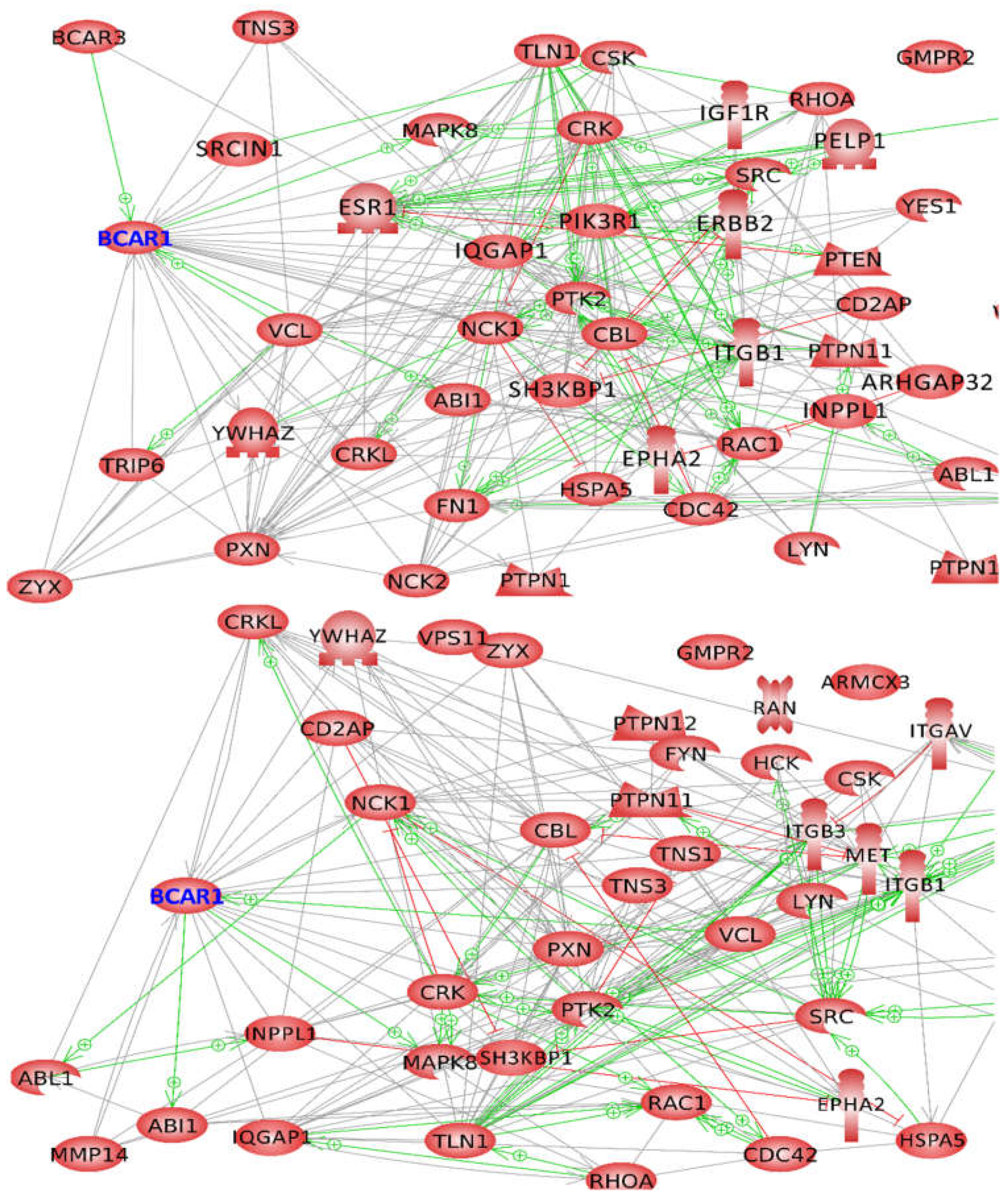
2.3.1. Proteins Binding to the Substrate Domain (SD) of BCAR1
2.3.2. Proteins Binding to other BCAR1 Domains
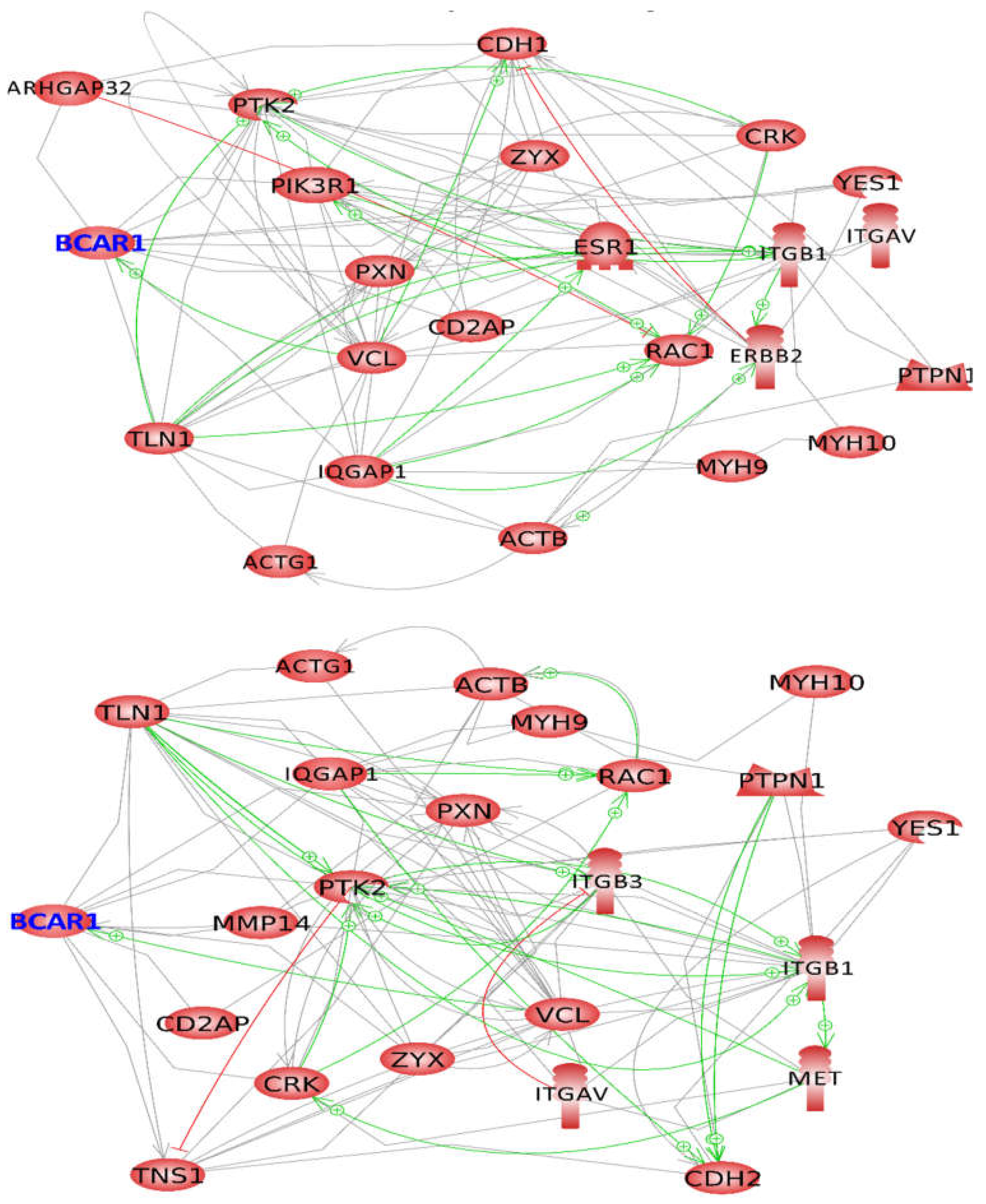
2.4. BCAR1, a Chain Link between Focal Adhesion and Cytoskeletal Proteins
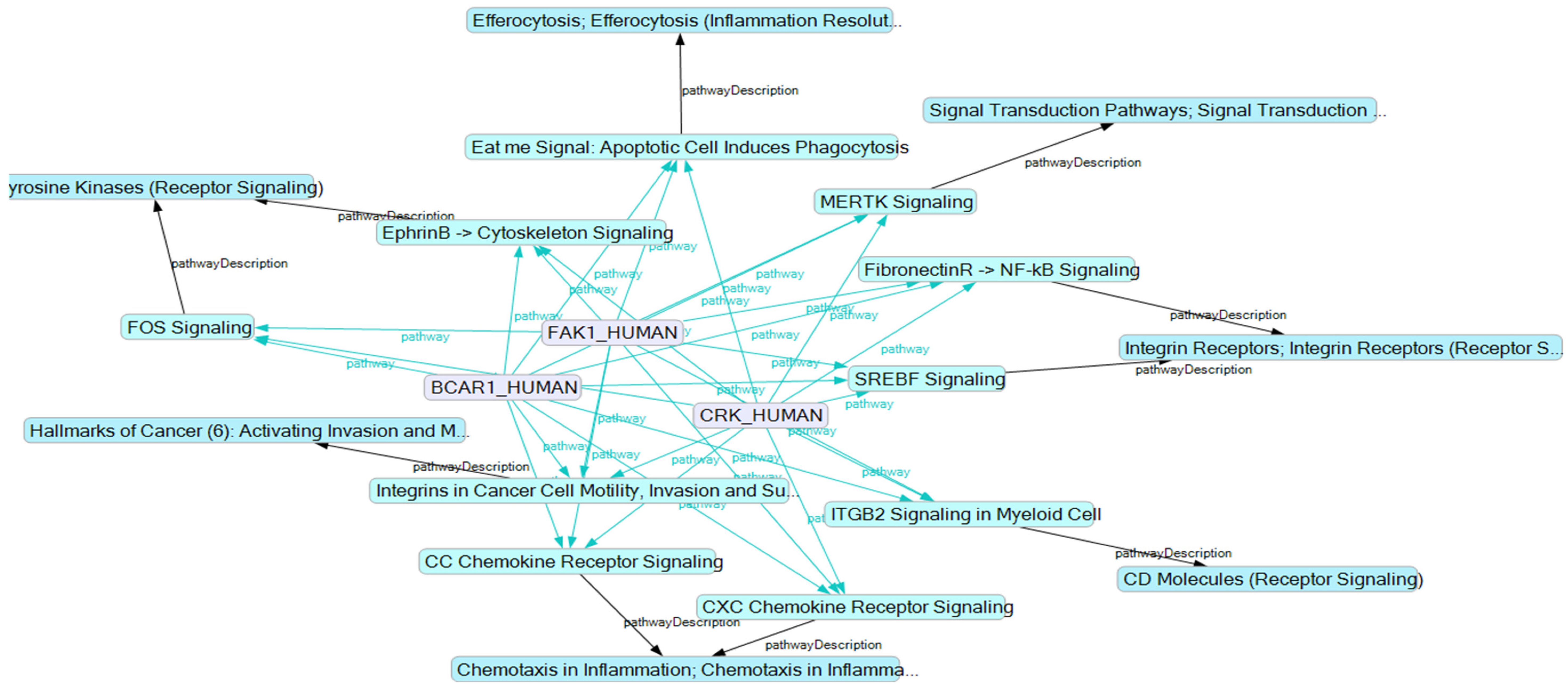
2.5. BCAR1’s Belonging to Cellular Processes
3. Discussion
4. Materials and Methods
4.1. Proteome Data
4.2. Searching Proteins Interacting with BCAR1
4.3. Creation of a Semantic Network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LOD | linked open data; |
| SKB | semantic knowledgebase; |
| KE | knowledge explorer; |
| RDF | resource description framework; |
| SPARQL | semantic protocol and RDF query language; |
| RPM | random positioning machine; |
| FA | focal adhesion complex; |
| AD | adherent cells; |
| 3D | three dimensional; |
| CM | consensus motif; |
| PK | protein kinase, |
| PP | protein phosphatase, |
| RCCS | Rotary Cell Culture System, |
| ROS | reactive oxygen species; |
| pN | piconewton; |
| SD | substrate domain; |
| Cas | CRK-associated substrate; |
| BCAR1 | breast cancer anti-estrogen resistance protein 1; |
| SwissProt AC# | SwissPro accession number. |
References
- Strauch, S.; Grimm, D.; Corydon, T.J.; Krüger, M.; Bauer, J.; Lebert, M.; Wise, P.; Infanger, M.; Richter, P. Current knowledge about the impact of microgravity on the proteome. Expert Rev. Proteom. 2019, 16, 5–16. [Google Scholar] [CrossRef]
- Warnke, E.; Pietsch, J.; Kopp, S.; Bauer, J.; Sahana, J.; Wehland, M.; Krüger, M.; Hemmersbach, R.; Infanger, M.; Lützenberg, R.; et al. Cytokine Release and Focal Adhesion Proteins in Normal Thyroid Cells Cultured on the Random Positioning Machine. Cell. Physiol. Biochem. 2017, 43, 257–270. [Google Scholar] [CrossRef]
- Bauer, J.; Wehland, M.; Infanger, M.; Grimm, D.; Gombocz, E. Semantic Analysis of Posttranslational Modification of Proteins Accumulated in Thyroid Cancer Cells Exposed to Simulated Microgravity. Int. J. Mol. Sci. 2018, 19, 2257. [Google Scholar] [CrossRef] [PubMed]
- Sawada, Y.; Tamada, M.; Dubin-Thaler, B.J.; Cherniavskaya, O.; Sakai, R.; Tanaka, S.; Sheetz, M.P. Force sensing by mechanical extenion of the Src family kinase substrate p130Cas. Cell 2006, 127, 1015–1026. [Google Scholar] [CrossRef]
- Goldmann, W.H. Vinculin-p130 Cas interaction is critical for focal adhesion dynamics and mechano-transduction. Cell Biol. Int. 2013, 38, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Karabulut, N.P.; Frishman, D. Sequence- and Structure-Based Analysis of Tissue-Specific Phosphorylation Sites. PLoS ONE 2016, 11, e0157896. [Google Scholar] [CrossRef]
- Hunter, T. Protein kinases and phosphatases: The Yin and Yang of protein phosphorylation and signaling. Cell 1995, 80, 225–236. [Google Scholar] [CrossRef]
- Vandermarliere, E.; Martens, L. Protein structure as a means to triage proposed PTM sites. Proteomics 2013, 13, 1028–1035. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, P.S.L.; Ferraz, F.A.N.; Pena, D.A.; Pramio, D.T.; Morais, F.A.; Schechtman, D. Revisiting protein kinase–substrate interactions: Toward therapeutic development. Sci. Signal. 2016, 9, re3. [Google Scholar] [CrossRef] [PubMed]
- Karasev, D.A.; Veselova, D.A.; Veselovsky, A.V.; Sobolev, B.N.; Zgoda, V.G.; Archakov, A.I. Spatial features of proteins related to their phosphorylation and associated structural changes. Proteins Struct. Funct. Bioinform. 2017, 86, 13–20. [Google Scholar] [CrossRef]
- Uppala, J.K.; Ghosh, C.; Sathe, L.; Dey, M. Phosphorylation of translation initiation factor eIF 2α at Ser51 depends on site- and context-specific information. FEBS Lett. 2018, 592, 3116–3125. [Google Scholar] [CrossRef]
- Ithychanda, S.S.; Fang, X.; Mohan, M.L.; Zhu, L.; Tirupula, K.C.; Naga Prasad, S.; Wang, Y.X.; Karnik, S.S.; Qin, J. A mechnism of global shape dependent recognition and phosphorylation of filamin by protein kinase A. J. Biol. Chem. 2015, 290, 8527–8538. [Google Scholar] [CrossRef]
- Su, M.-G.; Weng, J.T.-Y.; Hsu, J.B.-K.; Huang, K.-Y.; Chi, Y.-H.; Lee, T.-Y. Investigation and identification of functional post-translational modification sites associated with drug binding and protein-protein interactions. BMC Syst. Biol. 2017, 11, 69–80. [Google Scholar] [CrossRef]
- Pelaseyed, T.; Viswanatha, R.; Sauvanet, C.; Filter, J.J.; Goldberg, M.L.; Bretscher, A. Ezrin activation by LOK phosphorylation involves a PIP2-dependent wedge mechanism. eLife 2017, 6, 22759. [Google Scholar] [CrossRef]
- Acebrón, I.; Righetto, R.D.; Schoenherr, C.; De Buhr, S.; Redondo, P.; Culley, J.; Rodríguez, C.F.; Daday, C.; Biyani, N.; Llorca, O.; et al. Structural basis of Focal Adhesion Kinase activation on lipid membranes. EMBO J. 2020, 39, 104743. [Google Scholar] [CrossRef] [PubMed]
- Travers, T.; Shao, H.; Joughin, B.A.; Lauffenburger, D.A.; Wells, A.; Camacho, C.J. Tandem phosphorylation within an intrin-sically disordered region regulates ACTN4 function. Sci. Signal. 2015, 8, ra51. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.L.; McBeath, E.; Thomas, T.N.; Chiu, Y.J.; Clark, R.L.; Fujiwara, K. Mechanotransduction properties of the cytoplasmic tail of PECAM-1. Biol. Cell 2017, 109, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Röper, J.C.; Mitrossilis, D.; Stirnemann, G.; Waharte, F.; Brito, I.; Fernandez-Sanchez, M.E.; Baaden, M.; Salamero, J.; Farge, E. The major β-catenin/E-cadherin junctional binding site is a primary molecular mechano-transductor of differentiation in vivo. eLife 2018, 7, e33381. [Google Scholar] [CrossRef] [PubMed]
- Defilippi, P.; Di Stefano, P.; Cabodi, S. p130Cas: A versatile scaffold in signaling networks. Trends Cell Biol. 2006, 16, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Wisniewska, M.; Bossenmaier, B.; Georges, G.; Hesse, F.; Dangl, M.; Künkele, K.-P.; Ioannidis, I.; Huber, R.; Engh, R.A. The 1.1Å Resolution Crystal Structure of the p130cas SH3 Domain and Ramifications for Ligand Selectivity. J. Mol. Biol. 2005, 347, 1005–1014. [Google Scholar] [CrossRef]
- Mace, P.D.; Wallez, Y.; Dobaczewska, M.K.; Lee, J.J.; Robinson, H.; Pasquale, E.B.; Riedl, S.J. NSP-Cas protein structures reveal a promiscuous interaction module in cell signaling. Nat. Struct. Mol. Biol. 2011, 18, 1381–1387. [Google Scholar] [CrossRef]
- Hotta, K.; Ranganathan, S.; Liu, R.; Wu, F.; Machiyama, H.; Gao, R.; Hirata, H.; Soni, N.; Ohe, T.; Hogue, C.; et al. Biophysical Properties of Intrinsically Disordered p130Cas Substrate Domain—Implication in Mechanosensing. PLoS Comput. Biol. 2014, 10, e1003532. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Wu, F.; Qiu, W.; Liu, R. P130Cas substrate domain is intrinsically disordered as characterized by single-molecule force measurements. Biophys. Chem. 2013, 180–181, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, X.; Zhu, G.; Lu, L.; Yang, D. A method for determining structure ensemble of large disordered protein: Application to a mechanosensing protein. J. Am. Chem. Soc. 2018, 140, 11276–11285. [Google Scholar] [CrossRef] [PubMed]
- Schiller, H.B.; Fässler, R. Mechanosensitivity and compositional dynamics of cell-matrix adhesions. EMBO Rep. 2013, 14, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Donato, D.M.; Ryzhova, L.M.; Meenderink, L.M.; Kaverina, I.; Hanks, S.K. Dynamics and Mechanism of p130Cas Localization to Focal Adhesions. J. Biol. Chem. 2010, 285, 20769–20779. [Google Scholar] [CrossRef]
- Janostiak, R.; Brábek, J.; Auernheimer, V.; Tatarova, Z.; Lautscham, L.A.; Dey, T.; Gemperle, J.; Merkel, R.; Goldmann, W.H.; Fabry, B.; et al. CAS directly interacts with vinculin to control mechanosensing and focal adhesion dynamics. Cell. Mol. Life Sci. 2013, 71, 727–744. [Google Scholar] [CrossRef]
- Geiger, B.; Berdshadsky, A. Exploring the neighborhood: Adhesion-coupled cell mechanosensors. Cell 2002, 110, 139–142. [Google Scholar] [CrossRef]
- Barrett, A.; Pellet-Many, C.; Zachary, I.C.; Evans, I.M.; Frankel, P. p130Cas: A key signalling node in health and disease. Cell. Signal. 2013, 25, 766–777. [Google Scholar] [CrossRef]
- Cabodi, S.; Camacho-Leal, M.D.P.; Di Stefano, P.; Defilippi, P. Integrin signalling adaptors: Not only figurants in the cancer story. Nat. Rev. Cancer 2010, 10, 858–870. [Google Scholar] [CrossRef]
- Ratushnyy, A.Y.; Buravkova, L.B. Expression of focal adhesion genes in mesenchymal stem cells under simulated nicrogravity. Dokl. Biochem. Biophys. 2017, 477, 354–356. [Google Scholar]
- Tang, H.; Hao, Q.; Fitzgerald, T.; Sasaki, T.; Landon, E.J.; Inagami, T. Pyk2/CAK Tyrosine Kinase Activity-mediated An-giogenesis of Pulmonary Vascular Endothelial Cells. J. Biol. Chem. 2002, 277, 5441–5447. [Google Scholar] [CrossRef] [PubMed]
- Nassef, M.; Melnik, D.; Kopp, S.; Sahana, J.; Infanger, M.; Lützenberg, R.; Relja, B.; Wehland, M.; Grimm, D.; Krüger, M. Breast Cancer Cells in Microgravity: New Aspects for Cancer Research. Int. J. Mol. Sci. 2020, 21, 7345. [Google Scholar] [CrossRef]
- Konstantinovsky, S.; Davidson, B.; Reich, R. Ezrin and BCAR1/p130Cas mediate breast cancer growth as 3-D spheroids. Clin. Exp. Metastasis 2012, 29, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Sahana, J.; Nassef, M.Z.; Wehland, M.; Kopp, S.; Krüger, M.; Corydon, T.J.; Infanger, M.; Bauer, J.; Grimm, D. Decreased E-Cadherin in MCF7 Human Breast Cancer Cells Forming Multicellular Spheroids Exposed to Simulated Microgravity. Proteomics 2018, 18, e1800015. [Google Scholar] [CrossRef]
- Bauer, J.; Kopp, S.; Schlagberger, E.M.; Grosse, J.; Sahana, J.; Riwaldt, S.; Wehland, M.; Luetzenberg, R.; Infanger, M.; Grimm, D. Proteome Analysis of Human Follicular Thyroid Cancer Cells Exposed to the Random Positioning Machine. Int. J. Mol. Sci. 2017, 18, 546. [Google Scholar] [CrossRef]
- Gombocz, E.A.; Stanley, R.A.; Rockey, C.; Nishimura, T. Data Integration Framework for Discovery and Validation: Smart Merging of Experimental and Public Data Across Ontologies and Taxonomies. In Proceedings of the 2018 BIO-IT World conference, Boston, MA, USA, 16–18 April 2018; Available online: https://de.slideshare.net/crockey/data-integration-framework-for-discovery-and-validation (accessed on 1 March 2020).
- Soule, H.D.; Vazquez, J.; Long, A.; Albert, S.; Brennan, M. A Human Cell Line from a Pleural Effusion Derived from a Breast Carcinoma. J. Natl. Cancer Inst. 1973, 51, 1409–1416. [Google Scholar] [CrossRef]
- Kang, H.; Rho, J.G.; Kim, C.; Tak, H.; Lee, H.; Ji, E.; Ahn, S.; Shin, A.-R.; Cho, H.-I.; Huh, Y.H.; et al. The miR-24-3p/p130Cas: A novel axis regulating the migration and invasion of cancer cells. Sci. Rep. 2017, 7, 44847. [Google Scholar] [CrossRef]
- Goretzki, P.E.; Frilling, A.; Simon, D.; Roeher, H.-D. Growth Regulation of Normal Thyroids and Thyroid Tumors in Man. Methods Mol. Biol. 1990, 118, 48–63. [Google Scholar] [CrossRef]
- Simon, D.; Goretzki, P.E.; Gorelev, V.; Ebling, B.; Reishaus, E.; Lyons, J.; Haubruck, H.; Röher, H.D. Significance of P53 in human thyroid tumors. World J. Surg 1994, 18, 535–541. [Google Scholar] [CrossRef]
- Pietsch, J.; Sickmann, A.; Weber, G.; Bauer, J.; Egli, M.; Wildgruber, R.; Infanger, M.; Grimm, D. A proteomic approach to analysing spheroid formation of two human thyroid cell lines cultured on a random positioning machine. Proteomics 2011, 11, 2095–2104. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Pietsch, J.; Wehland, M.; Schulz, H.; Saar, K.; Hübner, N.; Bauer, J.; Braun, M.; Schwarzwälder, A.; Segerer, J.; et al. Differential gene expression profile and altered cytokine secretion of thyroid cancer cells in space. FASEB J. 2014, 28, 813–835. [Google Scholar] [CrossRef] [PubMed]
- Murakami, H.; Iwashita, T.; Asai, N.; Iwata, Y.; Narumiya, S.; Takahashi, M. Rho-dependent and independent tyrosine phos-phorylation of focal adhesion kinase, paxillin and p130Cas mediated by Ret kinase. Oncogene 1999, 18, 1975–1982. [Google Scholar] [CrossRef]
- Kodama, H.; Fukuda, K.; Takahashi, E.; Tahara, S.; Tomita, Y.; Ieda, M.; Kimura, K.; Owada, K.M.; Vuori, K.; Ogawa, S. Selective Involvement of p130Cas/Crk/Pyk2/c-Src in Endothelin-1-Induced JNK Activation. Hypertension 2003, 41, 1372–1379. [Google Scholar] [CrossRef][Green Version]
- Salgia, R.; Pisick, E.; Sattler, M.; Li, J.-L.; Uemura, N.; Wong, W.-K.; Burky, S.A.; Hirai, H.; Chen, L.B.; Griffin, J.D. p130CAS Forms a Signaling Complex with the Adapter Protein CRKL in Hematopoietic Cells Transformed by the BCR/ABL Oncogene. J. Biol. Chem. 1996, 271, 25198–25203. [Google Scholar] [CrossRef]
- Graf, R.; Barbero, S.; Keller, N.; Chen, L.; Uryu, S.; Schlaepfer, D.; Stupak, D. Src-inducible association of CrkL with pro-caspase-8 promotes cell migration. Cell Adhes. Migr. 2013, 7, 362–369. [Google Scholar] [CrossRef]
- Kain, K.H.; Klemke, R.L. Inhibition of Cell Migration by Abl Family Tyrosine Kinases through Uncoupling of Crk-CAS Complexes. J. Biol. Chem. 2001, 276, 16185–16192. [Google Scholar] [CrossRef]
- Hoffman, L.M.; Jensen, C.C.; Chaturvedi, A.; Yoshigi, M.; Beckerle, M.C. Stretch-induced actin remodeling requires targeting of zyxin to stress fibers and recruitment of actin regulators. Mol. Biol. Cell 2012, 23, 1846–1859. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Kloeker, S.; Jensen, C.C.; Bockholt, S.; Honda, H.; Hirai, H.; Beckerle, M.C. Members of the Zyxin Family of LIM Proteins Interact with Members of the p130Cas Family of Signal Transducers. J. Biol. Chem. 2002, 277, 9580–9589. [Google Scholar] [CrossRef] [PubMed]
- Prasad, N.; Topping, R.S.; Decker, S.J. SH2-Containing Inositol 5′-Phosphatase SHIP2 Associates with the p130 Cas Adapter Protein and Regulates Cellular Adhesion and Spreading. Mol. Cell. Biol. 2001, 21, 1416–1428. [Google Scholar] [CrossRef]
- Li, E.; Stupack, D.G.; Brown, S.L.; Klemke, R.; Schlaepfer, D.D.; Nemerow, G.R. Association of p130CAS with phosphati-dylinositol-3-OH kinase mediates adenovirus cell entry. J. Biol. Chem. 2000, 275, 14729–14735. [Google Scholar] [CrossRef]
- Buday, L.; Wunderlich, L.; Tamás, P. The Nck family of adapter proteins: Regulators of actin cytoskeleton. Cell. Signal. 2002, 14, 723–731. [Google Scholar] [CrossRef]
- Zhao, C.; Ma, H.; Bossy-Wetzel, E.; Lipton, S.A.; Zhang, Z.; Feng, G.S. GC-GAP, aRho family GTPase-activating protein that interacts with signalling adapters Gab1 and Gab2. J. Biol. Chem. 2003, 278, 34641–34653. [Google Scholar] [CrossRef] [PubMed]
- Rivera, G.M.; Antoku, S.; Gelkop, S.; Shin, N.Y.; Hanks, S.K.; Pawson, T.; Mayer, B.J. Requirement of Nck adaptors for actin dynamics and cell migration stimulated by platelet-derived growth factor B. Proc. Natl. Acad. Sci. USA 2006, 103, 9536–9541. [Google Scholar] [CrossRef]
- Ngoenkam, J.; Paensuwan, P.; Preechanukul, K.; Khamsri, B.; Yiemwattana, I.; Beck-García, E.; Minguet, S.; Schamel, W.W.; Pongcharoen, S. Non-overlapping functions of Nck1 and Nck2 adaptor proteins in T cell activation. Cell Commun. Signal. 2014, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Buvall, L.; Rashmi, P.; Lopez-Rivera, E.; Andreeva, S.; Weins, A.; Wallentin, H.; Greka, A.; Mundel, P. Proteasomal degra-dation of Nck1 but not Nck2 regulates RhoA activation and actin dynamics. Nat. Commun. 2013, 4, 2863. [Google Scholar] [CrossRef]
- Dikic, I.; Szymkiewicz, I.; Soubeyran, P. Cbl signaling networks in the regulation of cell function. Cell. Mol. Life Sci. 2003, 60, 1805–1827. [Google Scholar] [CrossRef]
- Howlett, C.J.; Robbins, S.M. Membrane-anchored CBL suppresses HCK protein-tyrosine kinase mediated cellular transformation. Oncogene 2002, 21, 1707–1716. [Google Scholar] [CrossRef]
- Bureau, J.F.; Cassonnet, P.; Grange, L.; Dessapt, J.; Jones, L.; Demeret, C.; Sakuntabhai, A.; Jacob, Y. The SRC-family tyrosine kinase HCK shapes the landscape of KAP2 interactome. Oncotarget 2018, 9, 13102–13115. [Google Scholar] [CrossRef]
- Kaabeche, K.; Leminnier, J.; Mee, S.L.; Caverzasio, J.; Marie, P.J. Cbl-mediated degradation of Lyn and Fyn induced by con-stitutive fibroblast growth factor receptor-2 activation supports osteoblast differentiation. J. Biol. Chem. 2004, 279, 36259–36267. [Google Scholar] [CrossRef]
- Gonsior, C.; Binamé, F.; Frühbeis, C.; Bauer, N.M.; Hoch-Kraft, P.; Luhmann, H.J.; Trotter, J.; White, R. Oligodendroglial p130Cas Is a Target of Fyn Kinase Involved in Process Formation, Cell Migration and Survival. PLoS ONE 2014, 9, e89423. [Google Scholar] [CrossRef]
- Hochgraefe, F.; Zhang, L.; O’Toole, S.A.; Browne, B.C.; Pinese, M.; Cubas, A.P.; Lehrbach, G.M.; Croucher, D.; Rickwood, D.; Boulghourjian, A.; et al. Tyrosine Phosphorylation Profiling Reveals the Signaling Network Characteristics of Basal Breast Cancer Cells. Cancer Res. 2010, 70, 9391–9401. [Google Scholar] [CrossRef]
- Watanabe, S.; Take, H.; Takeda, K.; Yu, Z.X.; Iwata, N.; Kajigaya, S. Characterization of the Cin85 adaptor protein and identification of components involved in CIN85 complexes. Biochem. Biophys. Res. Commun. 2000, 278, 167–174. [Google Scholar] [CrossRef]
- Verdier, F.; Valovka, T.; Zhyvoloup, A.; Drobot, L.B.; Buchman, V.; Waterfield, M.; Gout, I. Ruk is ubiquitinated but not degraded by the proteasome. JBIC J. Biol. Inorg. Chem. 2002, 269, 3402–3408. [Google Scholar] [CrossRef] [PubMed]
- Maia, V.; Ortiz-Rivero, S.; Sanz, M.; Gutierrez-Berzal, J.; Álvarez-Fernández, I.; Gutierrez-Herrero, S.; De Pereda, J.M.; Porras, A.; Guerrero, C. C3G forms complexes with Bcr-Abl and p38α MAPK at the focal adhesions in chronic myeloid leukemia cells: Implication in the regulation of leukemic cell adhesion. Cell Commun. Signal. 2013, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.J.; Hemmeryckx, B.; Groffen, J.; Heisterkamp, N. Interaction of Bcr/Abl with C3G, an exchange factor for the small GTPase Rap1, through the adapter protein Crkl. Biochem. Biophys. Res. Commun. 2005, 333, 1276–1283. [Google Scholar] [CrossRef]
- Kirsch, K.H.; Georgescu, M.M.; Shishido, T.; Langdon, W.Y.; Birge, R.B.; Hanafusa, H. The adapttertype protein CMS/CD2AP binds to the proto-oncogenic protein c-CBL through a tyrosine phosphorylation-regulated Src homology 3 domain interaction. J. Biol. Chem. 2001, 276, 4957–4963. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, K.H.; Georgescu, M.M.; Ishimaru, S.; Hanafusa, H. CMS: An adapter molecule involved in cytoskeletal rearrangements. Proc. Natl. Acad. Sci. USA 1999, 96, 6211–6216. [Google Scholar] [CrossRef] [PubMed]
- Evans, I.M.; Kennedy, S.A.; Paliashvili, K.; Santra, T.; Yamaji, M.; Lovering, R.C.; Britton, G.; Franke, V.; Kolch, W.; Zachary, I.C. Vascular endothelial growth factor (VEGF) promotes assembly to the p130Cas interactome to drive endothlial chemotatic signalling and angiognesis. Mol. Cell. Proteom. 2017, 16, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Gingras, D.; Michaud, M.; Di Tomasso, G.; Beliveau, E.; Nyalendo, C.; Beliveau, R. Sphingosine-1- phosphate induces the as-sociation of membrane-type 1 matrix metalloproteinase with p130Cas in endothelial cells. FEBS Lett. 2008, 582, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Ballestrem, C.; Erez, N.; Kirchner, J.; Kam, Z.; Bershadsky, A.; Geiger, B. Molecular mapping of tyrosine-phosphorylated proteins in focal adhesions using fluorescence resonance energy transfer. J. Cell Sci. 2006, 119, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Janoštiak, R.; Tolde, O.; Brůhová, Z.; Novotný, M.; Hanks, S.K.; Rösel, D.; Brábek, J. Tyrosine phosphorylation within the SH3 domain regulates CAS subcellular localization, cell migration, and invasiveness. Mol. Biol. Cell 2011, 22, 4256–4267. [Google Scholar] [CrossRef]
- Qian, X.; Li, G.; Vass, W.C.; Papageorge, A.; Walker, R.C.; Asnaghi, L.; Steinbach, P.J.; Tosato, G.; Hunter, K.; Lowy, D.R. The Tensin-3 Protein, Including its SH2 Domain, Is Phosphorylated by Src and Contributes to Tumorigenesis and Metastasis. Cancer Cell 2009, 16, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Miller, D.J.; Guibao, C.D.; Donato, D.M.; Hanks, S.K.; Zheng, J.J. Structural and functional insights into the interaction between the Cas family scaffolding protein p130Cas and the focal adhesion-associated protein paxillin. J. Biol. Chem. 2017, 292, 18281–18289. [Google Scholar] [CrossRef] [PubMed]
- McGowan, S.E.; McCoy, D.M. Platelet-derived growth factor receptor-α and Ras-related C3 botulinum toxin substrate-1 regulate mechano-responsiveness of lung fibroblasts. Am. J. Physiol. Cell. Mol. Physiol. 2017, 313, L1174–L1187. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.G.; Dolfi, F.; Russelo, M.; Vuori, K. Cell adhesion regulates the interaction between the docking protein p130cas and 14-3-3 proteins. J. Biol. Chem. 1999, 274, 5762–5768. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, P.; Cabodi, S.; Erba, E.B.; Margaria, V.; Bergatto, E.; Giuffrida, M.G.; Silengo, L.; Tarone, G.; Turco, E.; Defilippi, P. p130Cas-associated Protein (p140Cap) as a New Tyrosine-phosphorylated Protein Involved in Cell Spreading. Mol. Biol. Cell 2004, 15, 787–800. [Google Scholar] [CrossRef]
- Weissbach, L.; Bernardsb, A.; Herion, D.W. Binding of Myosin Essential Light Chain to the Cytoskeleton-Associated Protein IQGAP. Biochem. Biophys. Res. Commun. 1998, 251, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Briggs, M.W.; Sacks, D. IQGAP1 as signal integrator: Ca2+, calmodulin, Cdc42 and the cytoskeleton. FEBS Lett. 2003, 542, 7–11. [Google Scholar] [CrossRef]
- Sit, B.; Gutmann, D.; Iskratsch, T. Costameres, dense plaques and podosomes: The cell matrix adhesions in cardiovascular mechanosensing. J. Muscle Res. Cell Motil. 2019, 40, 197–209. [Google Scholar] [CrossRef]
- Lee, H.S.; Bellin, R.M.; Walker, D.I.; Patel, B.; Powers, P.; Liu, H.; Garcia-Alareez, B.; de Pereda, J.M.; Liddington, R.C.; Volkmann, N.; et al. Characterization of an actin-binding site within the talin FERM do-main. J. Mol. Biol. 2004, 343, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Steenblock, C.; Heckel, T.; Czupalla, C.; Santo, A.I.E.; Niehage, C.; Sztacho, M.; Hoflack, B. The Cdc42 Guanine Nucleotide Exchange Factor FGD6 Coordinates Cell Polarity and Endosomal Membrane Recycling in Osteoclasts. J. Biol. Chem. 2014, 289, 18347–18359. [Google Scholar] [CrossRef] [PubMed]
- Bauer, T.J.; Gombocz, E.; Krüger, M.; Sahana, J.; Corydon, T.J.; Bauer, J.; Infanger, M.; Grimm, D. Augmenting cancer cell proteomics with cellular images—A semantic approach to understand focal adhesion. J. Biomed. Inform. 2019, 100, 103320. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Suzuki, K. Regulation of protein phosphatase 2A-mediated recruitment of IQGAP1 to beta1 integrin by EGF through activation of Ca2+/calmodulin-dependent protein kinase II. J. Cell. Physiol. 2006, 208, 213–219. [Google Scholar] [CrossRef]
- Leal, M.D.P.C.; Pincini, A.; Tornillo, G.; Fiorito, E.; Bisaro, B.; De Luca, E.; Turco, E.; Defilippi, P.; Cabodi, S. p130Cas Over-Expression Impairs Mammary Branching Morphogenesis in Response to Estrogen and EGF. PLoS ONE 2012, 7, e49817. [Google Scholar] [CrossRef]
- Cabodi, S.; Moro, L.; Baj, G.; Smeriglio, M.; Di Stefano, P.; Gippone, S.; Surico, N.; Silengo, L.; Turco, E.; Tarone, G.; et al. p130Cas interacts with estrogen receptor α and modulates non-genomic estrogen signaling in breast cancer cells. J. Cell Sci. 2004, 117, 1603–1611. [Google Scholar] [CrossRef]
- Okabe, T.; Nakamura, T.; Nishimura, Y.N.; Kohu, K.; Ohwada, S.; Morishita, Y.; Akiyama, T. RICS, a Novel GTPase-activating Protein for Cdc42 and Rac1, Is Involved in the β-Catenin-N-cadherin and N-Methyl-d-aspartate Receptor Signaling. J. Biol. Chem. 2003, 278, 9920–9927. [Google Scholar] [CrossRef]
- Calautti, E.; Li, J.; Saoncella, S.; Brissette, J.L.; Goetinck, P.F. Phosphoinositide 3-Kinase Signaling to Akt Promotes Keratinocyte Differentiation Versus Death. J. Biol. Chem. 2005, 280, 32856–32865. [Google Scholar] [CrossRef]
- Lengyel, C.G.; Altuna, S.C.; Habeeb, B.; Trapani, D.; Khan, S.Z.; Lengyel, C.D. The Potential of PI3K/AKT/mTOR Signaling as a Druggable Target for Endometrial and Ovarian Carcinomas. Curr. Drug Targets 2020, 21, 946–961. [Google Scholar] [CrossRef]
- Nilsson, G.M.A.; Akhtar, N.; Kannius-Jkanson, M.; Baeckström, D. Loss of E-cadherin expression is not a prerequisite for c-erbB2-induced epithelial-mesenchymal transition. Int. J. Oncol. 2014, 45, 82–94. [Google Scholar] [CrossRef]
- Lo, S.H.; Yu, Q.-C.; Degenstein, L.; Chen, L.B.; Fuchs, E. Progressive Kidney Degeneration in Mice Lacking Tensin. J. Cell Biol. 1997, 136, 1349–1361. [Google Scholar] [CrossRef] [PubMed]
- Lefort, C.T.; Wojciechowski, K.; Hocking, D.C. N-cadherin cell-cell adhesion complexes are regulated by fibronectin matrix asembly. J. Biol. Chem. 2011, 286, 3149–3160. [Google Scholar] [CrossRef]
- Gálvez, B.G.; Matías-Román, S.; Yáñez-Mó, M.; Sánchez-Madrid, F.; Arroyo, A.G. ECM regulates MT1-MMP localization with β1 or αvβ3 integrins at distinct cell compartments modulating its internalization and activity on human endothelial cells. J. Biol. Chem. 2002, 159, 509–521. [Google Scholar] [CrossRef]
- Riordan, S.M.; Lidder, S.; Williams, R.; Skouteris, G.G. The beta-subunit of the hepatocyte growth factor/scatter factor (HGF/SF) receptor phosphorylates and associates with CrkII: Expression of CrkII enhances HGF/SF-induced mitogenesis. Biochem. J. 2000, 350, 925–932. [Google Scholar] [CrossRef]
- Riwaldt, S.; Pietsch, J.; Sickmann, A.; Bauer, J.; Braun, M.; Segerer, J.; Schwarzwälder, A.; Aleshcheva, G.; Corydon, T.J.; Infanger, M.; et al. Identification of proteins involved in inhibition of spheroid formation under microgravity. Proteomics 2015, 15, 2945–2952. [Google Scholar] [CrossRef] [PubMed]
- Spisni, E.; Toni, M.; Strillacci, A.; Galleri, G.; Santi, S.; Griffoni, C.; Tomasi, V. Caveolae and caveolae constituents in mech-anosensing: Effect of modeled microgravity on cultured human endothelial cells. Cell. Biochem. Biophys. 2006, 46, 155–164. [Google Scholar]
- Shi, F.; Zhao, T.-Z.; Wang, Y.-C.; Cao, X.-S.; Yang, C.-B.; Gao, Y.; Li, C.-F.; Zhao, J.-D.; Zhang, S.; Sun, X.-Q. The Impact of Simulated Weightlessness on Endothelium-Dependent Angiogenesis and the Role of Caveolae/Caveolin-1. Cell. Physiol. Biochem. 2016, 38, 502–513. [Google Scholar] [CrossRef]
- Saxena, R.; Pan, G.; McDonald, J.M. Osteoblast and Osteoclast Differentiation in Modeled Microgravity. Ann. N. Y. Acad. Sci. 2007, 1116, 494–498. [Google Scholar] [CrossRef]
- Koaykul, C.; Kim, M.-H.; Kawahara, Y.; Yuge, L.; Kino-Oka, M. Alterations in Nuclear Lamina and the Cytoskeleton of Bone Marrow-Derived Human Mesenchymal Stem Cells Cultured Under Simulated Microgravity Conditions. Stem Cells Dev. 2019, 28, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Brungs, S.; Kolanus, W.; Hemmersbach, R. Syk phosphorylation—A gravisensitive step in macrophage signalling. Cell Commun. Signal. 2015, 13, 1–9. [Google Scholar] [CrossRef]
- Dai, Z.; Guo, F.; Wu, F.; Xu, H.; Yang, C.; Li, J.; Liang, P.; Zhang, H.; Qu, L.; Tan, Y.; et al. Integrin αvβ3 mediates the synergetic regulation of core-binding factor α1 transcriptional activity by gravity and insulin-like growth factor-1 through phosphoinositide 3-kinase signaling. Bone 2014, 69, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, S.A.; Lang, C.; Zhang, Y.; Paul, E.M.; Laufenberg, L.J.; Lewis, G.; Donahue, H.J. Interdependence of Muscle Atrophy and Bone Loss Induced by Mechanical Unloading. J. Bone Miner. Res. 2014, 29, 1118–1130. [Google Scholar] [CrossRef] [PubMed]
- Bekker-Jensen, D.B.; Kelstrup, C.D.; Batth, T.S.; Larsen, S.C.; Haldrup, C.; Bramsen, J.B.; Sørensen, K.D.; Høyer, S.; Ørntoft, T.F.; Andersen, C.L.; et al. An Optimized Shotgun Strategy for the Rapid Generation of Comprehensive Human Proteomes. Cell Syst. 2017, 4, 587–599.e4. [Google Scholar] [CrossRef]
- Higashibata, A.; Imamizo-Sato, M.; Seki, M.; Yamazaki, T.; Ishioka, N. Influence of simulated microgravity on the activation of the small GTPase Rho involved in cytoskeletal formation—Molecular cloning and sequencing of bovine leukemia-associated guanine nucleotide exchange factor. BMC Biochem. 2006, 7, 19. [Google Scholar] [CrossRef]
- Sarkar, S.; Wise, K.C.; Manna, S.K.; Ramesh, V.; Yamauchi, K.; Thomas, R.L.; Wilson, B.L.; Kulkarni, A.D.; Pellis, N.R.; Ramesh, G.T. Activation of activator protein-1 in mouse brain regions exposed to simulated microgravity. In Vitro Cell Dev. Biol. Anim. 2006, 42, 96–99. [Google Scholar] [CrossRef]
- Ethiraj, P.; Ottinger, A.M.; Singh, T.; Singh, A.; Haire, K.M.; Reddy, S.V. Proteasome inhibition suppress microgravity elevated RANK signaling during osteoclast differentiation. Cytokine 2020, 125, 154821. [Google Scholar] [CrossRef]
- Koaykul, C.; Kim, M.-H.; Kawahara, Y.; Yuge, L.; Kino-Oka, M. Maintenance of Neurogenic Differentiation Potential in Passaged Bone Marrow-Derived Human Mesenchymal Stem Cells Under Simulated Microgravity Conditions. Stem Cells Dev. 2019, 28, 1552–1561. [Google Scholar] [CrossRef]
- Lai, Y.-J.; Chen, C.-S.; Lin, W.-C.; Lin, F.-T. c-Src-Mediated Phosphorylation of TRIP6 Regulates Its Function in Lysophosphatidic Acid-Induced Cell Migration. Mol. Cell. Biol. 2005, 25, 5859–5868. [Google Scholar] [CrossRef] [PubMed]
- Willier, S.; Butt, E.; Richter, G.H.; Burdach, S.; Grunewald, T.G. Defining the role of TRIP6 in cell physiology and cancer. Biol. Cell 2011, 103, 573–591. [Google Scholar] [CrossRef] [PubMed]
- Kopp, S.; Sahana, J.; Islam, T.; Petersen, A.G.; Bauer, J.; Corydon, T.J.; Schulz, H.; Saar, K.; Huebner, N.; Slumstrup, L.; et al. The role of NFκB in spheroid formation of human breast cancer cells cultured on the Random Positioning Machine. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhong, B. Regulation of Cellular Antiviral Signaling by Modifications of Ubiquitin and Ubiquitin-like Molecules. Immune Netw. 2018, 18, 4. [Google Scholar] [CrossRef] [PubMed]
- Uchida, T.; Sakashita, Y.; Kitahata, K.; Yamashita, Y.; Tomida, C.; Kimori, Y.; Komatsu, A.; Hirasaka, K.; Ohno, A.; Nakao, R.; et al. Reactive oxygen species upregulate expression of muscle atrophy-associated ubiquitin ligase Cbl-b in rat L6 skeletal muscle cells. Am. J. Physiol. Physiol. 2018, 314, C721–C731. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, I. Muscle fiber atrophy. Rinsho Shinkeigaku 2012, 52, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Li, Y.; Zhou, H.; Cai, M.; Liu, J.; Gao, S.; Yang, J.; Tong, L.; Wang, J.; Zhou, S.; et al. Simulated microgravity reduces intracellular-free calcium concentration by inhibiting calcium channels in primary mouse osteoblasts. J. Cell. Biochem. 2019, 120, 4009–4020. [Google Scholar] [CrossRef]
- Kriegs, B.; Theisen, R.; Schnabl, H. Inositol 1,4,5-trisphosphate and Ran expression during simulated and real microgravity. Protoplasma 2006, 229, 163–174. [Google Scholar] [CrossRef]
- Bandyopadhyay, C.; Veettil, M.V.; Dutta, S.; Chandran, B. p130Cas Scaffolds the Signalosome to Direct Adaptor-Effector Cross Talk during Kaposi’s Sarcoma-Associated Herpesvirus Trafficking in Human Microvascular Dermal Endothelial Cells. J. Virol. 2014, 88, 13858–13878. [Google Scholar] [CrossRef]
- Zhang, S.; Yuan, M.; Cheng, C.; Xia, D.-H.; Wu, S. Chinese Herbal Medicine Effects on Muscle Atrophy Induced by Simulated Microgravity. Aerosp. Med. Hum. Perform. 2018, 89, 883–888. [Google Scholar] [CrossRef]
- Hinohara, K.; Kobayashi, S.; Kanauchi, H.; Shimizu, S.; Nishioka, K.; Tsuji, E.; Tada, K.; Umezawa, K.; Mori, M.; Ogawa, T.; et al. ErbB receptor tyrosine kinase/NF-κB signaling controls mammosphere formation in human breast cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 6584–6589. [Google Scholar] [CrossRef]
- Parsons, M.J.; Patel, P.; Brat, D.J.; Colbert, L.; Vertino, P.M. Silencing of TMS1/ASC Promotes Resistance to Anoikis in Breast Epithelial Cells. Cancer Res. 2009, 69, 1706–1711. [Google Scholar] [CrossRef] [PubMed]
- Louis, F.; Deroanne, C.; Nusgens, B.; Vico, L.; Guignandon, A. RhoGTPases as Key Players in Mammalian Cell Adaptation to Microgravity. BioMed. Res. Int. 2015, 2015, 1–17. [Google Scholar] [CrossRef]
- Kashirini, D.N.; Kononikhin, A.S.; Marina, I.M.; Buravkova, L.B. Secretome of Cultured Human Endothelial Cells in Simulated Microgravity. Bull. Exp. Biol. Med. 2019, 167, 35–38. [Google Scholar] [CrossRef]
- Deng, B.; Liu, R.; Tian, X.; Han, Z.; Chen, J. Simulated microgravity inhibits the viability and migration of glioma via FAK/RhoA/Rock and FAK/Nek2 signaling. In Vitro Cell. Dev. Biol. Anim. 2019, 55, 260–271. [Google Scholar] [CrossRef]
- Xue, L.; Li, Y.; Chen, J. Duration of simulated microgravity affects the differentiation of mesenchymal stem cells. Mol. Med. Rep. 2017, 15, 3011–3018. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Xu, A.; Zhao, T.; Zhao, Q.; Zhang, J.; Fan, C.; Deng, Y.; Freywald, A.; Genth, H.; Xiang, J. Simulated microgravity inhibits cell focal adhesions leading to reduced melanoma cell proliferation and metastasis via FAK/RhoA-regulated mTORC1 and AMPK pathways. Sci. Rep. 2018, 8, 3769. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Li, R.; Tan, X.; Zhang, J.; Fan, C.; Zhao, Q.; Deng, Y.; Xu, A.; Lukong, K.E.; Genth, H.; et al. Simulated Microgravity Reduces Focal Adhesions and Alters Cytoskeleton and Nuclear Positioning Leading to Enhanced Apoptosis via Suppressing FAK/RhoA-Mediated mTORC1/NF-κB and ERK1/2 Pathways. Int. J. Mol. Sci. 2018, 19, 1994. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.C.; Gou, G.H.; Hsia, C.W.; Ho, C.W.; Huang, K.L.; Wu, Y.F.; Lee, S.Y.; Chen, Y.H. Simulated Microgravity Disrupts Cytoskeleton Organization and Increases Apoptosis of Rat Neural Crest Stem Cells Via Upregulating CXCR4 Expression and RhoA-ROCK1-p38 MAPK-p53. Signaling. Stem Cells Dev. 2016, 25, 1172–1193. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Wang, Y.C.; Hu, Z.B.; Xu, H.Y.; Sun, J.; Gao, Y.; Li, X.T.; Yang, C.B.; Xie, C.; Li, C.F.; et al. Simulated Microgravity Promotes Angiogenesis through RhoA- Dependent Rearrangement of the Actin Cytoskeleton. Cell. Physiol. Biochem. 2017, 41, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.; Grimm, D.; Strauch, S.M.; Erzinger, G.S.; Corydon, T.J.; Lebert, M.; Magnusson, N.E.; Infanger, M.; Richter, P.; Krüger, M. Influence of Microgravity on Apoptosis in Cells, Tissues, and Other Systems In Vivo and In Vitro. Int. J. Mol. Sci. 2020, 21, 9373. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhang, K.; Wei, D.; Tian, Y.; Gao, Y.; Chen, Z.; Qian, A. The Impact of Spaceflight and Simulated Microgravity on Cell Adhesion. Int. J. Mol. Sci. 2020, 21, 3031. [Google Scholar] [CrossRef]
- Smith, J.K. Osteoclasts and Microgravity. Life 2020, 10, 207. [Google Scholar] [CrossRef]
- Romswinkel, A.; Infanger, M.; Dietz, C.; Strube, F.; Kraus, A. The Role of C-X-C Chemokine Receptor Type 4 (CXCR4) in Cell Adherence and Spheroid Formation of Human Ewing’s Sarcoma Cells under Simulated Microgravity. Int. J. Mol. Sci. 2019, 20, 6073. [Google Scholar] [CrossRef]
- Ahn, C.B.; Lee, J.H.; Han, G.D.; Kang, H.W.; Lee, S.H.; Lee, J.I.; Son, K.H.; Lee, J.W. Simulated microgravity with floating environment promotes migration of non-small cell lung cancers. Sci. Rep. 2019, 8, 14553. [Google Scholar] [CrossRef]
- Liao, W.; Elfrink, K.; Bähler, M. Head of Myosin IX Binds Calmodulin and Moves Processively toward the Plus-end of Actin Filaments. J. Biol. Chem. 2010, 285, 24933–24942. [Google Scholar] [CrossRef]
- Kerber, M.L.; Cheney, R.E. Myosin-X: A MyTH-FERM myosin at the tips of filopodia. J. Cell. Sci. 2011, 124, 3733–3741. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.; Grimm, D.; Hofstaedter, F.; Wieland, W. Techniques for studies on growth characteristics of human prostatic cancer cells. Biotechnol. Prog. 1992, 8, 494–500. [Google Scholar] [CrossRef]
- Herranz, R.; Anken, R.; Boonstra, J.; Braun, M.; Christianen, P.C.M.; De Geest, M.; Hauslage, J.; Hilbig, R.; Hill, R.J.A.; Lebert, M.; et al. Ground-Based Facilities for Simulation of Microgravity: Organism-Specific Recommendations for Their Use, and Recommended Terminology. Astrobiology 2013, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Nassef, M.Z.; Kopp, S.; Wehland, M.; Melnik, D.; Sahana, J.; Krüger, M.; Corydon, T.J.; Oltmann, H.; Schmitz, B.; Schütte, A.; et al. Real Microgravity Influences the Cytoskeleton and Focal Adhesions in Human Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 3156. [Google Scholar] [CrossRef]
- Pang, S.M.; LE, S.; Yan, J. Mechanical responses of the mechanosensitive unstructured domains in cardiac titin. Biol. Cell 2018, 110, 65–76. [Google Scholar] [CrossRef]
- Grimm, D.; Pietsch, J.; Wehland, M.; Richter, P.; Strauch, S.; Lebert, M.; Magnusson, N.E.; Wise, P.; Bauer, J. The impact of microgravity-based proteomics research. Expert Rev. Proteom. 2014, 11, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, N.; Kulak, N.A.; Cox, J.; Neuhauser, N.; Mayr, K.; Hoerning, O.; Vorm, O.; Mann, M. System-wide Perturbation Analysis with Nearly Complete Coverage of the Yeast Proteome by Single-shot Ultra HPLC Runs on a Bench Top Orbitrap. Mol. Cell. Proteom. 2012, 11, 111. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. Max Quant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and pro-teome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein—Protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Colinge, J. IsobarPTM: A software tool for the quantitative analysis of post-translationally modified proteins. J. Proteom. 2013, 90, 77–84. [Google Scholar] [CrossRef]
- Stanley, R.A.; Gombocz, E.A. System, Method, Software Architecture, and Business Model for Intelligent Object Based Information Platform. U.S. Patent 7,702,639, 20 April 2010. [Google Scholar]
- Hancock, W.S.; Wu, S.L.; Stanley, R.R.; Gombocz, E.A. Publishing large proteome datasets: Scientific policy meets emerging technologies. Trends Biotechnol. 2002, 20, s39–s44. [Google Scholar] [CrossRef]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.; Poux, S.; Bougueleret, L.; Xenarios, I.; Boutet, E. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol. Biol. 2016, 1374, 23–54. [Google Scholar] [CrossRef]
- Lindberg, D.A. Internet access to the National Library of Medicine. Eff. Clin. Pr. ECP 2000, 3, 256–260. [Google Scholar]
- Milacic, M.; Haw, R.; Rothfels, K.; Wu, G.; Croft, D.; Hermjakob, H.; D’Eustachio, P.; Stein, L. Annotating Cancer Variants and Anti-Cancer Therapeutics in Reactome. Cancers 2012, 4, 1180–1211. [Google Scholar] [CrossRef] [PubMed]



| Entry | Gene Name | SwissProt Accession No. | Protein Name | Activity/Function |
|---|---|---|---|---|
| ABI1_HUMAN | ABI1 | Q8IZP0 | Abl interactor 1 | Adapter |
| ABL1_HUMAN | ABL1 | P00519 | Tyrosine-protein kinase ABL1 | Enzyme |
| RHG32_HUMAN | ARHGAP32 | A7KAX9 * | Rho GTPase-activating protein 32 | Adapter |
| ARMX3_HUMAN | ARMCX3 | Q9UH62 | Armadillo repeat-containing X-linked protein3 | Binding protein |
| BCAR1_HUMAN | BCAR1 | P56945 | Breast cancer anti-estrogen resistance protein 1 or p130cas (CRK-associated substrate) | Binding protein |
| BCAR3_HUMAN | BCAR3 | O75815 * | Breast cancer anti-estrogen resistance protein 3 | Adapter |
| CBL_HUMAN | CBL | P22681 | E3 ubiquitin-protein ligase CBL | Enzyme |
| CD2AP_HUMAN | CD2AP | Q9Y5K6 | CD2-associated protein | Adapter |
| CDC42_HUMAN | CDC42 | P60953 | Cell division control protein 42 homolog | Enzyme |
| CIZ1_HUMAN | CIZ1 | Q9ULV3 * | Cip1-interacting zinc finger protein | Binding protein |
| CRK_HUMAN | CRK | P46108 | Adapter molecule crk | Adapter |
| CRKL_HUMAN | CRKL | P46109 | Crk-like protein | Adapter |
| CSK_HUMAN) | CSK | P41240 | Tyrosine-protein kinase CSK | Enzyme |
| EPHA2_HUMAN | EPHA2 | P29317 | Ephrin type-A receptor 2 | Enzyme |
| ERBB2_HUMAN | ERBB2 | P04626 * | Receptor tyrosine-protein kinase erbB-2 | Enzyme |
| ESR1_HUMAN | ESR1 | P03372 * | Estrogen receptor | Hormone receptor |
| FINC_HUMAN | FN1 | P02751 | Fibronectin | ECM protein |
| FYN_HUMAN | FYN | P06241 ** | Tyrosine-protein kinase Fyn | Enzyme |
| GLIS2_HUMAN | GLIS2 | Q9BZE0 ** | Zinc finger protein GLIS2 | Binding protein |
| GMPR2_HUMAN | GMPR2 | Q9P2T1 | GMP reductase 2 | Enzyme |
| HCK_HUMAN | HCK | P08631 ** | Tyrosine-protein kinase HCK | Enzyme |
| BIP_HUMAN | HSPA5 | P11021 | Endoplasmic reticulum chaperone BiP | Enzyme |
| IGF1R_HUMAN | IGF1R | P08069 * | Insulin-like growth factor 1 receptor | Enzyme |
| IQGA1_HUMAN | IQGA1 | P46940 | Ras GTPase-activating-like protein IQGAP1 | Binding protein |
| ITAV_HUMAN | ITGAV | P06756 | Integrin alpha-V | Binding protein |
| ITB1_HUMAN | ITGB1 | P05556 | Integrin beta-1 | Binding protein |
| ITB3_HUMAN | ITGB3 | P05106 ** | Integrin beta-3 | Binding protein |
| KI13A_HUMAN | KIF13A | Q9H1H9 * | Kinesin-like protein KIF13A | Motor protein |
| LYN_HUMAN | LYN | P07948 | Tyrosine-protein kinase Lyn | Enzyme |
| MK08_HUMAN | MAPK8 | P45983 | Mitogen-activated protein kinase 8 | Enzyme |
| MET_HUMAN | MET | P08581 ** | Hepatocyte growth factor receptor | Enzyme |
| MMP14_HUMAN | MMP14 | P50281 ** | Matrix metalloproteinase-14 | Enzyme |
| NCK1_HUMAN | NCK1 | P16333 | Cytoplasmic protein NCK1 | Adapter |
| NCK2_HUMAN | NCK2 | O43639 * | Cytoplasmic protein NCK2 | Adapter |
| PELP1_HUMAN | PELP1 | Q8IZL8 | Proline-, glutamic acid- and leucine-rich protein 1 | Binding protein |
| P85A_HUMAN | PIK3R1 | P27986 * | Phosphatidylinositol 3-kinase regulatory subunit alpha | Adapter |
| PTEN_HUMAN | PTEN | P60484 * | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | Enzyme |
| FAK1_HUMAN | PTK2 | Q05397 | Focal adhesion kinase 1 | Enzyme |
| PTN12_HUMAN | PTN12 | Q05209 | Tyrosine-protein phosphatase non-receptor type 12 | Enzyme |
| PTN1_HUMAN | PTPN1 | P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Enzyme |
| PTN11_HUMAN | PTPN11 | Q06124 | Tyrosine-protein phosphatase non-receptor type 11 | Enzyme |
| PAXI_HUMAN | PXN | P49023 | Paxillin | Cytoskeletal protein |
| RAC1_HUMAN | RAC1 | P63000 | Ras-related C3 botulinum toxin substrate 1 | Enzyme |
| RAN_HUMAN | RAN | P62826 | GTP-binding nuclear protein Ran | Binding protein |
| RHOA_HUMAN | RHOA | P61586 | Transforming protein RhoA | Enzyme |
| SH3K1_HUMAN | SH3KBP1 | Q96B97 | SH3 domain-containing kinase-binding protein 1 | Adapter |
| SHIP2_HUMAN | SHIP2 | O15357 | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | Enzyme |
| SRC_HUMAN | SRC | P12931 | Proto-oncogene tyrosine-protein kinase Src | Enzyme |
| SRCN1_HUMAN | SRCN1 | Q9C0H9 * | SRC kinase signaling inhibitor 1 | Inhibitor |
| TENS3_HUMAN | TNS3 | Q68CZ2 | Tensin-3 | Cytoskeletal protein |
| TLN1_HUMAN | TLN1 | Q9Y490 | Talin-1 | Cytoskeletal protein |
| TENS1_HUMAN | TNS1 | Q9HBL0 ** | Tensin-1 | Cytoskeletal protein |
| TRIP6_HUMAN | TRIP6 | Q15654 * | Thyroid receptor-interacting protein 6 | Binding protein |
| VINC_HUMAN | VCL | P18206 | Vinculin | Cytoskeletal protein |
| VEGFA_HUMAN | VEGFA | P15692 ** | VEGFA | Growth factor |
| VPS11_HUMAN | VPS11 | Q9H270 | Vacuolar protein sorting-associated protein 11 homolog | Binding protein |
| YES_HUMAN | YES1 | P07947 | Tyrosine-protein kinase Yes | Enzyme |
| 1433Z_HUMAN | YWHAZ | P63104 | 14-3-3 protein zeta/delta | Adapter |
| ZYX_HUMAN | ZYX | Q15942 | Zyxin | Binding protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauer, J.; Gombocz, E.; Schulz, H.; Hauslage, J.; Grimm, D. Interaction Network Provides Clues on the Role of BCAR1 in Cellular Response to Changes in Gravity. Computation 2021, 9, 81. https://doi.org/10.3390/computation9080081
Bauer J, Gombocz E, Schulz H, Hauslage J, Grimm D. Interaction Network Provides Clues on the Role of BCAR1 in Cellular Response to Changes in Gravity. Computation. 2021; 9(8):81. https://doi.org/10.3390/computation9080081
Chicago/Turabian StyleBauer, Johann, Erich Gombocz, Herbert Schulz, Jens Hauslage, and Daniela Grimm. 2021. "Interaction Network Provides Clues on the Role of BCAR1 in Cellular Response to Changes in Gravity" Computation 9, no. 8: 81. https://doi.org/10.3390/computation9080081
APA StyleBauer, J., Gombocz, E., Schulz, H., Hauslage, J., & Grimm, D. (2021). Interaction Network Provides Clues on the Role of BCAR1 in Cellular Response to Changes in Gravity. Computation, 9(8), 81. https://doi.org/10.3390/computation9080081








