Conditional Variational Autoencoder for Learned Image Reconstruction
Abstract
:1. Introduction
2. Related Works
3. Preliminaries
3.1. Problem Formulation
3.2. Variational Inference and Variational Autoencoders
4. Proposed Framework
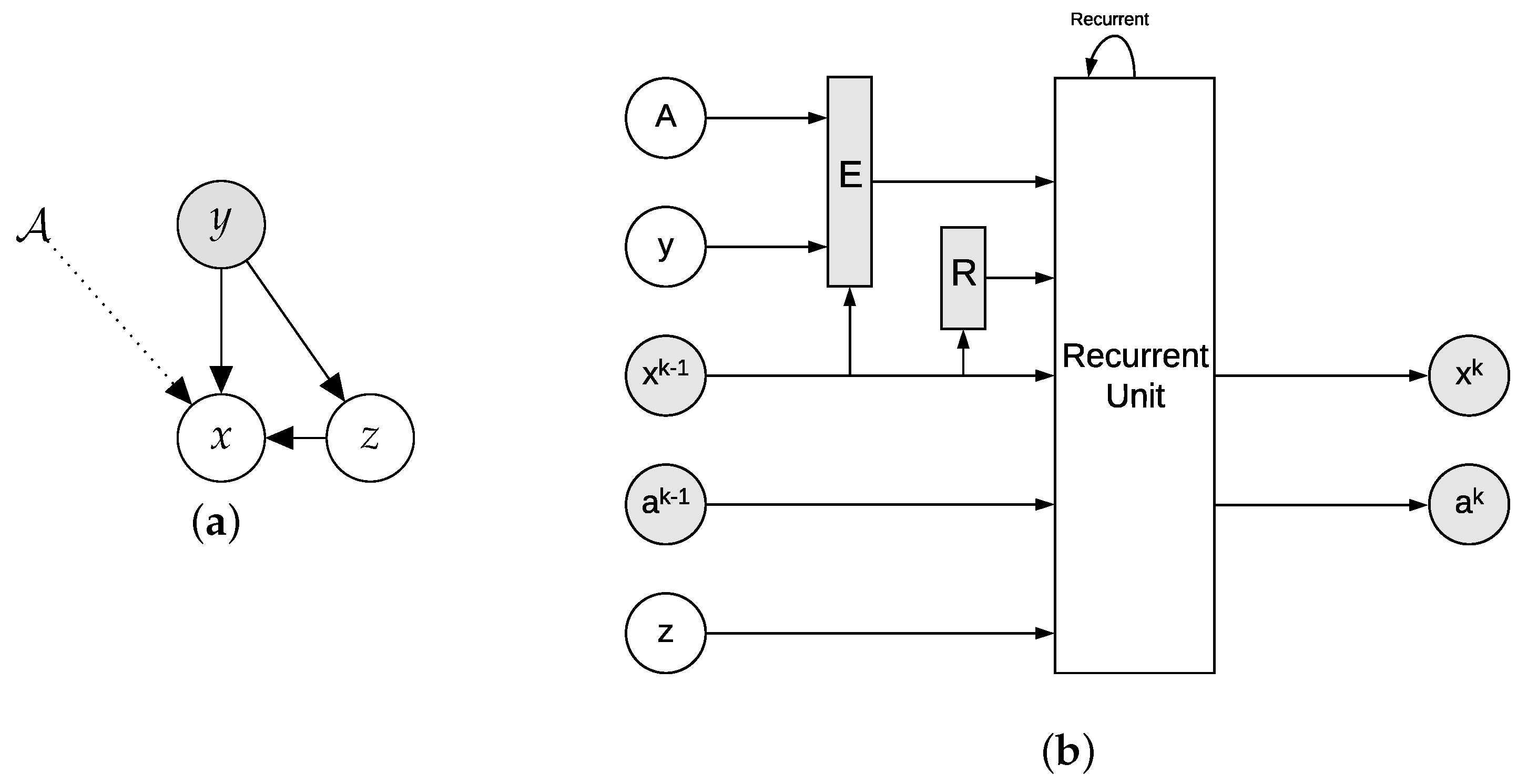
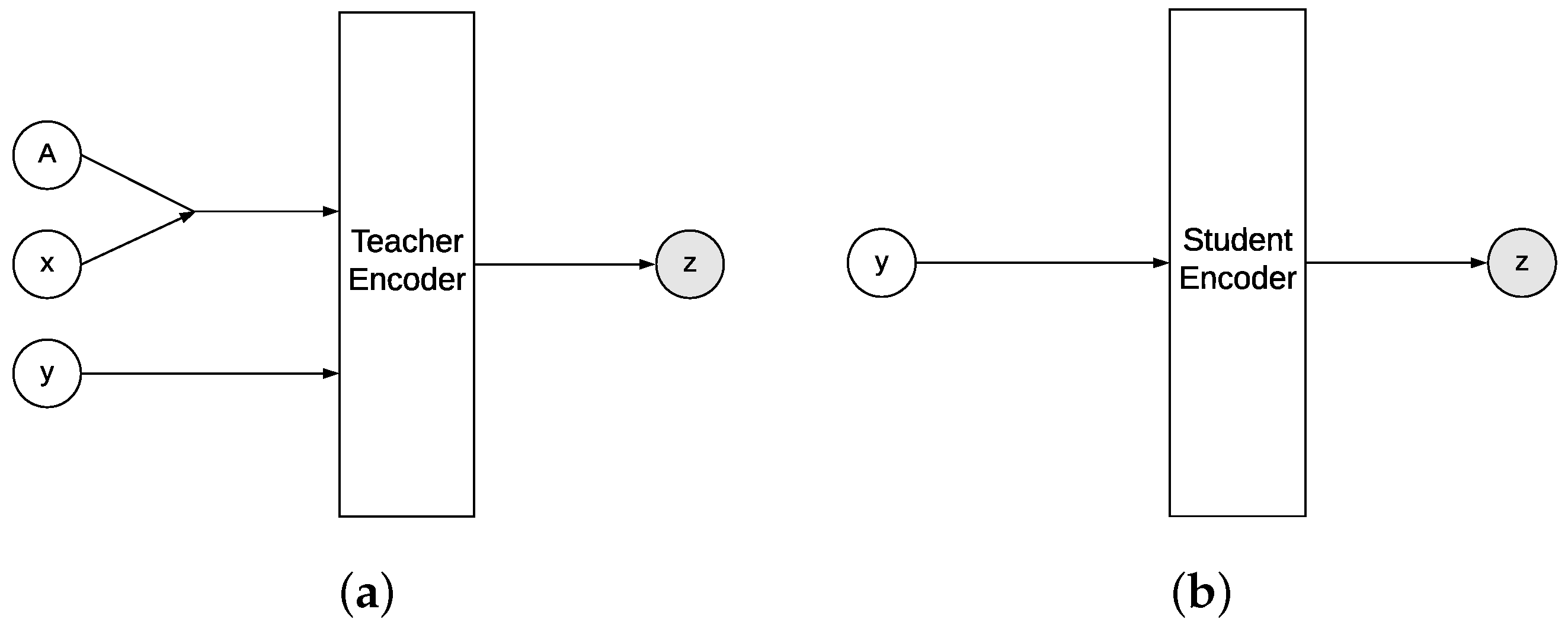
4.1. Conditional VAE as Approximate Inference
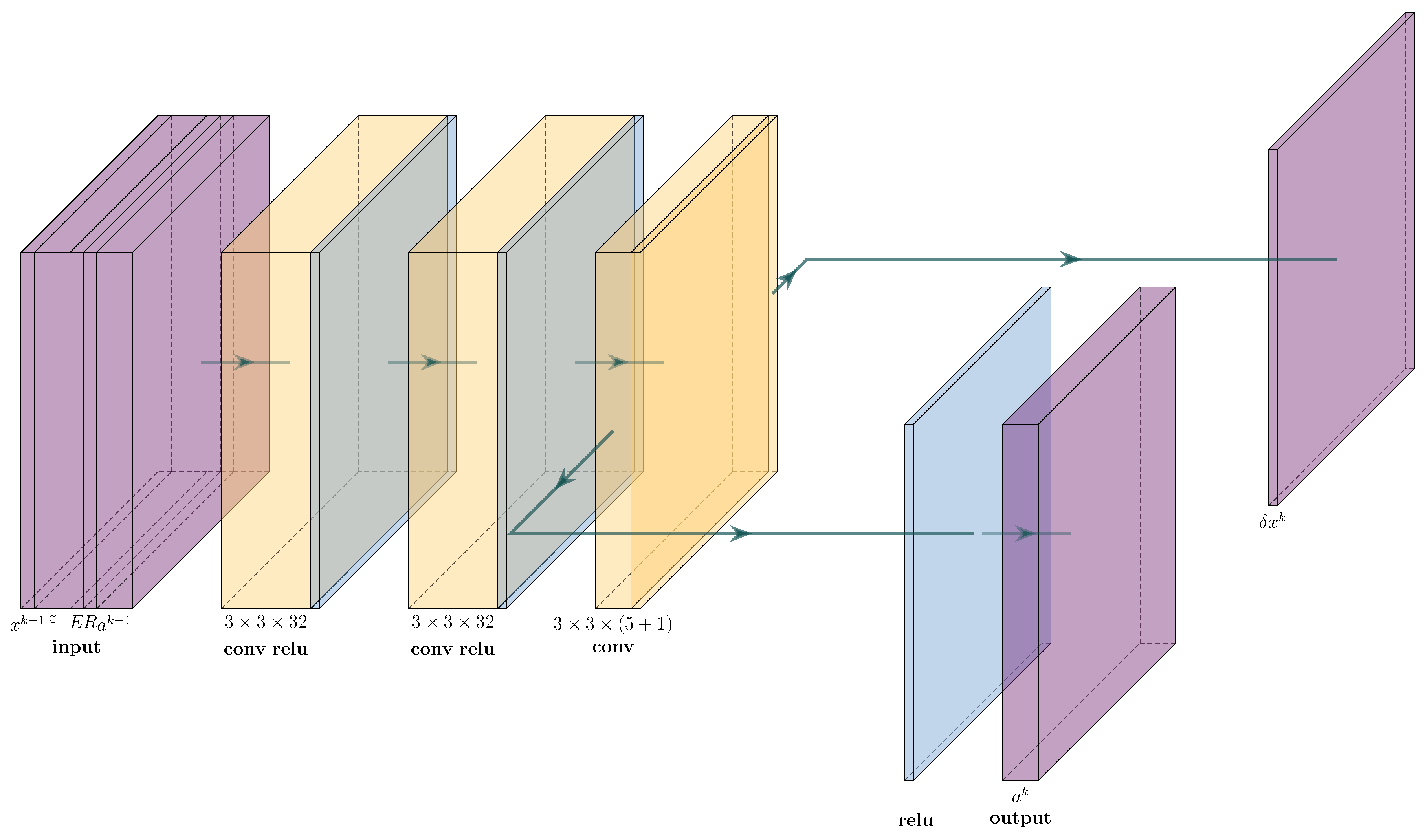
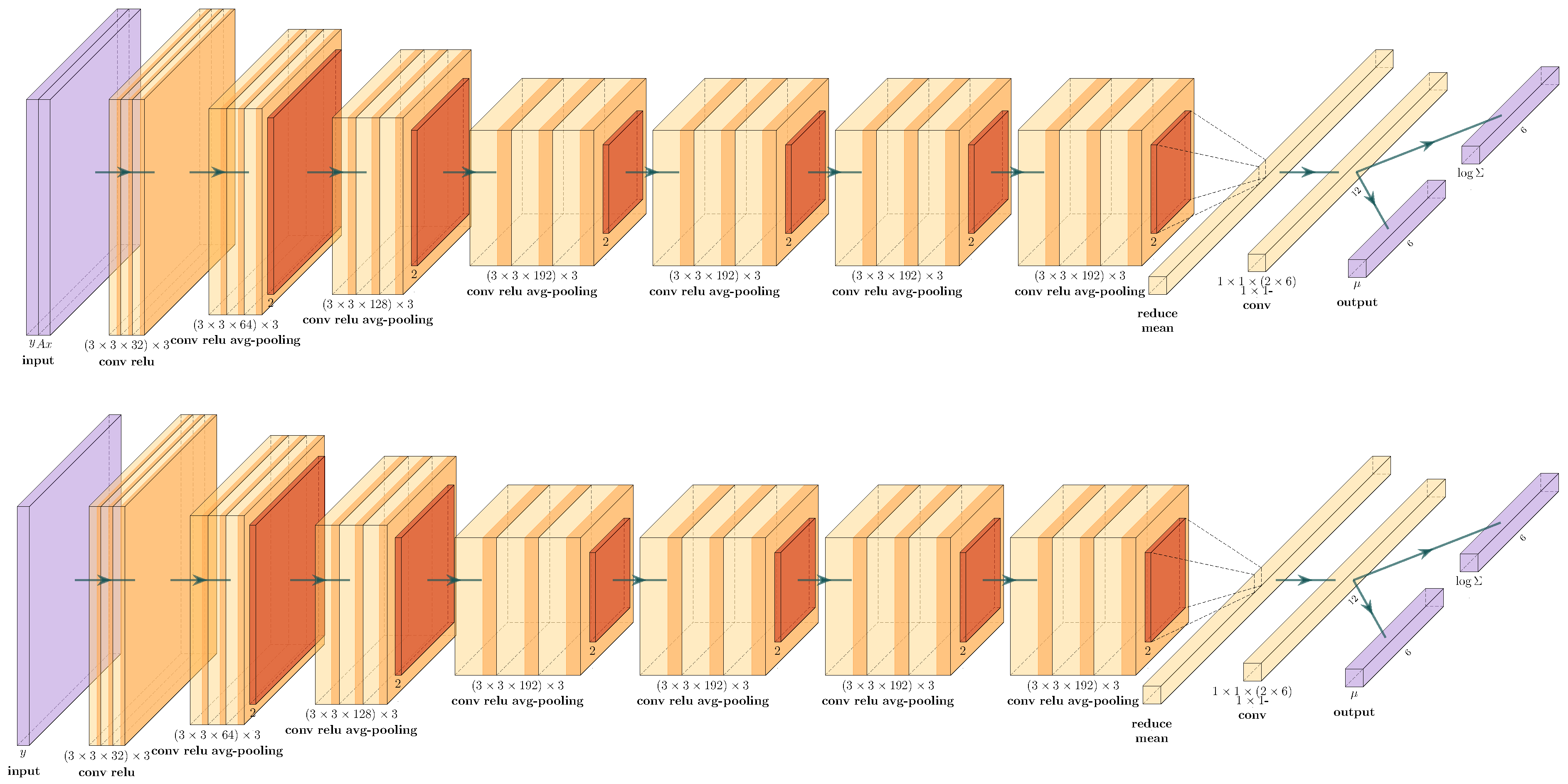
4.2. cVAE for Learned Reconstruction
| Algorithm 1 Training procedure |
|
| Algorithm 2 Inference procedure |
|
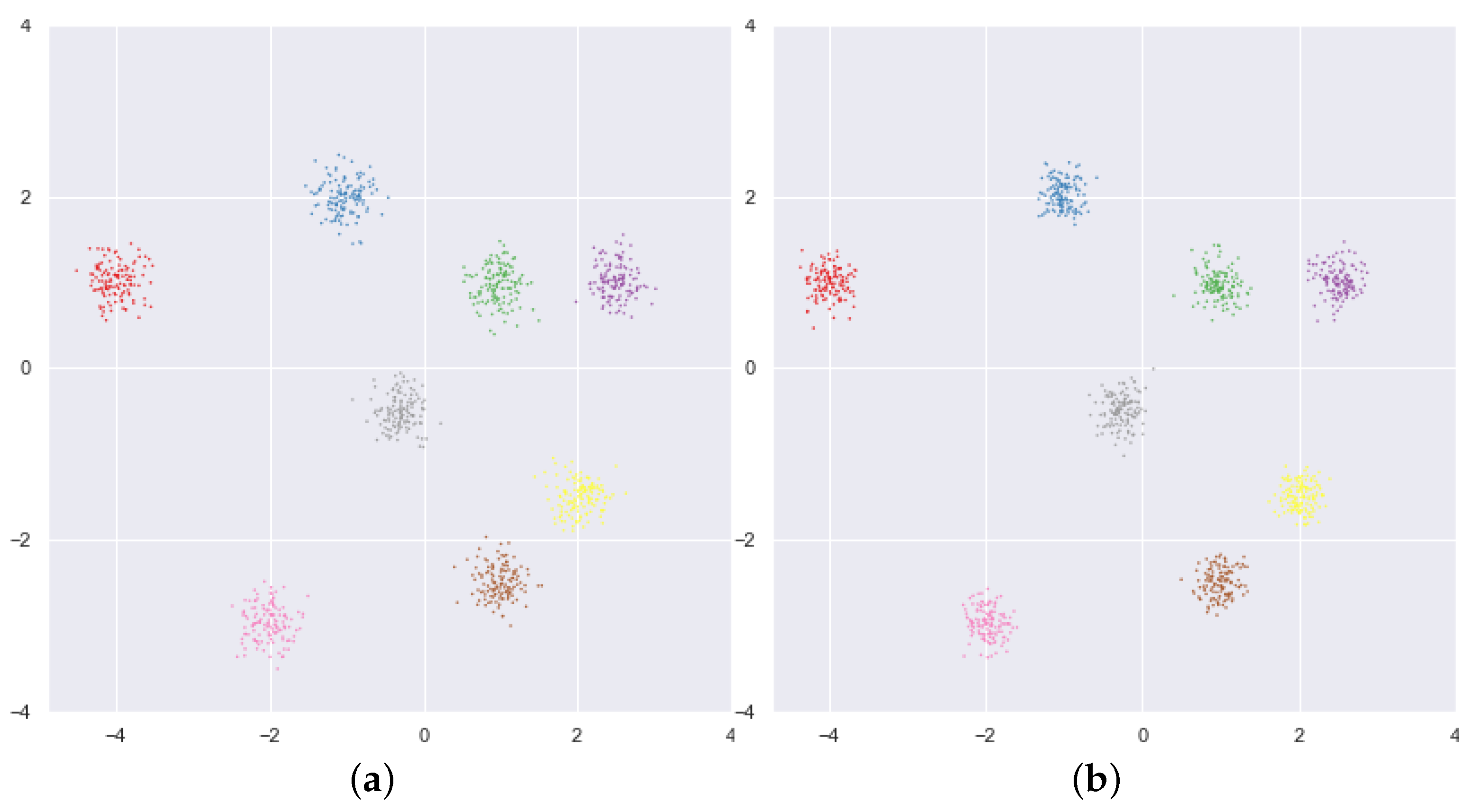
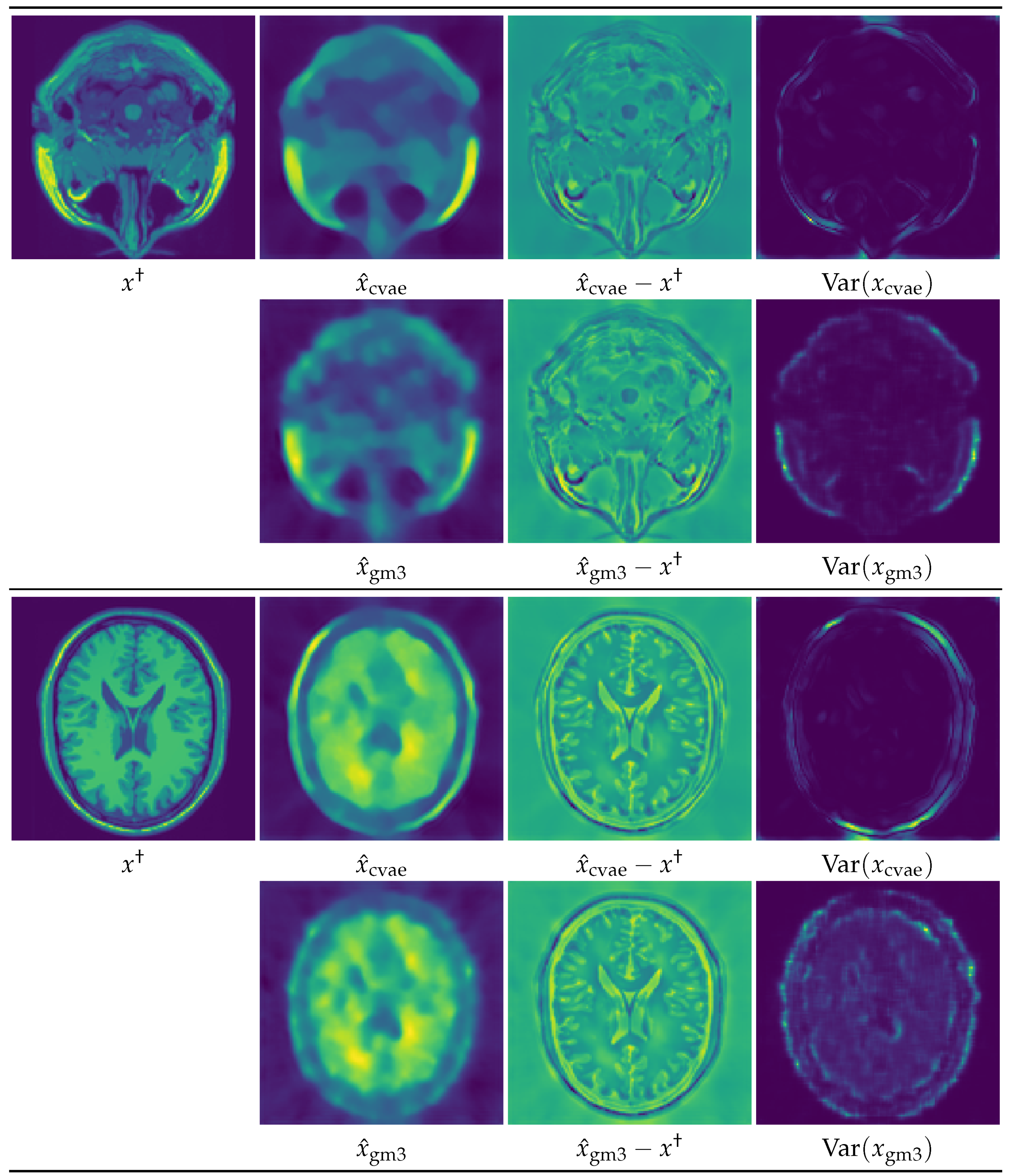
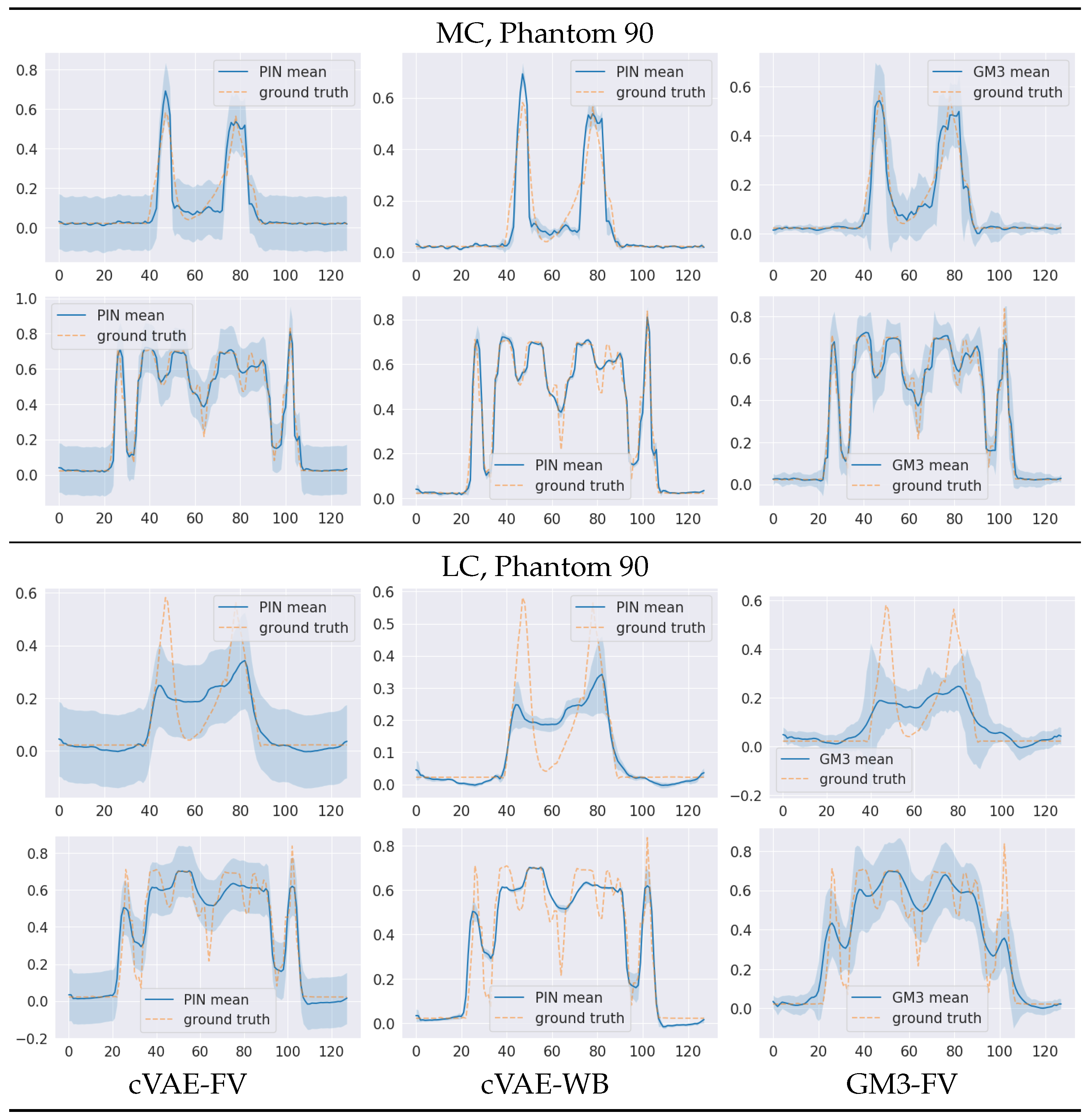
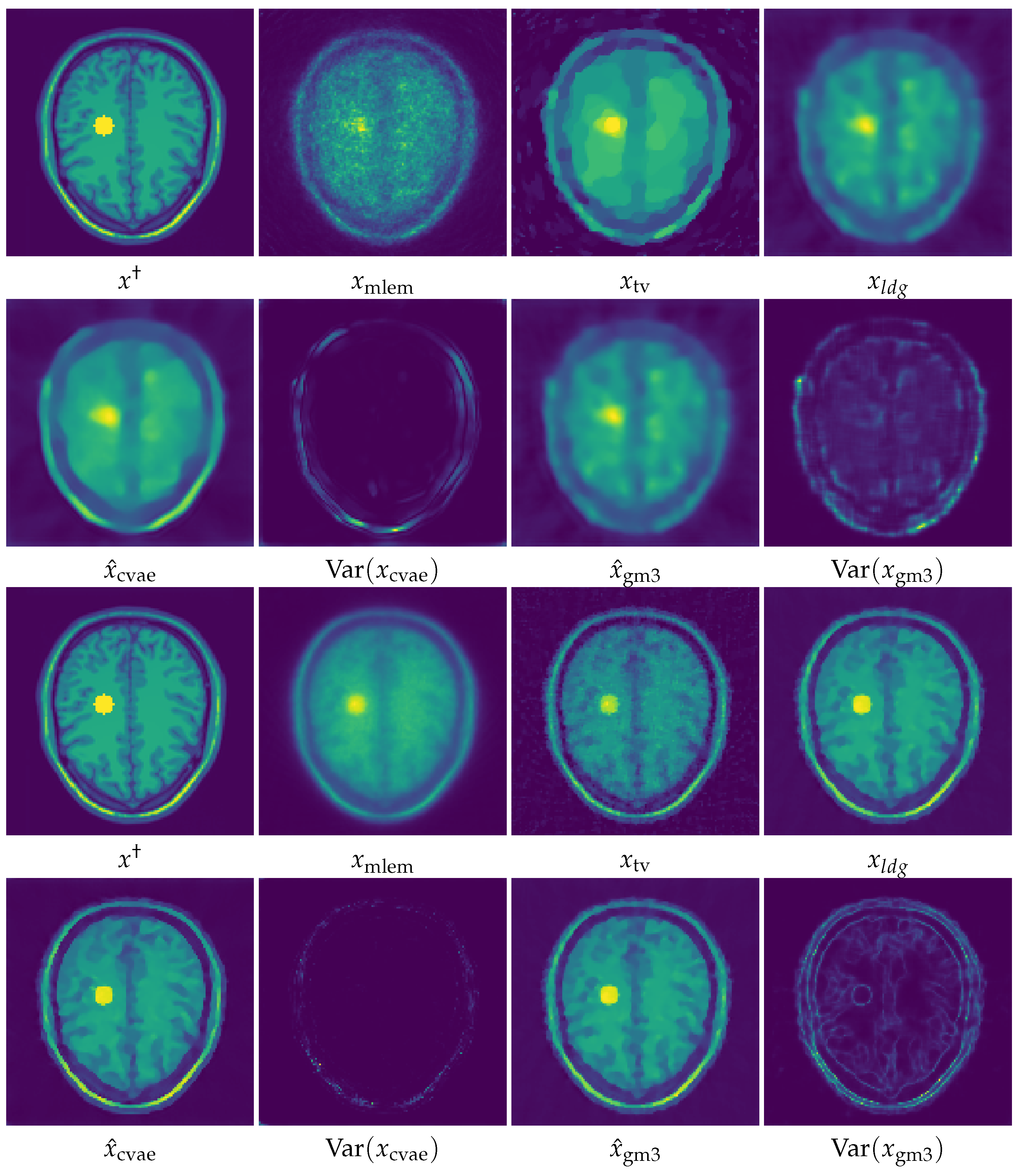
5. Numerical Experiments and Discussions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, K.; Zuo, W.; Chen, Y.; Meng, D.; Zhang, L. Beyond a Gaussian denoiser: Residual learning of deep CNN for image denoising. IEEE Trans. Imag. Proc. 2017, 26, 3142–3155. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Ren, J.S.; Liu, C.; Jia, J. Deep convolutional neural network for image deconvolution. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2014; pp. 1790–1798. [Google Scholar]
- Dong, C.; Loy, C.C.; He, K.; Tang, X. Learning a deep convolutional network for image super-resolution. In Proceedings of the ECCV 2014: Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 184–199. [Google Scholar]
- Kang, E.; Min, J.; Ye, J.C. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med. Phys. 2017, 44, e360–e375. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Zhang, Y.; Chen, Y.; Zhang, J.; Zhang, W.; Sun, H.; Lv, Y.; Liao, P.; Zhou, J.; Wang, G. LEARN: Learned experts’ assessment-based reconstruction network for sparse-data CT. IEEE Trans. Med. Imag. 2018, 37, 1333–1347. [Google Scholar] [CrossRef]
- Hyun, C.M.; Kim, H.P.; Lee, S.M.; Lee, S.; Seo, J.K. Deep learning for undersampled MRI reconstruction. Phys. Med. Biol. 2018, 63, 135007. [Google Scholar] [CrossRef]
- Kendall, A.; Gal, Y. What uncertainties do we need in Bayesian deep learning for computer vision? In Advances in Neural Information Processing Systems; Curran Associates Inc.: Red Hook, NY, USA, 2017; pp. 5574–5584. [Google Scholar]
- Barbano, R.; Arridge, S.; Jin, B.; Tanno, R. Uncertainty quantification in medical image synthesis. In Biomedical Image Synthesis and Simulations: Methods and Applications; Burgos, N., Svoboda, D., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; in press. [Google Scholar]
- Kaipio, J.; Somersalo, E. Statistical and Computational Inverse Problems; Springer: New York, NY, USA, 2005; p. xvi+339. [Google Scholar]
- Stuart, A.M. Inverse problems: A Bayesian perspective. Acta Numer. 2010, 19, 451–559. [Google Scholar] [CrossRef] [Green Version]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein GAN. arXiv 2017, arXiv:1701.07875. [Google Scholar]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Borcea, L. Electrical impedance tomography. Inverse Probl. 2002, 18, R99–R136. [Google Scholar] [CrossRef]
- Arridge, S.R.; Schotland, J.C. Optical tomography: Forward and inverse problems. Inverse Probl. 2009, 25, 123010. [Google Scholar] [CrossRef]
- Zhang, C.; Jin, B. Probabilistic residual learning for aleatoric uncertainty in image restoration. arXiv 2019, arXiv:1908.01010. [Google Scholar]
- Sohn, K.; Lee, H.; Yan, X. Learning structured output representation using deep conditional generative models. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2015; pp. 3483–3491. [Google Scholar]
- Monga, V.; Li, Y.; Eldar, Y.C. Algorithm unrolling: Interpretable, efficient deep learning for signal and image processing. IEEE Signal Proc. Mag. 2021, 38, 18–44. [Google Scholar] [CrossRef]
- Wainwright, M.J.; Jordan, M.I. Graphical models, exponential families, and variational inference. Found. Trends Mach. Learn. 2008, 1, 1–305. [Google Scholar] [CrossRef] [Green Version]
- Lakshminarayanan, B.; Pritzel, A.; Blundell, C. Simple and scalable predictive uncertainty estimation using deep ensembles. In Advances in Neural Information Processing Systems; Curran Associates Inc.: Red Hook, NY, USA, 2017; pp. 6402–6413. [Google Scholar]
- Gal, Y.; Ghahramani, Z. Dropout as a Bayesian approximation: Representing model uncertainty in deep learning. In Proceedings of the 33rd International Conference on Machine Learning, New York, NY, USA, 20–22 June 2016; pp. 1050–1059. [Google Scholar]
- Abdar, M.; Pourpanah, F.; Hussain, S.; Rezazadegan, D.; Liu, L.; Ghavamzadeh, M.; Fieguth, P.; Khosravi, A.; Acharya, U.R.; Makarenkov, V.; et al. A review of uncertainty quantification in deep learning: Techniques, applications and challenges. Inf. Fusion 2021, 76, 243–297. [Google Scholar] [CrossRef]
- Graves, A. Practical variational inference for neural networks. In Advances in Neural Information and Processing Systems; Curran Associates Inc.: Red Hook, NY, USA, 2011; pp. 2348–2356. [Google Scholar]
- Blundell, C.; Cornebise, J.; Kavukcuoglu, K.; Wierstra, D. Weight uncertainty in neural network. In Proceedings of the International Conference on Machine Learning, Lille, France, 7–9 July 2015; pp. 1613–1622. [Google Scholar]
- Barbano, R.; Zhang, C.; Arridge, S.; Jin, B. Quantifying model uncertainty in inverse problems via Bayesian deep gradient descent. In Proceedings of the 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 1392–1399. [Google Scholar]
- Barbano, R.; Kereta, Z.; Zhang, C.; Hauptmann, A.; Arridge, S.; Jin, B. Quantifying sources of uncertainty in deep learning-based image reconstruction. arXiv 2020, arXiv:2011.08413v2. [Google Scholar]
- Minka, T.P. Expectation propagation for approximate Bayesian inference. arXiv 2013, arXiv:1301.2294. [Google Scholar]
- Osawa, K.; Swaroop, S.; Jain, A.; Eschenhagen, R.; Turner, R.E.; Yokota, R.; Khan, M.E. Practical deep learning with Bayesian principles. arXiv 2019, arXiv:1906.02506v2. [Google Scholar]
- Qi, J.; Leahy, R.M. Iterative reconstruction techniques in emission computed tomography. Phys. Med. Biol. 2006, 51, R541–R578. [Google Scholar] [CrossRef]
- Zhang, C.; Arridge, S.R.; Jin, B. Expectation Propagation for Poisson Data. Inverse Probl. 2019, 35, 085006. [Google Scholar] [CrossRef] [Green Version]
- Filipović, M.; Barat, E.; Dautremer, T.; Comtat, C.; Stute, S. PET reconstruction of the posterior image probability, including multimodal images. IEEE Trans. Med. Imag. 2019, 38, 1643–1654. [Google Scholar] [CrossRef]
- Zhou, Q.; Yu, T.; Zhang, X.; Li, J. Bayesian inference and uncertainty quantification for medical image reconstruction with Poisson data. SIAM J. Imaging Sci. 2020, 13, 29–52. [Google Scholar] [CrossRef]
- Ongie, G.; Jalal, A.; Baraniuk, R.G.; Metzler, C.A.; Dimakis, A.G.; Willett, R. Deep learning techniques for inverse problems in imaging. IEEE J. Sel. Areas Inform. Theory 2020, 1, 39–56. [Google Scholar] [CrossRef]
- Gregor, K.; LeCun, Y. Learning fast approximations of sparse coding. In Proceedings of the International Conference on Machine Learning, Haifa, Israel, 21–24 June 2010; pp. 1–8. [Google Scholar]
- Putzky, P.; Welling, M. Recurrent inference machines for solving inverse problems. arXiv 2017, arXiv:1706.04008. [Google Scholar]
- Jordan, M.I.; Ghahramani, Z.; Jaakkola, T.S.; Saul, L.K. An introduction to variational methods for graphical models. Mach. Learn. 1999, 37, 183–233. [Google Scholar] [CrossRef]
- Opper, M.; Archambeau, C. The variational Gaussian approximation revisited. Neural Comput. 2009, 21, 786–792. [Google Scholar] [CrossRef]
- Arridge, S.R.; Ito, K.; Jin, B.; Zhang, C. Variational Gaussian approximation for Poisson data. Inverse Probl. 2018, 34, 025005. [Google Scholar] [CrossRef]
- MacKay, D.J.C. Information Theory, Inference and Learning Algorithms; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Chai, Y.; Liu, M.; Duffy, B.A.; Kim, H. Learning to Synthesize cortical morphological changes using graph conditional variational autoencoder. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021. [Google Scholar] [CrossRef]
- Hou, T.Y.; Lam, K.C.; Zhang, P.; Zhang, S. Solving Bayesian inverse problems from the perspective of deep generative networks. Comput. Mech. 2019, 64, 395–408. [Google Scholar] [CrossRef]
- Kullback, S.; Leibler, R.A. On information and sufficiency. Ann. Math. Stat. 1951, 22, 79–86. [Google Scholar] [CrossRef]
- Rezende, D.J.; Mohamed, S.; Wierstra, D. Stochastic backpropagation and approximate inference in deep generative models. In Proceedings of the 31st International Conference on International Conference on Machine Learning, Beijing, China, 21–26 June 2014; Volume 32, pp. 1278–1286. [Google Scholar]
- Kingma, D.P.; Welling, M. An introduction to variational autoencoders. arXiv 2019, arXiv:1906.02691. [Google Scholar] [CrossRef]
- Khemakhem, I.; Kingma, D.; Monti, R.; Hyvarinen, A. Variational autoencoders and nonlinear ica: A unifying framework. In Proceedings of the International Conference on Artificial Intelligence and Statistics, PMLR, Palermo, Italy, 3–5 June 2020; pp. 2207–2217. [Google Scholar]
- Walker, J.; Doersch, C.; Gupta, A.; Hebert, M. An uncertain future: Forecasting from static images using variational autoencoders. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; pp. 835–851. [Google Scholar]
- Liu, J.S. Monte Carlo Strategies in Scientific Computing; Springer: New York, NY, USA, 2001; p. xvi+343. [Google Scholar]
- Ito, K.; Jin, B. Inverse Problems: Tikhonov Theory and Algorithms; World Scientific Publishing Co. Pte. Ltd.: Hackensack, NJ, USA, 2015; p. x+318. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. In Proceedings of the 3rd International Conference on Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Cocosco, C.A.; Kollokian, V.; Kwan, R.K.S.; Pike, G.B.; Evans, A.C. Brainweb: Online interface to a 3D MRI simulated brain database. NeuroImage 1997, 5, S425. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Dillon, J.V.; Langmore, I.; Tran, D.; Brevdo, E.; Vasudevan, S.; Moore, D.; Patton, B.; Alemi, A.; Hoffman, M.; Saurous, R.A. Tensorflow distributions. arXiv 2017, arXiv:1711.10604. [Google Scholar]
- Shepp, L.A.; Vardi, Y. Maximum likelihood reconstruction for emission tomography. IEEE Trans. Med. Imag. 1982, 1, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Rudin, L.I.; Osher, S.; Fatemi, E. Nonlinear total variation based noise removal algorithms. Phys. D. Nonlinear Phenom. 1992, 60, 259–268. [Google Scholar] [CrossRef]
- Adler, J.; Öktem, O. Solving ill-posed inverse problems using iterative deep neural networks. Inverse Probl. 2017, 33, 124007. [Google Scholar] [CrossRef] [Green Version]
- He, B.; Lakshminarayanan, B.; Teh, Y.W. Bayesian deep ensembles via the neural tangent kernel. arXiv 2020, arXiv:2007.05864. [Google Scholar]
- Nix, D.A.; Weigend, A.S. Estimating the mean and variance of the target probability distribution. In Proceedings of the 1994 IEEE International Conference on Neural Networks (ICNN’94), Orlando, FL, USA, 28 June–2 July 1994; Volume 1, pp. 55–60. [Google Scholar]
- Moeller, M.; Möllenhoff, T.; Cremers, D. Controlling neural networks via energy dissipation. arXiv 2019, arXiv:1904.03081. [Google Scholar]

| MLEM | TV | LGD | cVAE | |
|---|---|---|---|---|
| MC | 0.74/23.20 | 0.85/28.76 | 0.92/29.07 | 0.91/28.01 |
| LC | 0.64/21.55 | 0.62/22.58 | 0.59/21.68 | 0.64/23.10 |
| 10 | 20 | 30 | 50 | 70 | ||
| PNSR | MC | 27.66/28.05 | 27.14/27.48 | 27.25/27.43 | 27.25/27.50 | 25.65/26.77 |
| LC | 22.60/21.86 | 22.09/21.35 | 22.30/21.09 | 22.14/20.69 | 20.87/19.32 | |
| SSIM | MC | 0.89/0.89 | 0.89/0.89 | 0.91/0.91 | 0.88/0.88 | 0.88/0.89 |
| LC | 0.65/0.62 | 0.65/0.61 | 0.69/0.62 | 0.54/0.50 | 0.46/0.45 | |
| 90 | 100 | 110 | 130 | 150 | ||
| PSNR | MC | 24.98/26.83 | 26.91/27.74 | 27.81/28.43 | 27.96/29.33 | 30.86/32.57 |
| LC | 20.52/19.02 | 21.39/19.74 | 22.22/20.61 | 22.48/21.32 | 25.78/23.67 | |
| SSIM | MC | 0.88/0.91 | 0.90/0.91 | 0.92/0.92 | 0.94/0.94 | 0.96/0.96 |
| LC | 0.60/0.53 | 0.62/0.55 | 0.61/0.57 | 0.60/0.57 | 0.73/0.67 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Barbano, R.; Jin, B. Conditional Variational Autoencoder for Learned Image Reconstruction. Computation 2021, 9, 114. https://doi.org/10.3390/computation9110114
Zhang C, Barbano R, Jin B. Conditional Variational Autoencoder for Learned Image Reconstruction. Computation. 2021; 9(11):114. https://doi.org/10.3390/computation9110114
Chicago/Turabian StyleZhang, Chen, Riccardo Barbano, and Bangti Jin. 2021. "Conditional Variational Autoencoder for Learned Image Reconstruction" Computation 9, no. 11: 114. https://doi.org/10.3390/computation9110114
APA StyleZhang, C., Barbano, R., & Jin, B. (2021). Conditional Variational Autoencoder for Learned Image Reconstruction. Computation, 9(11), 114. https://doi.org/10.3390/computation9110114






