Immune Response in Mice Immunized with Chimeric H1 Antigens
Abstract
:1. Introduction
2. Materials and Methods
2.1. HA Sequence Selection and Consensus Sequence cHA Construction In Silico
2.2. cHA Structural and Functional Predictions
2.3. cHA B-Cell Epitope and Antigenic Predictions
2.4. cHA Design
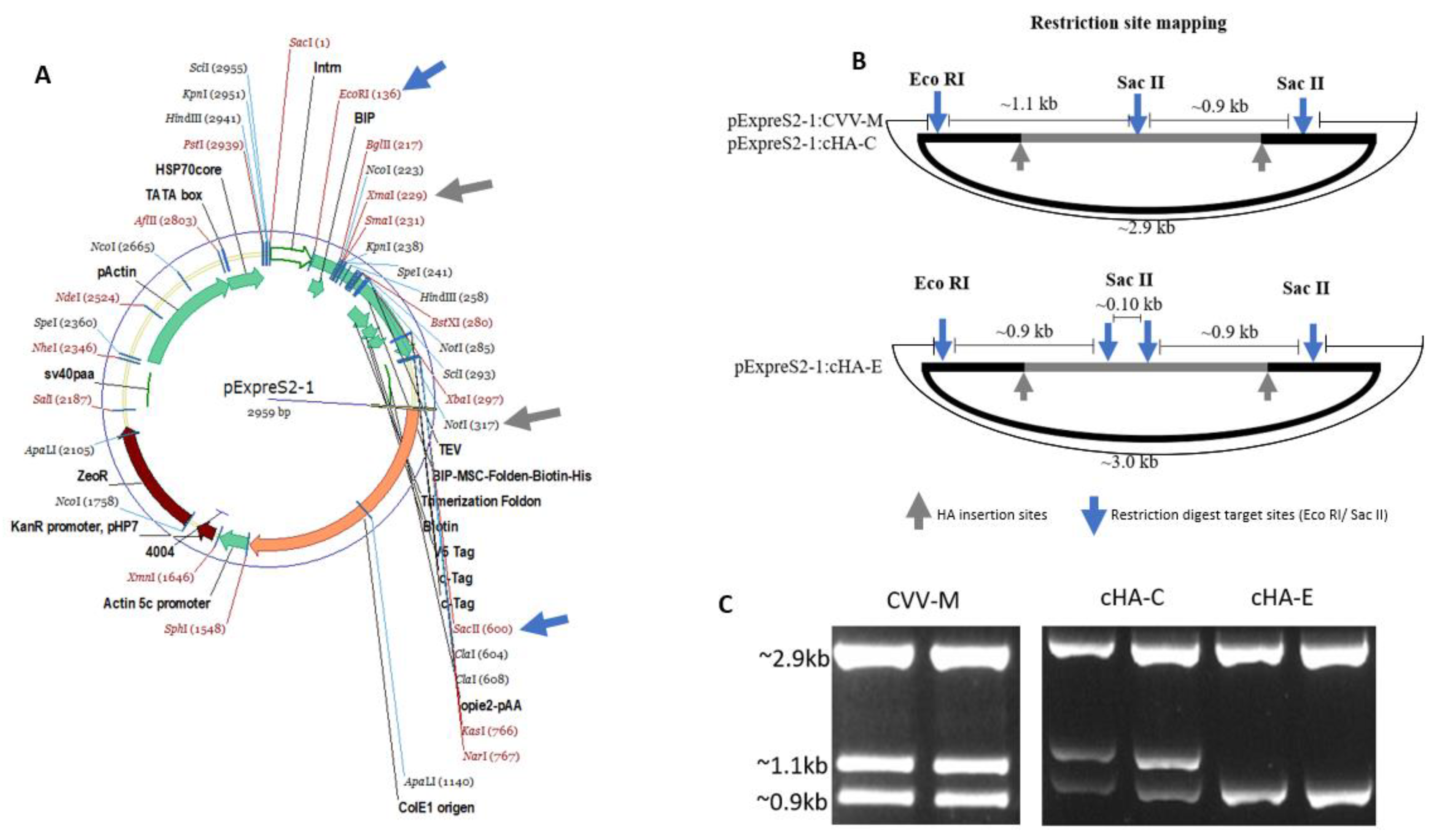
2.5. cHAs Cloning and Transfection
2.5.1. Purification of cHAs and Vaccine Preparation
2.5.2. HA Antigens’ Haemagglutination (HA) Activity Assessment
2.6. Haemagglutination Inhibition (HI) Assay
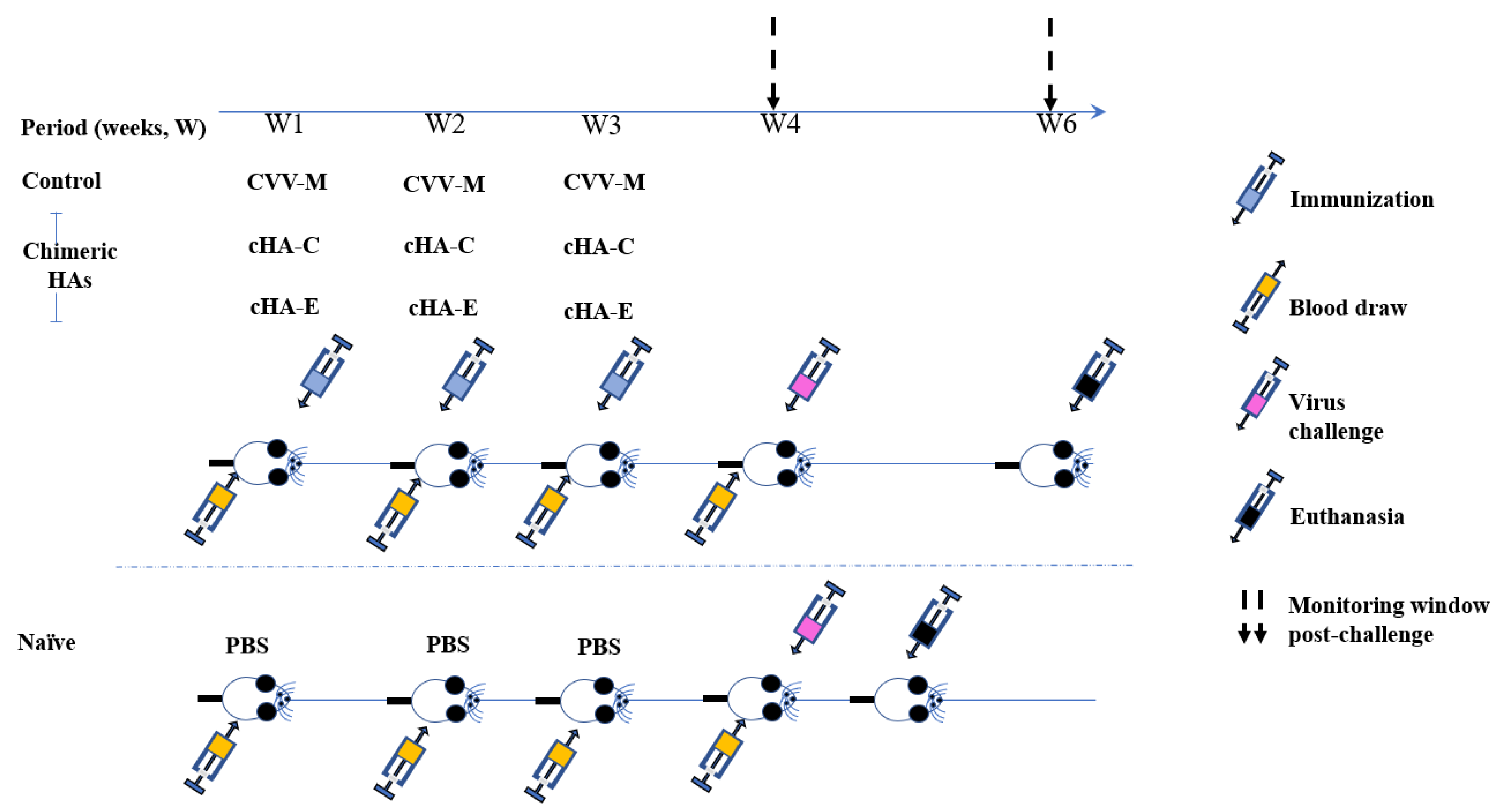
2.7. Immunization
Virus Challenge
3. Results
3.1. Conceptual Design of the H1-Based cHAs
3.2. cHAs Are Relatively Similar in Structure and Function to a Typical HA
3.3. cHA B-Cell Epitope and Antigenic Predictions
3.4. Recombinant HAs Had No Detectable Hemagglutination Activity
3.5. Mice Were Not Previously Exposed to at Least the H1 Strains of Influenza Used
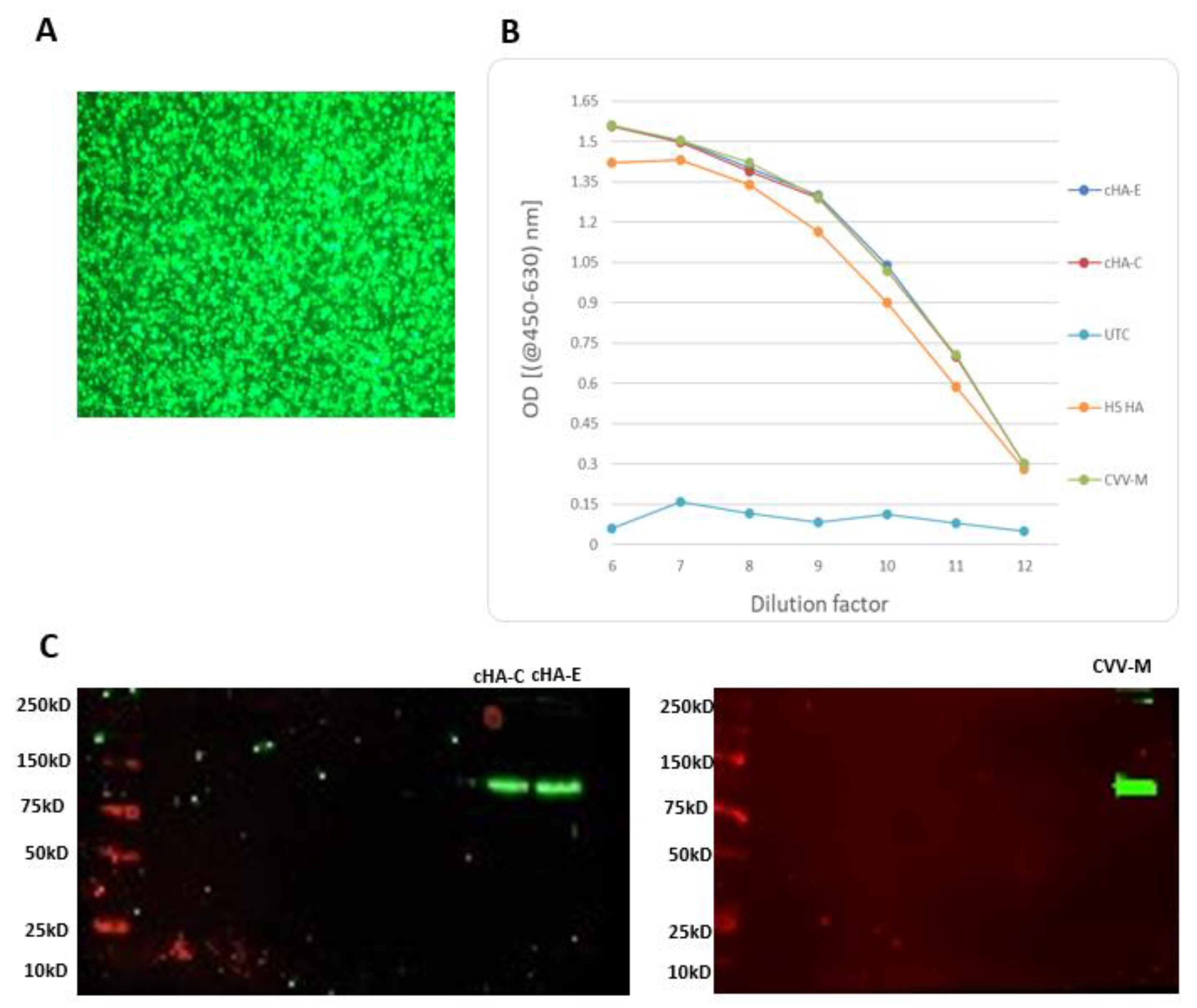
3.6. Stimulation of Mice with cHAs or CVV-M Induced Seroconversion
3.6.1. Anti-cHAs Antibodies Cross-React with Heterosubtypic H5N2 Virus
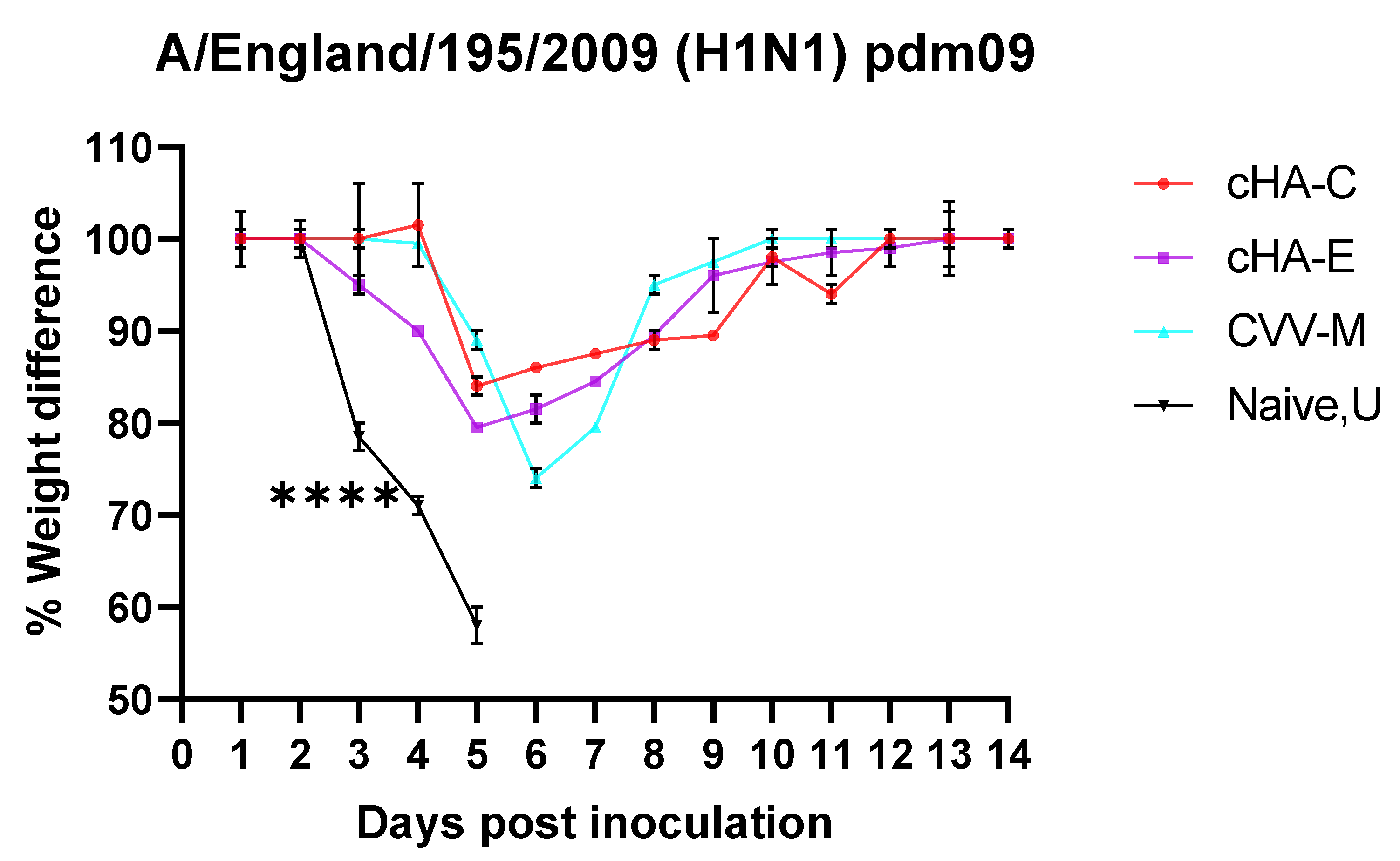
3.6.2. Mice Vaccinated with cHAs Subunit Vaccines Showed Reduced Morbidity (Weight Loss Rebound) against the Lethal Dose H1N1 Pdm09 Virus Challenge
4. Discussions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef]
- Dawood, F.S.; Iuliano, A.D.; Reed, C.; Meltzer, M.I.; Shay, D.K.; Cheng, P.-Y.; Bandaranayake, D.; Breiman, R.F.; Brooks, W.A.; Buchy, P. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: A modelling study. Lancet Infect. Dis. 2012, 12, 687–695. [Google Scholar] [CrossRef] [Green Version]
- Hampson, A.; Barr, I.; Cox, N.; Donis, R.O.; Siddhivinayak, H.; Jernigan, D.; Katz, J.; McCauley, J.; Motta, F.; Odagiri, T. Improving the selection and development of influenza vaccine viruses–Report of a WHO informal consultation on improving influenza vaccine virus selection, Hong Kong SAR, China, 18–20 November 2015. Vaccine 2017, 35, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.C.; Wang, M.H.; Chen, Z.; Hui, D.S.; Kwok, A.K.; Yeung, A.C.; Liu, K.M.; Yeoh, Y.K.; Lee, N.; Chan, P.K. Frequent genetic mismatch between vaccine strains and circulating seasonal influenza viruses, Hong Kong, China, 1996–2012. Emerg. Infect. Dis. 2018, 24, 1825. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, E.; Qiu, X.; Wilson, P.C.; Bahl, J.; Krammer, F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep. 2018, 8, 10432. [Google Scholar] [CrossRef]
- Wu, N.C.; Wilson, I.A. Influenza hemagglutinin structures and antibody recognition. Cold Spring Harb. Perspect. Med. 2020, 10, a038778. [Google Scholar] [CrossRef] [Green Version]
- Huber, V.C.; Lynch, J.M.; Bucher, D.J.; Le, J.; Metzger, D.W. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J. Immunol. 2001, 166, 7381–7388. [Google Scholar] [CrossRef] [Green Version]
- Jegaskanda, S.; Reading, P.C.; Kent, S.J. Influenza-specific antibody-dependent cellular cytotoxicity: Toward a universal influenza vaccine. J. Immunol. 2014, 193, 469–475. [Google Scholar] [CrossRef] [Green Version]
- Krammer, F.; Palese, P. Universal influenza virus vaccines that target the conserved hemagglutinin stalk and conserved sites in the head domain. J. Infect. Dis. 2019, 219, S62–S67. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Guptill, J.; Naficy, A.; Nachbagauer, R.; Berlanda-Scorza, F.; Feser, J.; Wilson, P.C.; Solórzano, A.; Van der Wielen, M.; Walter, E.B. Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: Interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect. Dis. 2020, 20, 80–91. [Google Scholar] [CrossRef] [Green Version]
- Nachbagauer, R.; Feser, J.; Naficy, A.; Bernstein, D.I.; Guptill, J.; Walter, E.B.; Berlanda-Scorza, F.; Stadlbauer, D.; Wilson, P.C.; Aydillo, T. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2020, 27, 106–114. [Google Scholar] [CrossRef]
- Nachbagauer, R.; Kinzler, D.; Choi, A.; Hirsh, A.; Beaulieu, E.; Lecrenier, N.; Innis, B.L.; Palese, P.; Mallett, C.P.; Krammer, F. A chimeric haemagglutinin-based influenza split virion vaccine adjuvanted with AS03 induces protective stalk-reactive antibodies in mice. NPJ Vaccines 2016, 1, 16015. [Google Scholar] [CrossRef]
- Zhang, Y.; Aevermann, B.D.; Anderson, T.K.; Burke, D.F.; Dauphin, G.; Gu, Z.; He, S.; Kumar, S.; Larsen, C.N.; Lee, A.J. Influenza Research Database: An integrated bioinformatics resource for influenza virus research. Nucleic Acids Res. 2017, 45, D466–D474. [Google Scholar] [CrossRef] [Green Version]
- Hall, T. BioEdit v7. 2.5: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 4, 95–98. [Google Scholar]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.V.; Zmasek, C.M. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinform. 2009, 10, 356. [Google Scholar] [CrossRef] [Green Version]
- Krammer, F.; Palese, P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 2015, 14, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Nachbagauer, R.; Liu, W.-C.; Choi, A.; Wohlbold, T.J.; Atlas, T.; Rajendran, M.; Solórzano, A.; Berlanda-Scorza, F.; García-Sastre, A.; Palese, P.; et al. A universal influenza virus vaccine candidate confers protection against pandemic H1N1 infection in preclinical ferret studies. NPJ Vaccines 2017, 2, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cock, P.J.; Chilton, J.M.; Grüning, B.; Johnson, J.E.; Soranzo, N. NCBI BLAST+ integrated into Galaxy. Gigascience 2015, 4, s13742-015. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Zhang, C.; Bell, E.W.; Zhang, Y. I-TASSER gateway: A protein structure and function prediction server powered by XSEDE. Future Gener. Comput. Syst. 2019, 99, 73–85. [Google Scholar] [CrossRef]
- Schrodinger, L. The PyMOL Molecular Graphics System. Version 2010. Available online: https://pymol.org/2/%EF%BC%8C%E8%BF%99%E4%B8%AA%E6%98%AF%E5%AE%98%E7%BD%91 (accessed on 12 July 2021).
- Dhanda, S.K.; Mahajan, S.; Paul, S.; Yan, Z.; Kim, H.; Jespersen, M.C.; Jurtz, V.; Andreatta, M.; Greenbaum, J.A.; Marcatili, P. IEDB-AR: Immune epitope database—Analysis resource in 2019. Nucleic Acids Res. 2019, 47, W502–W506. [Google Scholar] [CrossRef] [Green Version]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Strelkov, S.V.; Mesyanzhinov, V.V.; Rossmann, M.G. Structure of bacteriophage T4 fibritin: A segmented coiled coil and the role of the C-terminal domain. Structure 1997, 5, 789–798. [Google Scholar] [CrossRef] [Green Version]
- Whiteley, E.M.; Hsu, T.A.; Betenbaugh, M.J. Modeling assembly, aggregation, and chaperoning of immunoglobulin G production in insect cells. Biotechnol. Bioeng. 1997, 56, 106–116. [Google Scholar] [CrossRef]
- Hsu, T.A.; Betenbaugh, M.J. Coexpression of molecular chaperone BiP improves immunoglobulin solubility and IgG secretion from Trichoplusia ni insect cells. Biotechnol. Prog. 1997, 13, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Ailor, E.; Betenbaugh, M.J. Modifying secretion and post-translational processing in insect cells. Curr. Opin. Biotechnol. 1999, 10, 142–145. [Google Scholar] [CrossRef]
- Ailor, E.; Betenbaugh, M.J. Overexpression of a cytosolic chaperone to improve solubility and secretion of a recombinant IgG protein in insect cells. Biotechnol. Bioeng. 1998, 58, 196–203. [Google Scholar] [CrossRef]
- WHO Manual on Animal Influenza Diagnosis and Surveillance. Available online: https://www.who.int/csr/resources/publications/influenza/whocdscsrncs20025rev.pdf (accessed on 12 July 2021).
- Gautam, A.; Park, B.K.; Kim, T.H.; Akauliya, M.; Kim, D.; Maharjan, S.; Park, J.; Kim, J.; Lee, H.; Park, M.-S.; et al. Peritoneal Cells Mediate Immune Responses and Cross-Protection Against Influenza A Virus. Front. Immunol. 2019, 10, 1160. [Google Scholar] [CrossRef] [Green Version]
- Shu, B.; Wu, K.-H.; Emery, S.; Villanueva, J.; Johnson, R.; Guthrie, E.; Berman, L.; Warnes, C.; Barnes, N.; Klimov, A. Design and performance of the CDC real-time reverse transcriptase PCR swine flu panel for detection of 2009 A (H1N1) pandemic influenza virus. J. Clin. Microbiol. 2011, 49, 2614–2619. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Lipman, D.J. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. USA 1990, 87, 5509–5513. [Google Scholar] [CrossRef] [Green Version]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Budd, A.; Blanton, L.; Grohskopf, L.; Campbell, A.; Dugan, V.; Wentworth, D.E.; Brammer, L. Manual for the Surveillance of Vaccine-Preventable Diseases; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2017.
- Marchi, J.; Lässig, M.; Mora, T. Multi-Lineage Evolution in Viral Populations Driven by Host Immune Systems. Pathogens 2019, 8, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 47–60. [Google Scholar] [CrossRef]
- Valkenburg, S.A.; Leung, N.H.L.; Bull, M.B.; Yan, L.-m.; Li, A.P.Y.; Poon, L.L.M.; Cowling, B.J. The Hurdles From Bench to Bedside in the Realization and Implementation of a Universal Influenza Vaccine. Front. Immunol. 2018, 9, 1479. [Google Scholar] [CrossRef] [Green Version]
- Erbelding, E.J.; Post, D.J.; Stemmy, E.J.; Roberts, P.C.; Augustine, A.D.; Ferguson, S.; Paules, C.I.; Graham, B.S.; Fauci, A.S. A universal influenza vaccine: The strategic plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018, 218, 347–354. [Google Scholar] [CrossRef]
- Paules, C.I.; Marston, H.D.; Eisinger, R.W.; Baltimore, D.; Fauci, A.S. The pathway to a universal influenza vaccine. Immunity 2017, 47, 599–603. [Google Scholar] [CrossRef] [Green Version]
- Carter, D.M.; Darby, C.A.; Lefoley, B.C.; Crevar, C.J.; Alefantis, T.; Oomen, R.; Anderson, S.F.; Strugnell, T.; Cortés-Garcia, G.; Vogel, T.U. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J. Virol. 2016, 90, 4720–4734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crevar, C.J.; Carter, D.M.; Lee, K.Y.; Ross, T.M. Cocktail of H5N1 COBRA HA vaccines elicit protective antibodies against H5N1 viruses from multiple clades. Hum. Vaccines Immunother. 2015, 11, 572–583. [Google Scholar] [CrossRef] [Green Version]
- Allen, J.D.; Jang, H.; DiNapoli, J.; Kleanthous, H.; Ross, T.M. Elicitation of protective antibodies against 20 years of future H3N2 cocirculating influenza virus variants in ferrets preimmune to historical H3N2 influenza viruses. J. Virol. 2019, 93, e00946-18. [Google Scholar] [CrossRef] [Green Version]
- Steel, J.; Lowen, A.C.; Wang, T.T.; Yondola, M.; Gao, Q.; Haye, K.; García-Sastre, A.; Palese, P. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010, 1, e00018-10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotey, E.; Lukosaityte, D.; Quaye, O.; Ampofo, W.; Awandare, G.; Iqbal, M. Current and novel approaches in influenza management. Vaccines 2019, 7, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broecker, F.; Liu, S.T.; Suntronwong, N.; Sun, W.; Bailey, M.J.; Nachbagauer, R.; Krammer, F.; Palese, P. A mosaic hemagglutinin-based influenza virus vaccine candidate protects mice from challenge with divergent H3N2 strains. NPJ Vaccines 2019, 4, 31. [Google Scholar] [CrossRef] [PubMed]



| HAs | Predictions | |
|---|---|---|
| Structural | Functional | |
| TM-Score | SIA-Binding C-Score | |
| cHA-C | 0.831 | 0.77 |
| cHA-E | 0.836 | 0.7 |
| A/Michigan/45/2015 (H1N1) pdm09-like virus (CVV-M) | 1 | 1 |
| cHA-C | cHA-E | CVV-M | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | Peptide | Length | Start | End | Peptide | Length | Start | End | Peptide | Length |
| 146 | 155 | SSGVSAACSY | 10 | 69 | 76 | LGDCCTAG | 8 | 93 | 98 | WSYIVE | 6 |
| 267 | 277 | APEYAFALVRG | 11 | 139 | 145 | SWPVHYA | 7 | 150 | 156 | TAACPHA | 7 |
| 347 | 352 | FGAIAG | 6 | 175 | 184 | YPTLAASYAN | 10 | 264 | 270 | NLVVPRY | 7 |
| 462 | 469 | LYEKVKLQ * | 8 | 247 | 253 | YWTLLRP | 7 | 289 | 295 | VHDCNTT | 7 |
| 287 | 297 | AVMCECEAKCQ | 11 | ||||||||
| 460 | 469 | KKLYEKVKAQ * | 10 | ||||||||
| Expressed HA/Virus | HA Titre | ||
|---|---|---|---|
| CVV-M | <2 | <2 | <2 |
| cHA-C | <2 | <2 | <2 |
| cHA-E | <2 | <2 | <2 |
| Lab H5N2 (A/pheasant/New Jersey/1355/1998) virus isolate [PNJ] | 128 | 128 | 128 |
| PBS | <2 | <2 | <2 |
| Serum | Baseline | 3D | ||||||
|---|---|---|---|---|---|---|---|---|
| Virus | cHA-C | cHA-E | CVV-M | U | cHA-C | cHA-E | CVV-M | U |
| X-275 | <20 | <20 | <20 | <20 | 320 | 1280 | 640 | <20 |
| A/England/195/2009 (H1N1) pdm09 | <20 | <20 | <20 | <20 | 320 | 320 | 320 | <20 |
| PNJ | <20 | <20 | <20 | <20 | 160 | 160 | 160 | <20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotey, E.N.; Ampofo, W.K.; Daines, R.; Sadeyen, J.-R.; Iqbal, M.; Quaye, O. Immune Response in Mice Immunized with Chimeric H1 Antigens. Vaccines 2021, 9, 1182. https://doi.org/10.3390/vaccines9101182
Kotey EN, Ampofo WK, Daines R, Sadeyen J-R, Iqbal M, Quaye O. Immune Response in Mice Immunized with Chimeric H1 Antigens. Vaccines. 2021; 9(10):1182. https://doi.org/10.3390/vaccines9101182
Chicago/Turabian StyleKotey, Erasmus Nikoi, William Kwabena Ampofo, Rebecca Daines, Jean-Remy Sadeyen, Munir Iqbal, and Osbourne Quaye. 2021. "Immune Response in Mice Immunized with Chimeric H1 Antigens" Vaccines 9, no. 10: 1182. https://doi.org/10.3390/vaccines9101182
APA StyleKotey, E. N., Ampofo, W. K., Daines, R., Sadeyen, J.-R., Iqbal, M., & Quaye, O. (2021). Immune Response in Mice Immunized with Chimeric H1 Antigens. Vaccines, 9(10), 1182. https://doi.org/10.3390/vaccines9101182







