Specific CD8+ TCR Repertoire Recognizing Conserved Antigens of SARS-CoV-2 in Unexposed Population: A Prerequisite for Broad-Spectrum CD8+ T Cell Immunity
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Subjects
2.2. Epitope Prediction and Peptides Synthesis
2.3. Peripheral Blood Mononuclear Cells (PBMCs) Isolation
2.4. IFN-γ Enzyme-Linked Immunospot (ELISPOT) Assay
2.5. Flow Cytometer Analysis and Sorting
2.6. TCR β Repertoire Sequencing
2.7. Statistics
3. Results
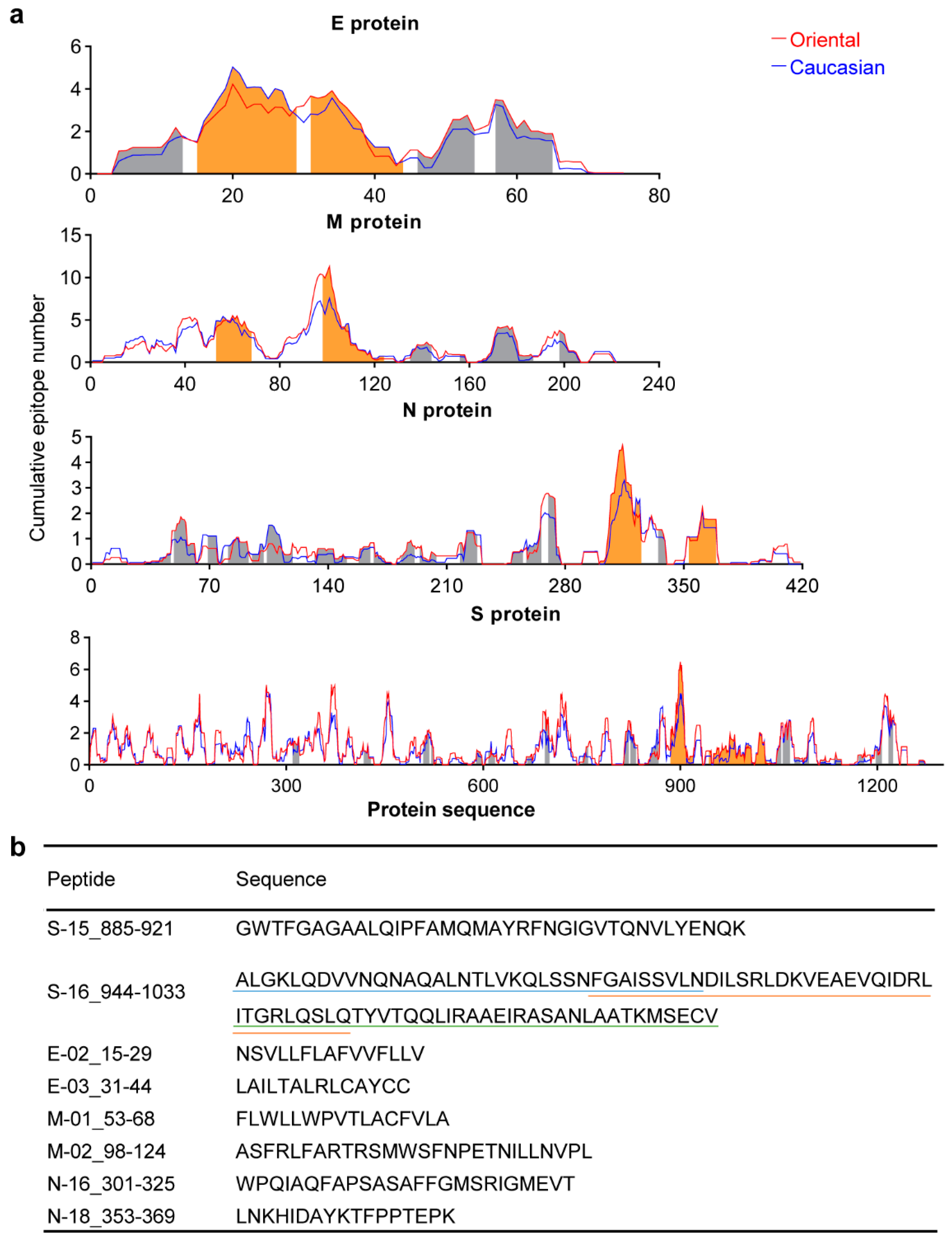
3.1. Conserved Antigens from the Structural Proteins
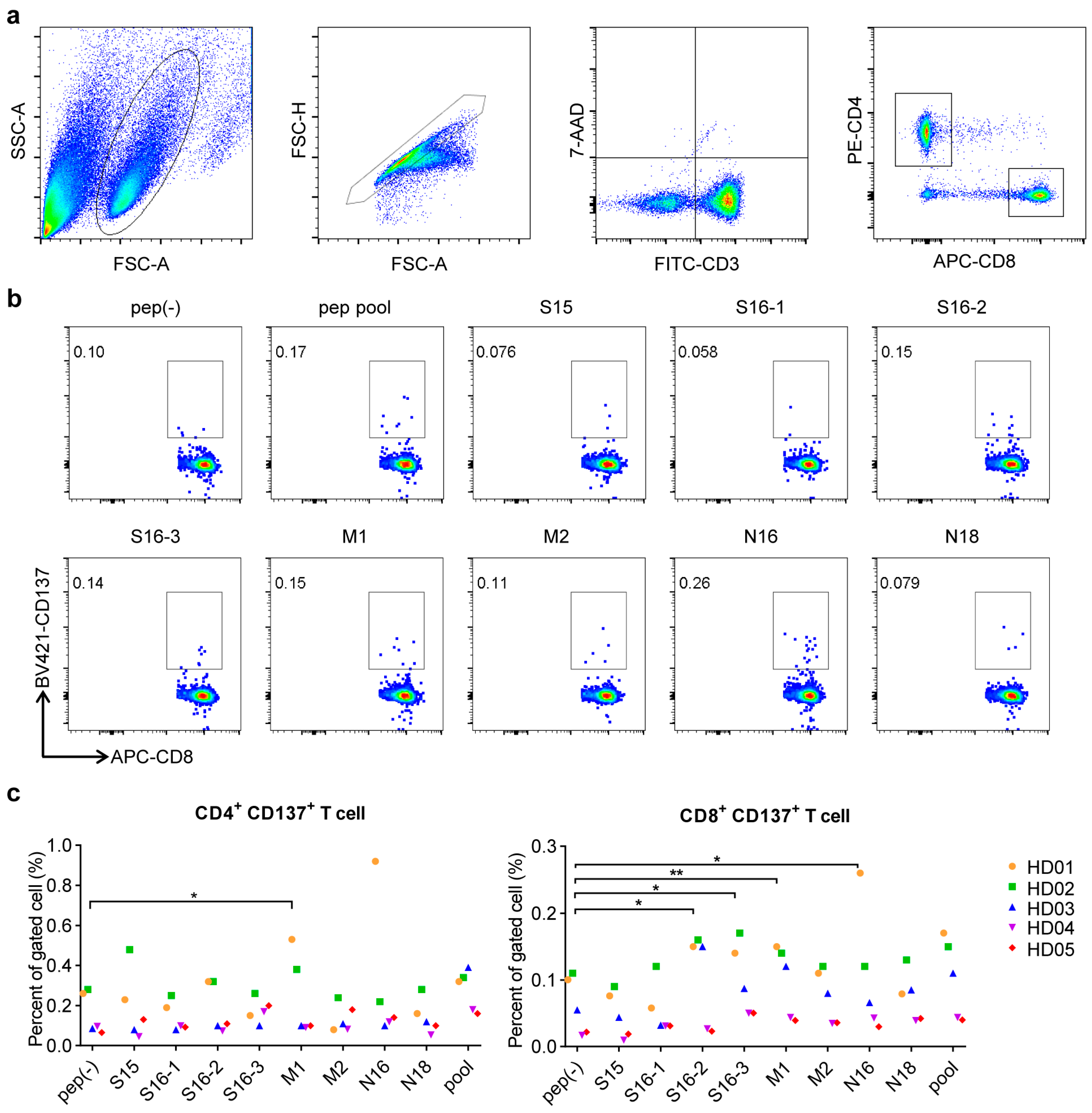
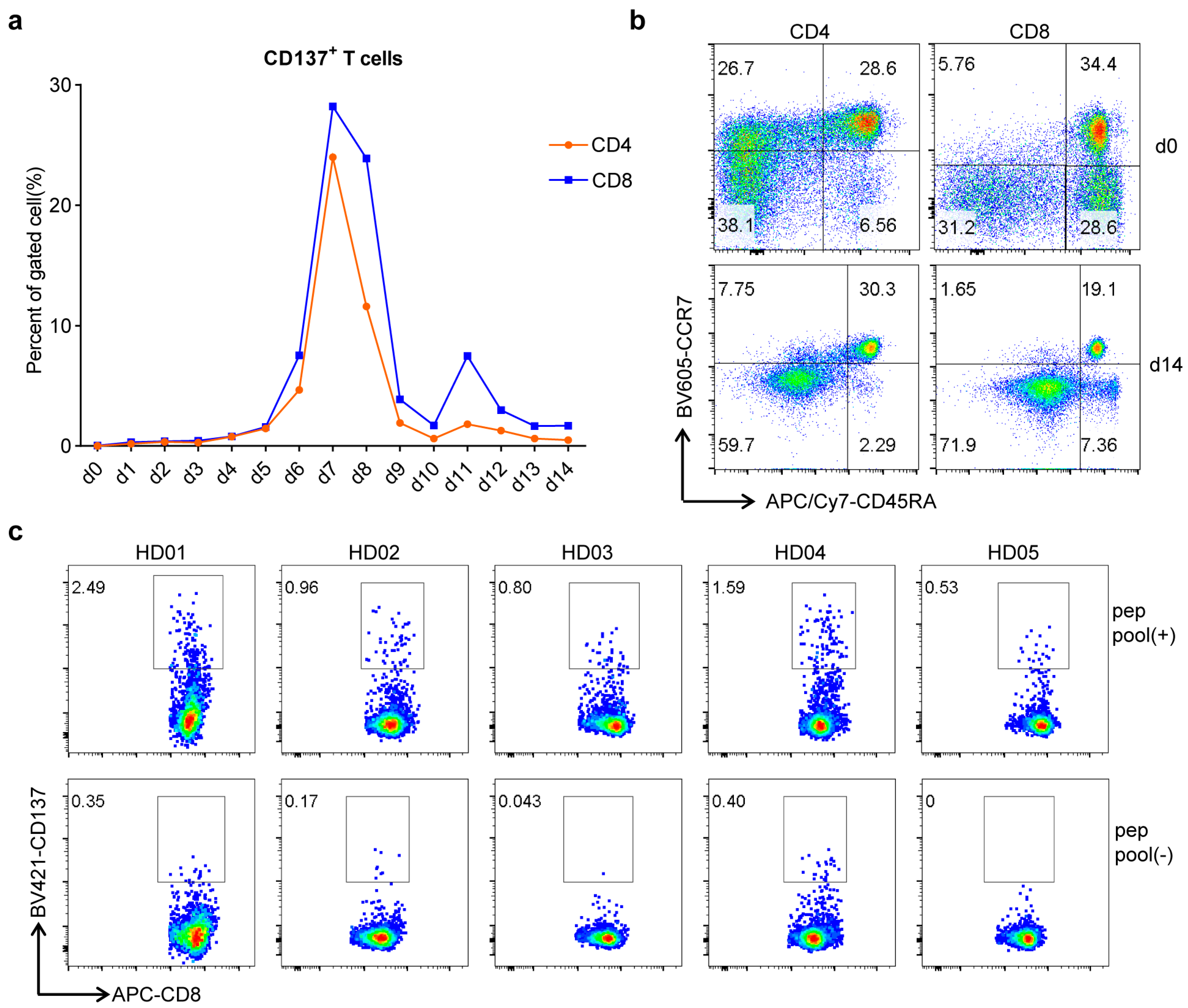
3.2. Conserved Antigen-Specific T Cell Responses
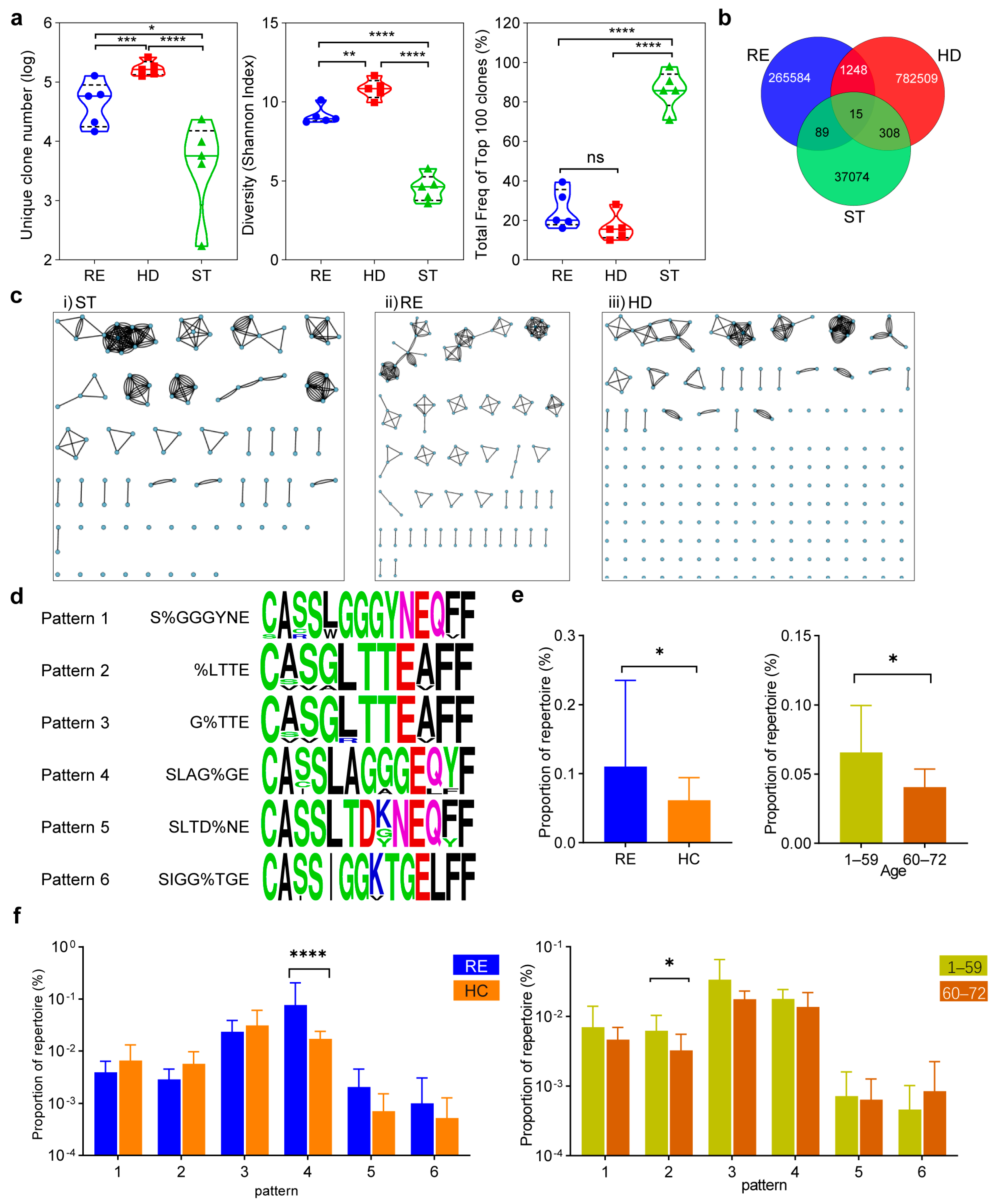
3.3. Characteristics of Conserved Epitope-Specific CD8+ TCR Repertoire
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bakhshandeh, B.; Jahanafrooz, Z.; Abbasi, A.; Goli, M.B.; Sadeghi, M.; Mottaqi, M.S.; Zamani, M. Mutations in SARS-CoV-2; Consequences in structure, function, and pathogenicity of the virus. Microb. Pathog. 2021, 154, 104831. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, Z.; Wang, C.; Ren, H.; Gao, L.; Peng, H.; Niu, Z.; Ren, H.; Huang, H.; Sun, Q. Bimodular effects of D614G mutation on the spike glycoprotein of SARS-CoV-2 enhance protein processing, membrane fusion, and viral infectivity. Signal Transduct. Target. Ther. 2020, 5, 268. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zheng, Y.; Niu, Z.; Jiang, X.; Sun, Q. The virological impacts of SARS-CoV-2 D614G mutation. J. Mol. Cell Biol. 2021, mjab045. [Google Scholar] [CrossRef] [PubMed]
- Supasa, P.; Zhou, D.; Dejnirattisai, W.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.; Nutalai, R.; Tuekprakhon, A.; et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell 2021, 184, 2201–2211.e7. [Google Scholar] [CrossRef]
- Zhou, D.; Dejnirattisai, W.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.; Tuekprakhon, A.; Nutalai, R.; et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 2021, 184, 2348–2361.e6. [Google Scholar] [CrossRef] [PubMed]
- Verghese, M.; Jiang, B.; Iwai, N.; Mar, M.; Sahoo, M.K.; Yamamoto, F.; Mfuh, K.O.; Miller, J.; Wang, H.; Zehnder, J.; et al. A SARS-CoV-2 Variant with L452R and E484Q Neutralization Resistance Mutations. J. Clin. Microbiol. 2021, 59, JCM-00741. [Google Scholar] [CrossRef]
- Luan, B.; Huynh, T. Insights into SARS-CoV-2’s Mutations for Evading Human Antibodies: Sacrifice and Survival. J. Med. Chem. 2021. [Google Scholar] [CrossRef]
- Wang, Z.; Fu, Y.; Guo, Z.; Li, J.; Li, J.; Cheng, H.; Lu, B.; Sun, Q. Transmission and prevention of SARS-CoV-2. Biochem. Soc. Trans. 2020, 48, 2307–2316. [Google Scholar] [CrossRef]
- Weber, S.; Ramirez, C.M.; Weiser, B.; Burger, H.; Doerfler, W. SARS-CoV-2 Worldwide Replication Drives Rapid Rise and Selection of Mutations across the Viral Genome: A Time-Course Study-Potential Challenge for Vaccines and Therapies. EMBO Mol. Med. 2021, 13, e14062. [Google Scholar] [CrossRef]
- Niu, Z.; Zhang, Z.; Gao, X.; Du, P.; Lu, J.; Yan, B.; Wang, C.; Zheng, Y.; Huang, H.; Sun, Q. N501Y mutation imparts cross-species transmission of SARS-CoV-2 to mice by enhancing receptor binding. Signal Transduct. Target. Ther. 2021, 6, 284. [Google Scholar] [CrossRef]
- Chaillon, A.; Wagner, G.A.; Hepler, N.L.; Little, S.J.; Pond, S.L.K.; Caballero, G.; Pacold, M.E.; Phung, P.; Wrin, T.; Richman, D.D.; et al. Dynamics of viral evolution and neutralizing antibody response after HIV-1 superinfection. J. Virol. 2013, 87, 12737–12744. [Google Scholar] [CrossRef][Green Version]
- Bar, K.J.; Tsao, C.Y.; Iyer, S.S.; Decker, J.M.; Yang, Y.; Bonsignori, M.; Chen, X.; Hwang, K.K.; Montefiori, D.C.; Liao, H.X.; et al. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog. 2012, 8, e1002721. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Bruel, T.; Grzelak, L.; Guivel-Benhassine, F.; Staropoli, I.; Porrot, F.; Planchais, C.; Buchrieser, J.; Rajah, M.M.; Bishop, E.; et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021, 27, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Sui, Z.; Dai, X.; Lu, Q.; Zhang, Y.; Huang, M.; Li, S.; Peng, T.; Xie, J.; Zhang, Y.; Wu, C.; et al. Viral dynamics and antibody responses in people with asymptomatic SARS-CoV-2 infection. Signal Transduct. Target Ther. 2021, 6, 181. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.G.; Ford, J.C.; Lewis, M.S.; Ventura, A.B.; Hughes, C.M.; Coyne-Johnson, L.; Whizin, N.; Oswald, K.; Shoemaker, R.; Swanson, T.; et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 2011, 473, 523–527. [Google Scholar] [CrossRef]
- Masopust, D.; Soerens, A.G. Tissue-Resident T Cells and Other Resident Leukocytes. Annu. Rev. Immunol. 2019, 37, 521–546. [Google Scholar] [CrossRef]
- Teijaro, J.R.; Turner, D.; Pham, Q.; Wherry, E.J.; Lefrancois, L.; Farber, D.L. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 2011, 187, 5510–5514. [Google Scholar] [CrossRef]
- Iijima, N.; Iwasaki, A. T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science 2014, 346, 93–98. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Charles, T.P.; Joag, V.; Bollimpelli, V.S.; Scott, M.K.D.; Wimmers, F.; Burton, S.L.; Labranche, C.C.; Petitdemange, C.; Gangadhara, S.; et al. T cell-inducing vaccine durably prevents mucosal SHIV infection even with lower neutralizing antibody titers. Nat. Med. 2020, 26, 932–940. [Google Scholar] [CrossRef]
- Zuo, J.; Dowell, A.C.; Pearce, H.; Verma, K.; Long, H.M.; Begum, J.; Aiano, F.; Amin-Chowdhury, Z.; Hoschler, K.; Brooks, T.; et al. Robust SARS-CoV-2-specific T cell immunity is maintained at 6 months following primary infection. Nat. Immunol. 2021, 22, 620–626. [Google Scholar] [CrossRef]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020, 587, 270–274. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.; Hafezi, M.; Chia, A.; Chng, M.H.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Mateus, J.; Grifoni, A.; Tarke, A.; Sidney, J.; Ramirez, S.I.; Dan, J.M.; Burger, Z.C.; Rawlings, S.A.; Smith, D.M.; Phillips, E.; et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020, 370, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Meckiff, B.J.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Kusnadi, A.; Simon, H.; Grifoni, A.; Pelosi, E.; Weiskopf, D.; Sette, A.; et al. Single-Cell Transcriptomic Analysis of SARS-CoV-2 Reactive CD4+ T Cells. SSRN 2020, 3641939. [Google Scholar] [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Strålin, K.; Gorin, J.-B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020, 183, 158–168.e14. [Google Scholar] [CrossRef] [PubMed]
- Weiskopf, D.; Schmitz, K.S.; Raadsen, M.P.; Grifoni, A.; Okba, N.M.; Endeman, H.; van den Akker, J.P.; Molenkamp, R.; Koopmans, M.P.; van Gorp, E.C.; et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020, 5, eabd2071. [Google Scholar] [CrossRef] [PubMed]
- Nelde, A.; Bilich, T.; Heitmann, J.S.; Maringer, Y.; Salih, H.R.; Roerden, M.; Lübke, M.; Bauer, J.; Rieth, J.; Wacker, M.; et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021, 22, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; He, M.; Li, L. HLA class I restricted epitopes prediction of common tumor antigens in white and East Asian populations: Implication on antigen selection for cancer vaccine design. PLoS ONE 2020, 15, e0229327. [Google Scholar] [CrossRef]
- Choi, B.K.; Lee, S.C.; Lee, M.J.; Kim, Y.H.; Ryu, K.-W.; Lee, J.-H.; Shin, S.M.; Suzuki, S.; Oh, H.S.; Kim, C.H.; et al. 4-1BB-based isolation and expansion of CD8+ T cells specific for self-tumor and non-self-tumor antigens for adoptive T-cell therapy. J. Immunother. 2014, 37, 225–236. [Google Scholar] [CrossRef]
- Lythgoe, K.A.; Hall, M.; Ferretti, L.; de Cesare, M.; MacIntyre-Cockett, G.; Trebes, A.; Andersson, M.; Otecko, N.; Wise, E.L.; Moore, N.; et al. SARS-CoV-2 within-host diversity and transmission. Science 2021, 372, eabg0821. [Google Scholar] [CrossRef]
- Chang, Y.M.; Wieland, A.; Li, Z.-R.; Im, S.J.; McGuire, D.; Kissick, H.T.; Antia, R.; Ahmed, R. T Cell Receptor Diversity and Lineage Relationship between Virus-Specific CD8 T Cell Subsets during Chronic Lymphocytic Choriomeningitis Virus Infection. J Virol. 2020, 94, e00935-20. [Google Scholar] [CrossRef]
- Glanville, J.; Huang, H.; Nau, A.; Hatton, O.; Wagar, L.E.; Rubelt, F.; Ji, X.; Han, A.; Krams, S.M.; Pettus, C.; et al. Identifying specificity groups in the T cell receptor repertoire. Nature 2017, 547, 94–98. [Google Scholar] [CrossRef]
- Grifoni, A.; Voic, H.; Dhanda, S.K.; Kidd, C.K.; Brien, J.D.; Buus, S.; Stryhn, A.; Durbin, A.P.; Whitehead, S.; Diehl, S.A.; et al. T Cell Responses Induced by Attenuated Flavivirus Vaccination Are Specific and Show Limited Cross-Reactivity with Other Flavivirus Species. J. Virol. 2020, 94, e00089-20. [Google Scholar] [CrossRef]
- Shomuradova, A.S.; Vagida, M.S.; Sheetikov, S.A.; Zornikova, K.V.; Kiryukhin, D.; Titov, A.; Peshkova, I.O.; Khmelevskaya, A.; Dianov, D.V.; Malasheva, M.; et al. SARS-CoV-2 Epitopes Are Recognized by a Public and Diverse Repertoire of Human T Cell Receptors. Immunity 2020, 53, 1245–1257.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zheng, Y.; Niu, Z.; Zhang, B.; Wang, C.; Yao, X.; Peng, H.; Franca, D.N.; Wang, Y.; Zhu, Y.; et al. SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination. Cell Death Differ. 2021, 28, 2765–2777. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Niu, Z.; Jiang, X.; Zhang, Z.; Zheng, Y.; Wang, Z.; Zhu, Y.; Gao, L.; Huang, H.; Wang, X.; et al. SARS-CoV-2 Targets by the pscRNA Profiling of ACE2, TMPRSS2 and Furin Proteases. iScience 2020, 23, 101744. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, Y.; Shao, C.; Huang, J.; Gan, J.; Huang, X.; Bucci, E.; Piacentini, M.; Ippolito, G.; Melino, G. COVID-19 infection: The perspectives on immune responses. Cell Death Differ. 2020, 27, 1451–1454. [Google Scholar] [CrossRef] [PubMed]
- Moderbacher, C.R.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996–1012.e19. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, W.; He, M.; Wang, X.; Sun, Q.; Kuang, M. Specific CD8+ TCR Repertoire Recognizing Conserved Antigens of SARS-CoV-2 in Unexposed Population: A Prerequisite for Broad-Spectrum CD8+ T Cell Immunity. Vaccines 2021, 9, 1093. https://doi.org/10.3390/vaccines9101093
Hu W, He M, Wang X, Sun Q, Kuang M. Specific CD8+ TCR Repertoire Recognizing Conserved Antigens of SARS-CoV-2 in Unexposed Population: A Prerequisite for Broad-Spectrum CD8+ T Cell Immunity. Vaccines. 2021; 9(10):1093. https://doi.org/10.3390/vaccines9101093
Chicago/Turabian StyleHu, Wei, Meifang He, Xiaoning Wang, Qiang Sun, and Ming Kuang. 2021. "Specific CD8+ TCR Repertoire Recognizing Conserved Antigens of SARS-CoV-2 in Unexposed Population: A Prerequisite for Broad-Spectrum CD8+ T Cell Immunity" Vaccines 9, no. 10: 1093. https://doi.org/10.3390/vaccines9101093
APA StyleHu, W., He, M., Wang, X., Sun, Q., & Kuang, M. (2021). Specific CD8+ TCR Repertoire Recognizing Conserved Antigens of SARS-CoV-2 in Unexposed Population: A Prerequisite for Broad-Spectrum CD8+ T Cell Immunity. Vaccines, 9(10), 1093. https://doi.org/10.3390/vaccines9101093





