Identification of Mycobacterium tuberculosis Antigens with Vaccine Potential Using a Machine Learning-Based Reverse Vaccinology Approach
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Novel Protective Antigens Predicted by Vaxign-ML
3.2. Antigens Belonging to Protein Families of Previously Established MTB PAgs
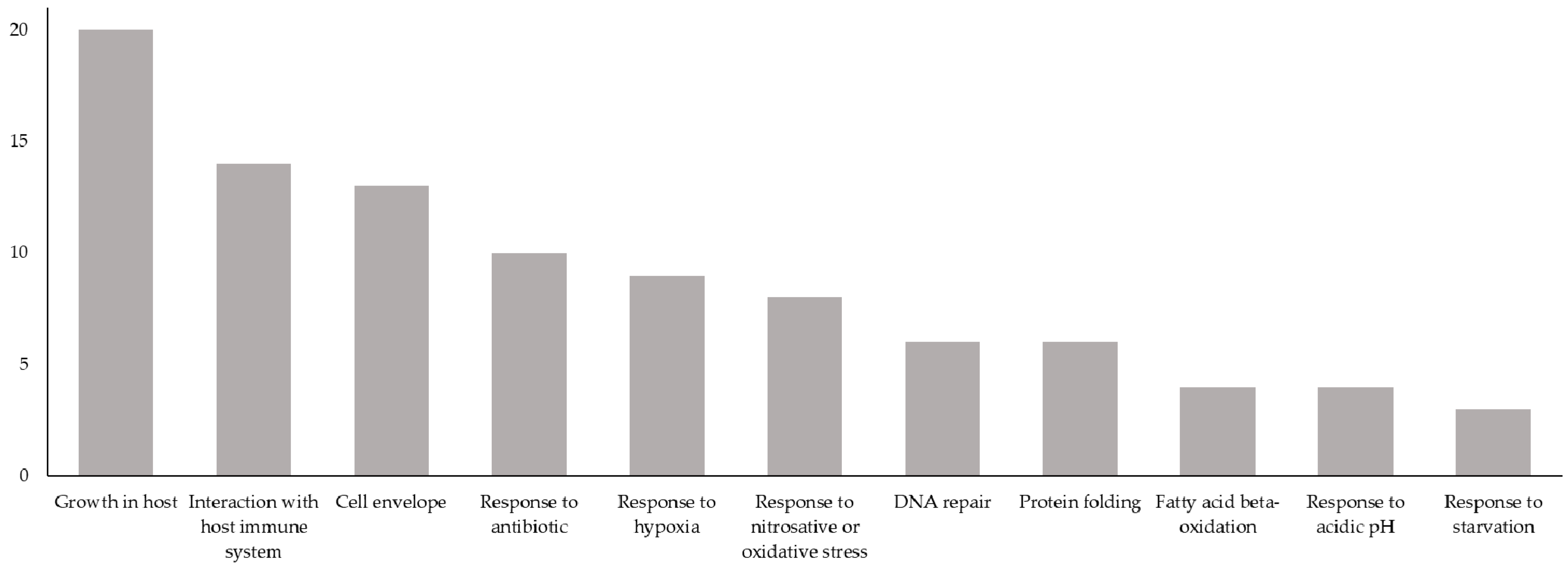
3.3. Antigens Having Biological Processes Associated with MTB Virulence or LTBI
3.4. Antigens with the Greatest Number of Promiscuous MHC-I and MHC-II Epitopes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Global Tuberculosis Report 2020; World Health Organization, 2020. Available online: https://apps.who.int/iris/bitstream/handle/10665/336069/9789240013131-eng.pdf (accessed on 1 February 2021).
- Daniel, T.M.; Bates, J.H.; Downes, K.A. History of Tuberculosis. In Tuberculosis; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1994; pp. 13–24. [Google Scholar] [CrossRef]
- Seung, K.J.; Keshavjee, S.; Rich, M.L. Multidrug-Resistant Tuberculosis and Extensively Drug-Resistant Tuberculosis. Cold Spring Harb. Perspect. Med. 2015, 5, a017863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, L.C.K.; Noursadeghi, M. Pathogenesis of HIV-1 and Mycobacterium tuberculosis co-infection. Nat. Rev. Genet. 2017, 16, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Uplekar, M.; Weil, D.; Lonnroth, K.; Jaramillo, E.; Lienhardt, C.; Dias, H.M.; Falzon, D.; Floyd, K.; Gargioni, G.; Getahun, H.; et al. WHO’s new End TB Strategy. Lancet 2015, 385, 1799–1801. [Google Scholar] [CrossRef]
- The End TB Strategy. World Health Organization, 2015. Available online: https://www.who.int/tb/End_TB_brochure.pdf (accessed on 7 February 2021).
- The Sustainable Development Goals Report 2020; United Nations, 2020. Available online: https://unstats.un.org/sdgs/report/2020/The-Sustainable-Development-Goals-Report-2020.pdf (accessed on 9 February 2021).
- Hmama, Z.; Peña-Díaz, S.; Joseph, S.; Av-Gay, Y. Immunoevasion and immunosuppression of the macrophage byMycobacterium tuberculosis. Immunol. Rev. 2015, 264, 220–232. [Google Scholar] [CrossRef]
- Ramakrishnan, L. Revisiting the role of the granuloma in tuberculosis. Nat. Rev. Immunol. 2012, 12, 352–366. [Google Scholar] [CrossRef]
- Cadena, A.M.; Fortune, S.M.; Flynn, J.L. Heterogeneity in tuberculosis. Nat. Rev. Immunol. 2017, 17, 691–702. [Google Scholar] [CrossRef]
- Flynn, J.L.; Chan, J. Tuberculosis: Latency and Reactivation. Infect. Immun. 2001, 69, 4195–4201. [Google Scholar] [CrossRef] [Green Version]
- Andersen, P.; Scriba, T.J. Moving tuberculosis vaccines from theory to practice. Nat. Rev. Immunol. 2019, 19, 550–562. [Google Scholar] [CrossRef]
- Ong, E.; He, Y.; Yang, Z. Epitope promiscuity and population coverage of Mycobacterium tuberculosis protein antigens in current subunit vaccines under development. Infect. Genet. Evol. 2020, 80, 104186. [Google Scholar] [CrossRef]
- McShane, H.; Pathan, A.A.; Sander, C.R.; Goonetilleke, N.P.; Fletcher, H.A.; Hill, A.V. Boosting BCG with MVA85A: The first candidate subunit vaccine for tuberculosis in clinical trials. Tuberculosis 2005, 85, 47–52. [Google Scholar] [CrossRef]
- Tameris, M.D.; Hatherill, M.; Landry, B.S.; Scriba, T.; Snowden, M.A.; Lockhart, S.; Shea, J.E.; McClain, J.B.; Hussey, G.D.; Hanekom, W.A.; et al. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: A randomised, placebo-controlled phase 2b trial. Lancet 2013, 381, 1021–1028. [Google Scholar] [CrossRef] [Green Version]
- Van Der Meeren, O.; Hatherill, M.; Nduba, V.; Wilkinson, R.; Muyoyeta, M.; Van Brakel, E.; Ayles, H.M.; Henostroza, G.; Thienemann, F.; Scriba, T.; et al. Phase 2b Controlled Trial of M72/AS01EVaccine to Prevent Tuberculosis. N. Engl. J. Med. 2018, 379, 1621–1634. [Google Scholar] [CrossRef]
- Aagaard, C.; Hoang, T.; Dietrich, J.; Cardona, P.-J.; Izzo, A.; Dolganov, G.; Schoolnik, G.K.; Cassidy, J.P.; Billeskov, R.; Andersen, P. A multistage tuberculosis vaccine that confers efficient protection before and after exposure. Nat. Med. 2011, 17, 189–194. [Google Scholar] [CrossRef]
- Luabeya, A.K.K.; Kagina, B.; Tameris, M.D.; Geldenhuys, H.; Hoff, S.T.; Shi, Z.; Kromann, I.; Hatherill, M.; Mahomed, H.; Hanekom, W.A.; et al. First-in-human trial of the post-exposure tuberculosis vaccine H56:IC31 in Mycobacterium tuberculosis infected and non-infected healthy adults. Vaccine 2015, 33, 4130–4140. [Google Scholar] [CrossRef]
- Bertholet, S.; Ireton, G.C.; Ordway, D.; Windish, H.P.; Pine, S.O.; Kahn, M.; Phan, T.; Orme, I.M.; Vedvick, T.S.; Baldwin, S.L.; et al. A Defined Tuberculosis Vaccine Candidate Boosts BCG and Protects against Multidrug-Resistant Mycobacterium tuberculosis. Sci. Transl. Med. 2010, 2, 53ra74. [Google Scholar] [CrossRef] [Green Version]
- Coler, R.N.; Day, T.A.; Ellis, R.; Piazza, F.M.; Beckmann, A.M.; Vergara, J.; Rolf, T.; Lu, L.; Alter, G.; Hokey, D.; et al. The TLR-4 agonist adjuvant, GLA-SE, improves magnitude and quality of immune responses elicited by the ID93 tuberculosis vaccine: First-in-human trial. NPJ Vaccines 2018, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H.E.; Dockrell, H.M.; Drager, N.; Ho, M.M.; McShane, H.; Neyrolles, O.; Ottenhoff, T.H.M.; Brij, B.; Roordink, D.; Spertini, F.; et al. TBVAC2020: Advancing Tuberculosis Vaccines from Discovery to Clinical Development. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rappuoli, R. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine 2001, 19, 2688–2691. [Google Scholar] [CrossRef]
- Ong, E.; Wang, H.; Wong, M.U.; Seetharaman, M.; Valdez, N.; He, Y. Vaxign-ML: Supervised machine learning reverse vaccinology model for improved prediction of bacterial protective antigens. Bioinformatics 2020, 36, 3185–3191. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.; Eiglmeier, K.; Gas, S.; Barry, C.E.; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef]
- Ong, E.; Cooke, M.F.; Huffman, A.; Xiang, Z.; Wong, M.U.; Wang, H.; Seetharaman, M.; Valdez, N.; He, Y. Vaxign2: The second generation of the first Web-based vaccine design program using reverse vaccinology and machine learning. Nucleic Acids Res. 2021, 49, W671–W678. [Google Scholar] [CrossRef]
- Yang, B.; Sayers, S.; Xiang, Z.; He, Y. Protegen: A web-based protective antigen database and analysis system. Nucleic Acids Res. 2010, 39, D1073–D1078. [Google Scholar] [CrossRef] [Green Version]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489. [Google Scholar] [CrossRef]
- Jarlier, V.; Nikaido, H. Mycobacterial cell wall: Structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 1994, 123, 11–18. [Google Scholar] [CrossRef]
- Brennan, P.J.; Nikaido, H. The Envelope of Mycobacteria. Annu. Rev. Biochem. 1995, 64, 29–63. [Google Scholar] [CrossRef]
- Dos Vultos, T.; Mestre, O.; Tonjum, T.; Gicquel, B. DNA repair inMycobacterium tuberculosisrevisited. FEMS Microbiol. Rev. 2009, 33, 471–487. [Google Scholar] [CrossRef] [Green Version]
- Bishai, W.R. Lipid lunch for persistent pathogen. Nature 2000, 406, 683–684. [Google Scholar] [CrossRef]
- Forrellad, M.A.; Klepp, L.I.; Gioffré, A.; García, J.S.Y.; Morbidoni, H.R.; Santangelo, M.D.L.P.; Cataldi, A.A.; Bigi, F. Virulence factors of theMycobacterium tuberculosiscomplex. Virulence 2013, 4, 3–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qamra, R.; Mande, S.C.; Coates, A.R.; Henderson, B. The unusual chaperonins of Mycobacterium tuberculosis. Tuberculosis 2005, 85, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L. Antibiotic resistance mechanisms in M. tuberculosis: An update. Arch. Toxicol. 2016, 90, 1585–1604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huynh, K.K.; Grinstein, S. Regulation of Vacuolar pH and Its Modulation by Some Microbial Species. Microbiol. Mol. Biol. Rev. 2007, 71, 452–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rustad, T.R.; Sherrid, A.M.; Minch, K.J.; Sherman, D.R. Hypoxia: A window intoMycobacterium tuberculosislatency. Cell. Microbiol. 2009, 11, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Betts, J.C.; Lukey, P.T.; Robb, L.C.; McAdam, R.A.; Duncan, K. Evaluation of a nutrient starvation model ofMycobacterium tuberculosispersistence by gene and protein expression profiling. Mol. Microbiol. 2002, 43, 717–731. [Google Scholar] [CrossRef] [PubMed]
- Sturniolo, T.; Bono, E.; Ding, J.; Raddrizzani, L.; Tuereci, O.; Sahin, U.; Braxenthaler, M.; Gallazzi, F.; Protti, M.P.; Sinigaglia, F.; et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat. Biotechnol. 1999, 17, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Dhanda, S.K.; Mahajan, S.; Paul, S.; Yan, Z.; Kim, H.; Jespersen, M.C.; Jurtz, V.; Andreatta, M.; Greenbaum, J.A.; Marcatili, P.; et al. IEDB-AR: Immune epitope database—analysis resource in 2019. Nucleic Acids Res. 2019, 47, W502–W506. [Google Scholar] [CrossRef] [Green Version]
- Weiskopf, D.; Angelo, M.A.; de Azeredo, E.L.; Sidney, J.; Greenbaum, J.A.; Fernando, A.N.; Broadwater, A.; Kolla, R.V.; De Silva, A.D.; de Silva, A.M.; et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA 2013, 110, E2046–E2053. [Google Scholar] [CrossRef] [Green Version]
- Moutaftsi, M.; Peters, B.; Pasquetto, V.; Tscharke, D.; Sidney, J.; Bui, H.-H.; Grey, H.M.; Sette, A. A consensus epitope prediction approach identifies the breadth of murine TCD8+-cell responses to vaccinia virus. Nat. Biotechnol. 2006, 24, 817–819. [Google Scholar] [CrossRef]
- Kotturi, M.F.; Peters, B.; Buendia-Laysa, F.; Sidney, J.; Oseroff, C.; Botten, J.; Grey, H.; Buchmeier, M.J.; Sette, A. The CD8 + T-Cell Response to Lymphocytic Choriomeningitis Virus Involves the L Antigen: Uncovering New Tricks for an Old Virus. J. Virol. 2007, 81, 4928–4940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Sidney, J.; Kim, Y.; Sette, A.; Lund, O.; Nielsen, M.; Peters, B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinform. 2010, 11, 568. [Google Scholar] [CrossRef] [Green Version]
- Jensen, K.K.; Andreatta, M.; Marcatili, P.; Buus, S.; Greenbaum, J.A.; Yan, Z.; Sette, A.; Peters, B.; Nielsen, M. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 2018, 154, 394–406. [Google Scholar] [CrossRef]
- Nielsen, M.; Lundegaard, C.; Lund, O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinform. 2007, 8, 238. [Google Scholar] [CrossRef] [Green Version]
- Sidney, J.; Assarsson, E.; Moore, C.; Ngo, S.; Pinilla, C.; Sette, A.; Peters, B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Greenbaum, J.; Sidney, J.; Chung, J.; Brander, C.; Peters, B.; Sette, A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics 2011, 63, 325–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Southwood, S.; Sidney, J.; Kondo, A.; Del Guercio, M.F.; Appella, E.; Hoffman, S.; Kubo, R.T.; Chesnut, R.W.; Grey, H.M.; Sette, A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 1998, 160. [Google Scholar]
- Singh, S.K.; Tripathi, D.K.; Singh, P.K.; Sharma, S.; Srivastava, K.K. Protective and survival efficacies of Rv0160c protein in murine model of Mycobacterium tuberculosis. Appl. Microbiol. Biotechnol. 2012, 97, 5825–5837. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Singh, L.K.; Kumari, S.; Hakiem, O.R.; Batra, J.K. ClpB is an essential stress regulator of Mycobacterium tuberculosis and endows survival advantage to dormant bacilli. Int. J. Med Microbiol. 2020, 310, 151402. [Google Scholar] [CrossRef] [PubMed]
- Behr, M.A.; Sherman, D.R. Mycobacterial virulence and specialized secretion: Same story, different ending. Nat. Med. 2007, 13, 286–287. [Google Scholar] [CrossRef] [PubMed]
- Be, N.A.; Lamichhane, G.; Grosset, J.; Tyagi, S.; Cheng, Q.; Kim, K.S.; Bishai, W.R.; Jain, S.K. Murine Model to Study the Invasion and Survival of Mycobacterium tuberculosis in the Central Nervous System. J. Infect. Dis. 2008, 198, 1520–1528. [Google Scholar] [CrossRef] [Green Version]


| Protein Family | Protein | Tuberculist ID |
|---|---|---|
| mycobacterial A85 antigen | FbpC (Ag85C) | Rv0129c |
| mycobacterial PE | PE_PGRS11 | Rv0754 |
| PE4 | Rv0160c | |
| PE26 | Rv2519 | |
| mycobacterial PPE | Hypothetical protein Rv3822 | Rv3822 |
| PPE28 | Rv1800 | |
| PPE8 | Rv0355c | |
| PPE12 | Rv0755c | |
| PPE30 | Rv1802 | |
| peptidase S1C | HtrA | Rv1223 |
| PepD | Rv0983 | |
| PstS | PstS2 | Rv0932c |
| RsiV | Hypothetical protein Rv3036c | Rv3036c |
| WXG100 | EsxI | Rv1037c |
| Biological Process Category | Proteins (Tuberculist IDs) |
|---|---|
| Cell envelope biogenesis and maintenance | PonA2 (Rv3682), PonA1 (Rv0050), FadD15 (Rv2187), LdtB (Rv2518c), PbpB (Rv2163c), FadD30 (Rv0404), FbpC (Ag85C) (Rv0129c), AccD4 (Rv3799c), FadD32 (Rv3801c), hypothetical protein Rv3811 (Rv3811), LprQ (Rv0483), PbpA (Rv0016c), FadD19 (Rv3515c) |
| DNA Repair | RecA (Rv2737c), HtpG (Rv2299c), UvrA (Rv1638), LigD (Rv0938), RecG (Rv2973c), UvrB (Rv1633) |
| Interaction with host immune system | FadE5 (Rv0244c), Mce1A (Rv0169), probable aldehyde dehydrogenase (Rv0458), CaeA (Rv2224c), LprA (Rv1270c), FadD30 (Rv0404), EccCa1 (Rv3870), Icl1 (Rv0467), PknH (Rv1266c), MmpL12 (Rv1522c), UvrB (Rv1633), EccB1 (Rv3869), halimadienyl diphosphate synthase (Rv3377c), FadD19 (Rv3515c) |
| Fatty acid beta-oxidation | FadB (Rv0860), FadA3 (Rv1074c), Ltp1 (Rv2790c), probable nonspecific lipid-transfer protein (Rv1627c) |
| Growth in host | FadD13 (Rv3089), Mce2C (Rv0591), Mce4A (Rv3499c), Mce1A (Rv0169), Mce1F (Rv0174), Tgs4 (Rv3088), Mce3C (Rv1968), Mce1C (Rv0171), Mce3D (Rv1969), Mce3A (Rv1966), EccCa (Rv3870), Mce4C (Rv3497c), Mce2F (Rv0594), Mce4D (Rv3496c), Mce2A (Rv0589), EccA1 (Rv3868), Mce2D (Rv0592), Mce1D (Rv0172), FadA (Rv0243), Mce4F (Rv3494c) |
| Protein folding | GroEL2 (Rv0440), Mpa (Rv2115c), GroEL1 (Rv3417c), ClpX (Rv2457c), HtpG (Rv2299c), ClpB (Rv0384c) |
| Response to antibiotic | PonA2 (Rv3682), GyrB (Rv0005), RecA (Rv2737c), RpoB (Rv0667), PonA1 (Rv0050), FbpC (Ag85C) (Rv0129c), possible penicillin-binding lipoprotein (Rv2864c), PepD (Rv0983), IleS (Rv1536), LprG (Rv1411c) |
| Response to acidic pH | FadD13 (Rv3089), Tgs4 (Rv3088), Icl1 (Rv0467), AccD4 (Rv3799c) |
| Response to hypoxia | GroEL2 (Rv0440), PonA1 (Rv0050), Tgs4 (Rv3088), Icl1 (Rv0467), AccD4 (Rv3799c), PE_PGRS11 (Rv0754), Tuf (Rv0685), SdhA (Rv3318), probable succinate dehydrogenase (Rv0248c) |
| Response to nitrosative or oxidative stress | Mpa (Rv2115c), FtsH (Rv3610c), HtpG (Rv2299c), Tgs4 (Rv3088), Mpt53 (Rv2878c), AccD4 (Rv3799c), UvrB (Rv1633), CysN (Rv1286) |
| Response to starvation | halimadienyl diphosphate synthase (Rv3377c), PknD (Rv0931c), CysN (Rv1286) |
| Protein | Tuberculist ID | GO Biological Process | Subcellular Location | MHC-I Promiscuous Epitopes | MHC-II Promiscuous Epitopes |
|---|---|---|---|---|---|
| Having highest numbers of both MHC-I and MHC-II promiscuous epitopes | |||||
| PPE8 | Rv0355c | not available | not available | 104 | 194 |
| IleS | Rv1536 | isoleucyl-tRNA aminoacylation, response to antibiotics | cytoplasm | 92 | 156 |
| MmpL12 | Rv1522c | response to host immune response | cell membrane, multi-pass membrane protein | 86 | 263 |
| UvrA | Rv1638 | cellular response to DNA damage stimulus, negative regulation of strand invasion, nucleotide-excision repair, SOS response | cytoplasm | 73 | 116 |
| RpoB | Rv0667 | response to antibiotic, DNA-templated transcription | cell wall, cytosol, plasma membrane | 72 | 109 |
| ClpB | Rv0384c | protein refolding, response to heat | cytoplasm | 62 | 104 |
| Having highest number of MHC-I promiscuous epitopes only | |||||
| PonA2 | Rv3682 | peptidoglycan biosynthetic process, response to antibiotic | not available | 60 | 88 |
| FadE5 | Rv0244c | response to host immune response | extracellular region, plasma membrane | 57 | 86 |
| Mce2D | Rv0592 | growth of symbiont in host, growth of symbiont in host vacuole | cell wall | 57 | 84 |
| FadD30 | Rv0404 | Actinobacterium-type cell wall biogenesis, fatty acid biosynthetic process, induction by symbiont of host immune response, lipid biosynthetic process | not available | 56 | 85 |
| EccCa1 | Rv3870 | evasion of host immune response, growth of symbiont in host, pathogenesis, protein secretion by the type VII secretion system | cell inner membrane, multi-pass membrane protein | 56 | 79 |
| Having highest number of MHC-II promiscuous epitopes only | |||||
| PPE28 | Rv1800 | not available | not available | 54 | 176 |
| PE4 | Rv0160c | not available | not available | 48 | 100 |
| FadD15 | Rv2187 | Actinobacterium-type cell wall biogenesis, fatty acid biosynthetic process, lipid biosynthetic process, long-chain fatty acid metabolic process | cell wall, plasma membrane | 48 | 113 |
| PknD | Rv0931c | cellular response to phosphate starvation, negative regulation of catalytic activity, negative regulation of fatty acid biosynthetic process, negative regulation of protein binding, pathogenesis, positive regulation of catalytic activity | cell membrane, single-pass membrane protein | 42 | 105 |
| Mce2A | Rv0589 | growth of symbiont in host, growth of symbiont in host vacuole | integral component of membrane | 36 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teahan, B.; Ong, E.; Yang, Z. Identification of Mycobacterium tuberculosis Antigens with Vaccine Potential Using a Machine Learning-Based Reverse Vaccinology Approach. Vaccines 2021, 9, 1098. https://doi.org/10.3390/vaccines9101098
Teahan B, Ong E, Yang Z. Identification of Mycobacterium tuberculosis Antigens with Vaccine Potential Using a Machine Learning-Based Reverse Vaccinology Approach. Vaccines. 2021; 9(10):1098. https://doi.org/10.3390/vaccines9101098
Chicago/Turabian StyleTeahan, Blaine, Edison Ong, and Zhenhua Yang. 2021. "Identification of Mycobacterium tuberculosis Antigens with Vaccine Potential Using a Machine Learning-Based Reverse Vaccinology Approach" Vaccines 9, no. 10: 1098. https://doi.org/10.3390/vaccines9101098
APA StyleTeahan, B., Ong, E., & Yang, Z. (2021). Identification of Mycobacterium tuberculosis Antigens with Vaccine Potential Using a Machine Learning-Based Reverse Vaccinology Approach. Vaccines, 9(10), 1098. https://doi.org/10.3390/vaccines9101098






