Design of Periodic Neural Networks for Computational Investigations of Nonlinear Hepatitis C Virus Model Under Boozing
Abstract
1. Introduction
- Computational investigations of the nonlinear mathematical model of HCV are effectively presented using a novel implementation of the integrated computing approach combining PNNs, GA, and IPA.
- The overlapping of the results from the designed PNNs-GA-IPA with the reference solutions from the RK-4 numerical solver for the nonlinear HCV model demonstrates its value through high accuracy and strong convergence indices.
- The performance and validation of the designed PNNs-GA-IPA is certified through statistical evaluations, including the mean absolute error (MAE) index, Theil’s inequality coefficient (TIC), and the root mean square error (RMSE) metric for multiple trails.
2. Mathematical Model
3. Proposed Methodology
3.1. Framework of PNNs-GA-IPA
- ,
- ,
- ,
- ,
- ,
- .
3.2. Optimization Procedure for GA-IPA
4. Statistical Measures
5. Results and Discussion
6. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bertino, G.; Ardiri, A.; Proiti, M.; Rigano, G.; Frazzetto, E.; Demma, S.; Ruggeri, M.I.; Scuderi, L.; Malaguarnera, G.; Bertino, N.; et al. Chronic hepatitis C: This and the new era of treatment. World J. Hepatol. 2016, 8, 92. [Google Scholar] [CrossRef] [PubMed]
- Platt, L.; Easterbrook, P.; Gower, E.; McDonald, B.; Sabin, K.; McGowan, C.; Yanny, I.; Razavi, H.; Vickerman, P. Prevalence and burden of HCV co-infection in people living with HIV: A global systematic review and meta-analysis. Lancet Infect. Dis. 2016, 16, 797–808. [Google Scholar] [CrossRef]
- WHO. Hepatitis C. 2021. Available online: https://www.who.int/publications/i/item/9789240053779 (accessed on 22 July 2022).
- WHO: Global Health Sector Strategy on Viral Hepatitis, 2016–2021. Available online: https://www.who.int/publications/i/item/WHO-HIV-2016.06 (accessed on 17 May 2016).
- Mohamed, A.A.; Elbedewy, T.A.; El-Serafy, M.; El-Toukhy, N.; Ahmed, W.; El Din, Z.A. Hepatitis C virus: A global view. World J. Hepatol. 2015, 7, 2676. [Google Scholar] [CrossRef]
- Huang, R.; Rao, H.; Yang, M.; Gao, Y.; Wang, J.; Jin, Q.; Ma, D.; Wei, L. Noninvasive measurements predict liver fibrosis well in hepatitis C virus patients after direct-acting antiviral therapy. Dig. Dis. Sci. 2020, 65, 1491–1500. [Google Scholar] [CrossRef]
- Fong, T.L.; Kanel, G.C.; Conrad, A.; Valinluck, B.; Charboneau, F.; Adkins, R.H. Clinical significance of concomitant hepatitis C infection in patients with alcoholic liver disease. Hepatology 1994, 19, 554–557. [Google Scholar] [CrossRef]
- Coelho-Little, M.E.; Jeffers, L.J.; Bernstein, D.E.; Goodman, J.J.; Reddy, K.R.; de Medina, M.; Li, X.; Hill, M.; La Rue, S.; Schiff, E.R. Hepatitis C virus in alcoholic patients with and without clinically apparent liver disease. Alcohol. Clin. Exp. Res. 1995, 19, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Sata, M.; Fukuizmi, K.; Uchimura, Y.; Nakano, H.; Ishii, K.; Kumashiro, R.; Mizokami, M.; Lau, J.Y.; Tanikawa, K. Hepatitis C virus in patients with clinically diagnosed alcoholic liver disease. J. Viral. Hepat. 1996, 3, 143–148. [Google Scholar] [CrossRef]
- Poynard, T.; Bedrossa, P.; Opolon, P.; for the OBSVIRIC, METAVIR, CLINIVIR, and DOSVIRIC groups. Natural history of liver fibrosis progression in patients with chronic hepatitis C. Lancet 1997, 349, 825–832. [Google Scholar] [CrossRef]
- Bellentani, S.; Pozzato, G.; Saccoccio, G.; Crovatto, M.; Croce, L.S.; Mazzoran, L.; Masutti, F.; Cristianini, G.; Tiribelli, C. Clinical course and risk factors of hepatitis C virus related liver disease in the general population: Report from the Dionysos study. Gut 1999, 44, 874–880. [Google Scholar] [CrossRef]
- Wiley, T.E.; McCarthy, M.; Breidi, L.; Layden, T.J. Impact of alcohol on the histological and clinical progression of hepatitis C infection. Hepatology 1998, 28, 805–809. [Google Scholar] [CrossRef]
- Thomas, D.L.; Astemborski, J.; Rai, R.M.; Anania, F.A.; Schaeffer, M.; Galai, N.; Nolt, K.; Nelson, K.E.; Strathdee, S.A.; Johnson, L.; et al. The natural history of hepatitis C virus infection: Host, viral, and environmental factors. JAMA 2000, 284, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.R.; Gonin, R.; Alter, H.J.; Wright, E.C.; Buskell, Z.J.; Hollinger, F.B.; Seeff, L.B. The relationship of acute transfusion-associated hepatitis to the development of cirrhosis in the presence of alcohol abuse. Ann. Intern. Med. 2001, 134, 120–124. [Google Scholar] [CrossRef]
- Ohnishi, K.; Matsuo, S.; Matsutani, K.; Itahashi, M.; Kakihara, K.; Suzuki, K.; Ito, S.; Fujiwara, K. Interferon therapy for chronic hepatitis C in habitual drinkers: Comparison with chronic hepatitis C in infrequent drinkers. Am. J. Gastroenterol. 1996, 91, 1374–1379. [Google Scholar] [PubMed]
- Loguercio, C.; Di Pierro, M.; Di Marino, M.P.; Federico, A.; Disalvo, D.; Crafa, E.; Tuccillo, C.; Baldi, F.; del VecchioBlanco, C. Drinking habits of subjects with hepatitis C virus-related chronic liver disease: Prevalence and effect on clinical, virological and pathological aspects. Alcohol Alcohol. 2000, 35, 296–301. [Google Scholar] [CrossRef]
- Mahdy, A.M.S.; Sweilam, N.H.; Higazy, M. Approximate solution for solving nonlinear fractional order smoking model. Alex. Eng. J. 2020, 59, 739–752. [Google Scholar] [CrossRef]
- Mahdy, A.M.S. Numerical studies for solving fractional integro-differential equations. J. Ocean Eng. Sci. 2018, 3, 127–132. [Google Scholar] [CrossRef]
- Mahdy, A.M.S.; Higazy, M.; Gepreel, K.A.; El-Dahdouh, A.A.A. Optimal control and bifurcation diagram for a model nonlinear fractional SIRC. Alex. Eng. J. 2020, 59, 3481–3501. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, C.; Yang, X.; Chu, Y.; Xia, Z. Efficient, non-iterative, and second-order accurate numerical algorithms for the anisotropic Allen–Cahn Equation with precise nonlocal mass conservation. J. Comput. Appl. Math. 2020, 363, 444–463. [Google Scholar] [CrossRef]
- Mahdy, A.M.S.; Lotfy, K.; Hassan, W.; El-Bary, A.A. Analytical solution of magneto-photothermal theory during variable thermal conductivity of a semiconductor material due to pulse heat flux and volumetric heat source. Waves Random Complex Media 2021, 31, 2040–2057. [Google Scholar] [CrossRef]
- Ul Rahman, J.; Danish, S.; Lu, D. Deep Neural Network-Based Simulation of Sel’kov Model in Glycolysis: A Comprehensive Analysis. Mathematics 2023, 11, 3216. [Google Scholar] [CrossRef]
- Ul Rahman, J.; Makhdoom, F.; Ali, A.; Danish, S. Mathematical modeling and simulation of biophysics systems using neural network. Int. J. Mod. Phys. B 2023, 38, 2450066. [Google Scholar] [CrossRef]
- Hassan, A.; Kamran, M.; Illahi, A.; Zahoor, R.M.A. Design of cascade artificial neural networks optimized with the memetic computing paradigm for solving the nonlinear Bratu system. Eur. Phys. J. Plus 2019, 134, 122. [Google Scholar] [CrossRef]
- Umar, M.; Sabir, Z.; Raja, M.A.Z. Intelligent computing for numerical treatment of nonlinear prey–predator models. Appl. Soft Comput. 2019, 80, 506–524. [Google Scholar] [CrossRef]
- Sabir, Z.; Raja, M.A.Z.; Umar, M.; Shoaib, M. Neuro-swarm intelligent computing to solve the second-order singular functional differential model. Eur. Phys. J. Plus 2020, 135, 474. [Google Scholar] [CrossRef]
- Sabir, Z.; Raja, M.A.Z.; Umar, M.; Shoaib, M. Design of neuro-swarming-based heuristics to solve the third-order nonlinear multi-singular Emden–Fowler equation. Eur. Phys. J. Plus 2020, 135, 410. [Google Scholar] [CrossRef]
- Cheema, T.N.; Raja, M.A.Z.; Ahmad, I.; Naz, S.; Ilyas, H.; Shoaib, M. Intelligent computing with Levenberg–Marquardt artificial neural networks for nonlinear system of COVID-19 epidemic model for future generation disease control. Eur. Phys. J. Plus 2020, 135, 932. [Google Scholar] [CrossRef]
- Umar, M.; Sabir, Z.; Raja, M.A.Z.; Sánchez, Y.G. A stochastic numerical computing heuristic of SIR nonlinear model based on dengue fever. Results Phys. 2020, 19, 103585. [Google Scholar] [CrossRef]
- Sabir, Z.; Saoud, S.; Raja, M.A.Z.; Wahab, H.A.; Arbi, A. Heuristic computing technique for numerical solutions of nonlinear fourth order Emden–Fowler equation. Math. Comput. Simul. 2020, 178, 534–548. [Google Scholar] [CrossRef]
- Sabir, Z.; Akkurt, N.; Said, S.B. A novel radial basis Bayesian regularization deep neural network for the Maxwell nanofluid applied on the Buongiorno model. Arab. J. Chem. 2023, 16, 104706. [Google Scholar] [CrossRef]
- Sabir, Z.; Said, S.B. Heuristic computing for the novel singular third order perturbed delay differential model arising in thermal explosion theory. Arab. J. Chem. 2023, 16, 104509. [Google Scholar] [CrossRef]
- Sabir, Z.; Said, S.B.; Guirao, J.L.G. A radial basis scale conjugate gradient deep neural network for the monkeypox transmission system. Mathematics 2023, 11, 975. [Google Scholar] [CrossRef]
- Raja, M.A.Z.; Umar, M.; Sabir, Z.; Khan, J.A.; Baleanu, D. A new stochastic computing paradigm for the dynamics of nonlinear singular heat conduction model of the human head. Eur. Phys. J. Plus 2018, 133, 364. [Google Scholar] [CrossRef]
- Bukhari, A.H.; Sulaiman, M.; Islam, S.; Shoaib, M.; Kumam, P.; Raja, M.A.Z. Neuro-fuzzy modeling and prediction of summer precipitation with application to different meteorological stations. Alex. Eng. J. 2020, 59, 101–116. [Google Scholar] [CrossRef]
- Sabir, Z.; Sadat, R.; Ali, M.R.; Said, S.B.; Azhar, M. A numerical performance of the novel fractional water pollution model through the Levenberg-Marquardt backpropagation method. Arab. J. Chem. 2023, 16, 104493. [Google Scholar] [CrossRef]
- Mustafa, N.; Ul Rahman, J.; Omame, A. Modelling of marburg virus transmission dynamics: A deep learning-driven approach with the effect of quarantine and health awareness interventions. Model. Earth Syst. Environ. 2024, 10, 7337–7357. [Google Scholar] [CrossRef]
- Ali, A.H.; Amir, M.; Rahman, J.U.; Raza, A.; Arif, G.E. Design of Morlet Wavelet Neural Networks for Solving the Nonlinear Van der Pol–Mathieu–Duffing Oscillator Model. Computers 2025, 14, 14. [Google Scholar] [CrossRef]
- Shah, N.H.; Shukla, N.; Satia, M.H.; Thakkar, F.A. Optimal Control of HCV transmission under liquoring. J. Theor. Biol. 2019, 465, 27–33. [Google Scholar] [CrossRef]
- Wei, H.; Bao, H.; Ruan, X. Genetic algorithm-driven discovery of unexpected thermal conductivity enhancement by disorder. Nano Energy 2020, 71, 104619. [Google Scholar] [CrossRef]
- Podvalny, S.L.; Chizhov, M.I.; Gusev, P.Y.; Gusev, K.Y. The crossover operator of a genetic algorithm as applied to the task of a production planning. Procedia Comput. Sci. 2019, 150, 603–608. [Google Scholar] [CrossRef]
- Gani, M.M.; Islam, M.S.; Ullah, M.A. Optimal PID tuning for controlling the temperature of electric furnace by genetic algorithm. SN Appl. Sci. 2019, 1, 880. [Google Scholar] [CrossRef]
- Parinam, S.; Kumar, M.; Kumari, N.; Karar, V.; Sharma, A.L. An improved optical parameter optimisation approach using Taguchi and genetic algorithm for high transmission optical filter design. Optik 2019, 182, 382–392. [Google Scholar] [CrossRef]
- Wambacq, J.; Ulloa, J.; Lombaert, G.; François, S. Interior-point methods for the phase-field approach to brittle and ductile fracture. Comput. Methods Appl. Mech. Eng. 2021, 375, 113612. [Google Scholar] [CrossRef]
- Zanelli, A.; Domahidi, A.; Jerez, J.; Morari, M. FORCES NLP: An efficient implementation of interior-point methods for multistage nonlinear nonconvex programs. Int. J. Control 2020, 93, 13–29. [Google Scholar] [CrossRef]
- Umar, M.; Sabir, Z.; Raja, M.A.Z.; Amin, F.; Saeed, T.; Guerrero-Sanchez, Y. Integrated neuro-swarm heuristic with interior-point for nonlinear SITR model for dynamics of novel COVID-19. Alex. Eng. J. 2021, 60, 2811–2824. [Google Scholar] [CrossRef]
- Bleyer, J. Advances in the simulation of viscoplastic fluid flows using interior-point methods. Comput. Methods Appl. Mech. Eng. 2018, 330, 368–394. [Google Scholar] [CrossRef]



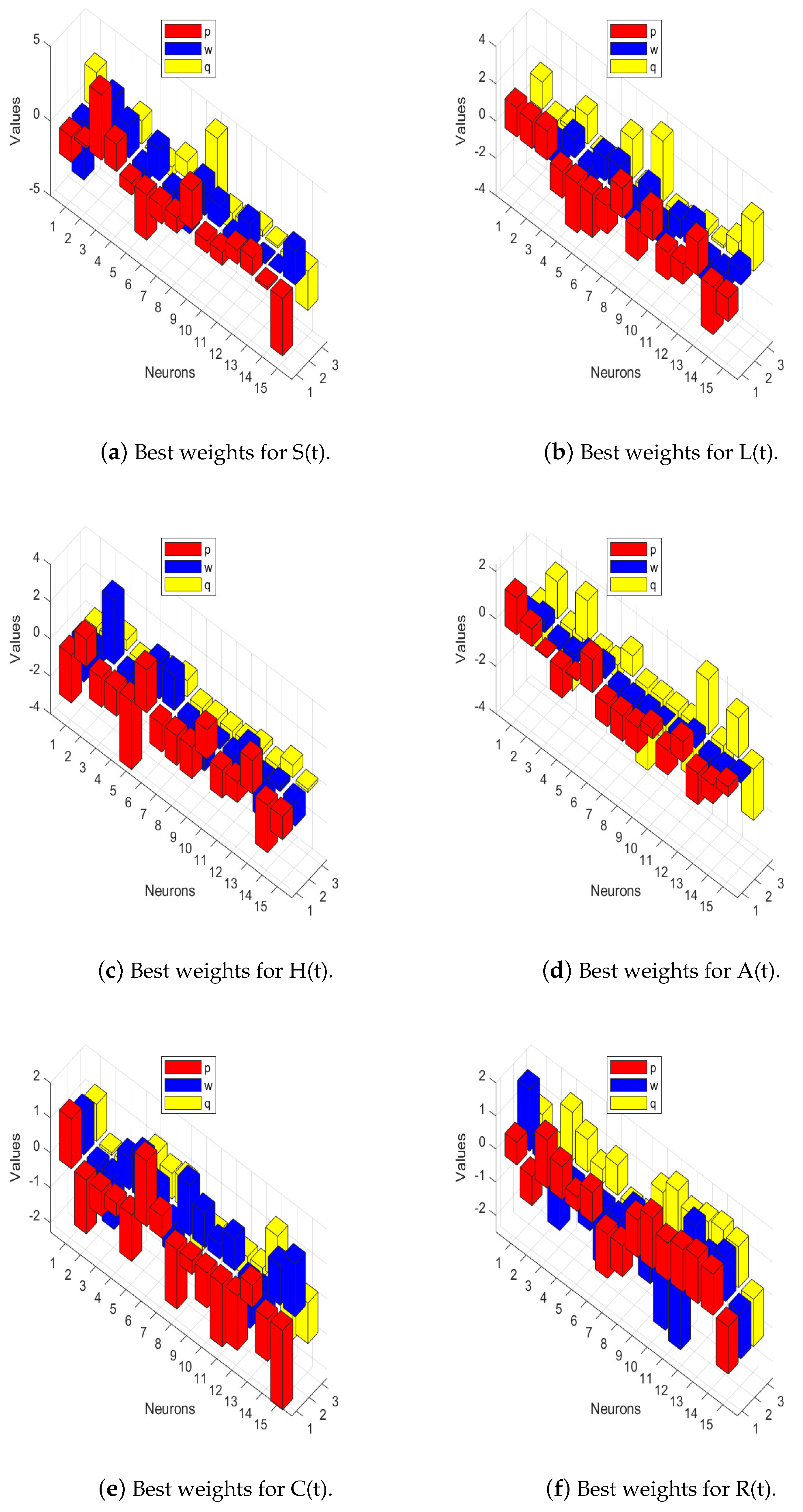


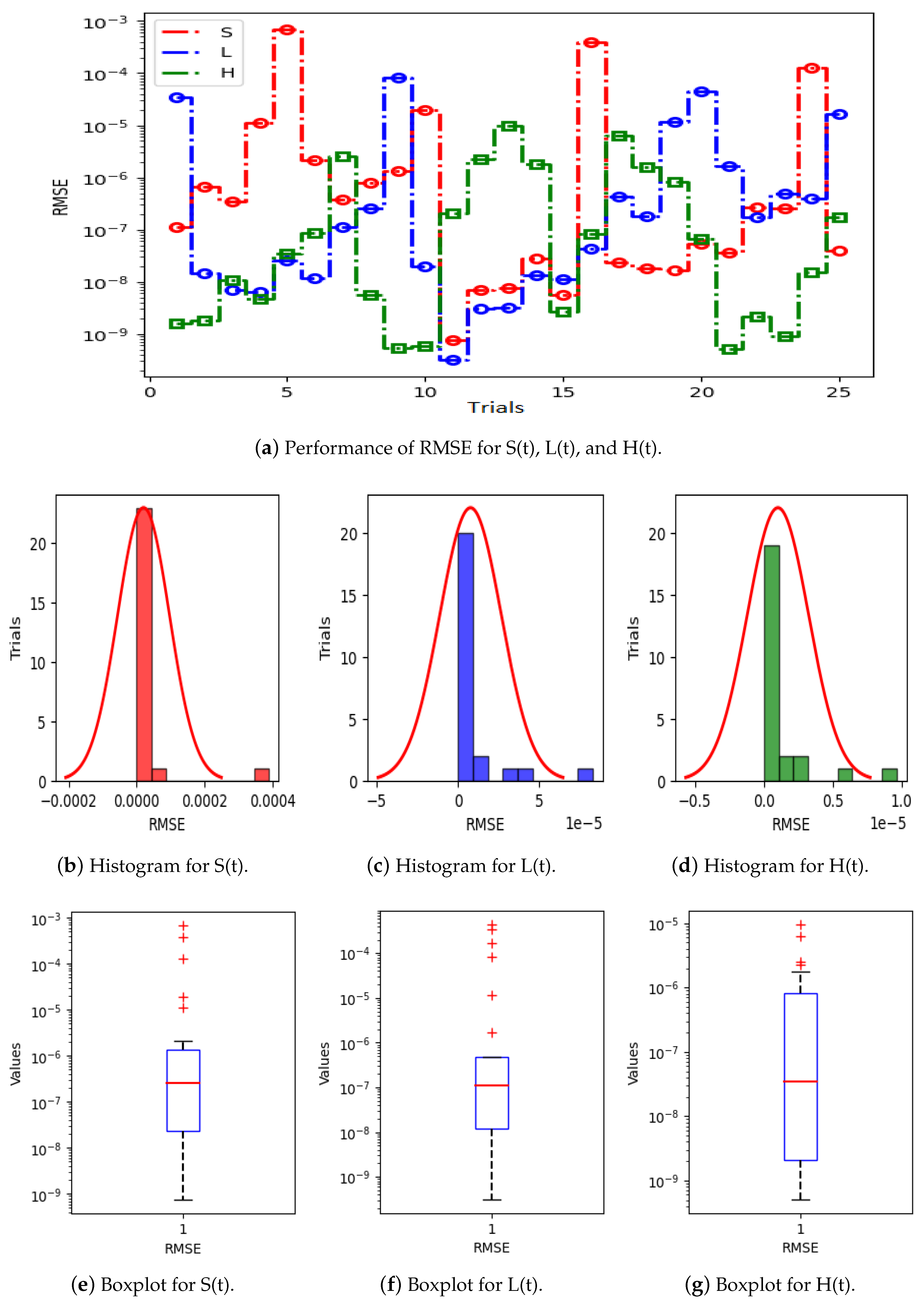


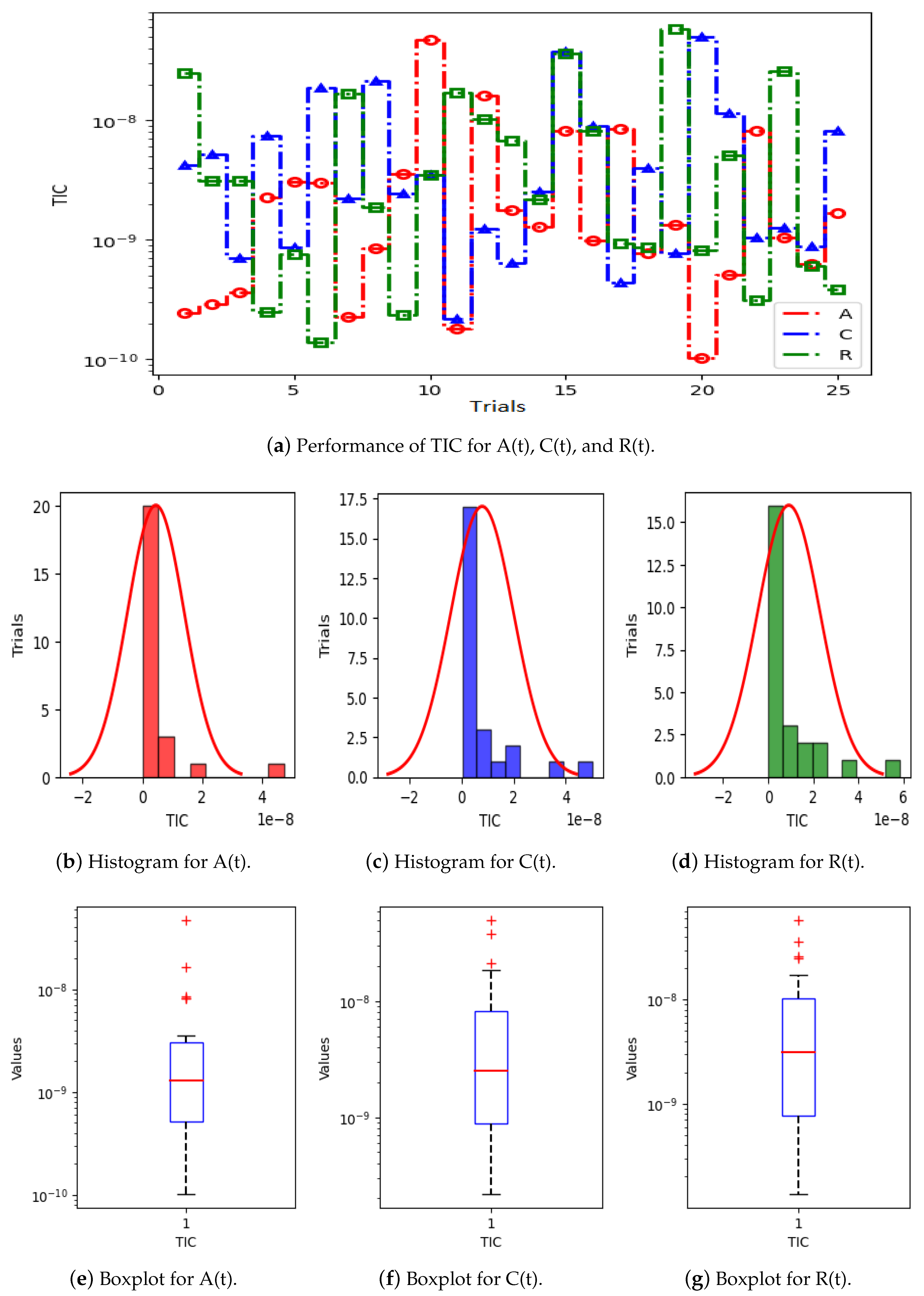
| Parameter | Value | Parameter | Value | Parameter | Value | Parameter | Value |
|---|---|---|---|---|---|---|---|
| D | 0.2 | 0.35 | 0.6 | 1 | |||
| 0.5 | 0.1 | 0.3 | 0.4807 | ||||
| 0.6 | 0.5 | 0.1 | 0.3121 | ||||
| 0.2 | 0.5 | 0.02 | 0.0371 | ||||
| 0.5 | 0.3 | 0.40 | 0.2690 |
| GA Process | |
|---|---|
| Inputs | Parameters of the PNNs . |
| Parameters | All parameters described as: . |
| Outputs | GA weights are denoted as . |
| Initialization | Adjust . |
| Stopping Criteria | Terminate if any of the criteria is achieved . |
| Ranking | Rank precise in the specific population based on loss L. |
| Save | Store with time, trails, and loss. |
| GA Procedure Stop | — |
| Start the IPA | |
| Inputs | is taken as initial point. |
| Initialization | Initialize , assignments, generations, and other standards. |
| Terminating Standards | Stop if any of the following criteria are achieved: . |
| Evaluation of loss | Find the value of W and L taking by Equations (6)–(13). |
| Tuning | Apply ‘fmincon’, update W and find loss L taking into consideration Equations (6)–(13). |
| Save | Store , loss L, function counts, and iterations with time. |
| IPA procedure | — |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mannan, A.; Ul Rahman, J.; Iqbal, Q.; Zulfiqar, R. Design of Periodic Neural Networks for Computational Investigations of Nonlinear Hepatitis C Virus Model Under Boozing. Computation 2025, 13, 66. https://doi.org/10.3390/computation13030066
Mannan A, Ul Rahman J, Iqbal Q, Zulfiqar R. Design of Periodic Neural Networks for Computational Investigations of Nonlinear Hepatitis C Virus Model Under Boozing. Computation. 2025; 13(3):66. https://doi.org/10.3390/computation13030066
Chicago/Turabian StyleMannan, Abdul, Jamshaid Ul Rahman, Quaid Iqbal, and Rubiqa Zulfiqar. 2025. "Design of Periodic Neural Networks for Computational Investigations of Nonlinear Hepatitis C Virus Model Under Boozing" Computation 13, no. 3: 66. https://doi.org/10.3390/computation13030066
APA StyleMannan, A., Ul Rahman, J., Iqbal, Q., & Zulfiqar, R. (2025). Design of Periodic Neural Networks for Computational Investigations of Nonlinear Hepatitis C Virus Model Under Boozing. Computation, 13(3), 66. https://doi.org/10.3390/computation13030066






