Abstract
Tick-borne diseases affecting humans and animals are on the rise worldwide. Vaccines constitute an effective control measure, but very few are available. We selected Lyme borreliosis, a bacterial infection transmitted by the hard tick Ixodes, to validate a new concept to identify vaccine candidates. This disease is the most common tick-borne disease in the Northern Hemisphere. Although attempts to develop a vaccine exist, none have been successfully marketed. In tick-borne diseases, the skin constitutes a very specific environment encountered by the pathogen during its co-inoculation with tick saliva. In a mouse model, we developed a proteomic approach to identify vaccine candidates in skin biopsies. We identified 30 bacterial proteins after syringe inoculation or tick inoculation of bacteria. Discovery proteomics using mass spectrometry might be used in various tick-borne diseases to identify pathogen proteins with early skin expression. It should help to better develop sub-unit vaccines based on a cocktail of several antigens, associated with effective adjuvant and delivery systems of antigens. In all vector-borne diseases, the skin deserves further investigation to better define its role in the elaboration of protective immunity against pathogens.
1. Introduction
Vector-borne diseases (VBDs) represent a major burden for human and animal health. Ticks are hematophagous ectoparasites that transmit a large panel of pathogens such as bacteria, viruses, and parasites. The bacterial infection called Lyme borreliosis is the most prevalent vector-borne infectious disease in the Northern Hemisphere [1]. The disease is a zoonosis that affects wild and domestic animals; humans constitute accidental hosts. Currently, no vaccine is available to prevent the disease in humans, although various vaccines have been developed in the past, mainly based on Borrelia lipoproteins, outer surface protein C (OspC) and OspA [2]. OspC, first considered as an ideal vaccine candidate, was selected. OspC is synthetized within the tick during the migration of the spirochete from the gut to the salivary glands [3]. It is also an essential lipoprotein for transmission and dissemination of Borrelia in the vertebrate host [4]. However, due to its diversity, protection was mainly strain-specific. Improvement in the vaccine design, using epitopes from different OspC stains, did not lead to development of a vaccine in humans thus far [5]. OspA on the surface of the spirochete is essential for it to bind to the tick midgut using the specific receptor TROSPA (Tick receptor for outer surface protein A). Upon tick blood meal, spirochetes detach from the midgut receptor and switch their surface lipoproteins from OspA to OspC [6]. The idea emerged that by immunizing mice with OspA, specific antibodies against OspA could neutralize the bacteria within the tick, thus blocking transmission to the vertebrate host [7]. Further, the use of this vaccine in human clinical trials proved that the concept of a “transmission-blocking vaccine” could be used to neutralize a pathogen within the tick vector. Although effective, this vaccine tested in the general population was associated with adverse effects in a few patients and the vaccine was withdrawn from the market in 2002 [8]. Recently, a new formulation of OspA vaccine without the epitopes associated with potential autoimmunity problem and containing six different serotypes has been developed. It is presently tested in a phase 2 human clinical trial [9]. Other Borrelia burgdorferi proteins were also tested as vaccine candidates, such as decorin-binding protein A (DbpA) and fibronectin-binding protein (BBK32) [10,11].
Other vaccine concepts have been developed to reduce the incidence of Lyme disease in animal reservoir. As this disease is a zoonosis with the bacterium circulating in numerous hosts, vaccines have also been tested in the main animal reservoirs. The wild white-footed mouse, Peromyscus leucopus, has been immunized with various formulations of recombinant OspA to reduce nymphal infection prevalence. Various routes of inoculation have been tested, either subcutaneously on small populations of rodents [12] or on a larger scale with baited oral vaccination strategy [13]. Targeting the tick vector by immunization of the vertebrate host with tick saliva proteins is also explored. As the blood meal of hard ticks lasts for several days, tick saliva is essential to this long feeding process. Tick proteins target the pharmacology (coagulation, itching, and pain) and the immunology (complement pathway, cells of innate and adaptive immunity) of the vertebrate host to allow for blood meal completion. Several candidates have been Rhipicephalus identified; among them are Subolesin [14], Salp15 (Salivary protein of 15 kDa) [15], ISAC (Ixodes scapularis anticomplement) [16], Ixolaris [17], sialostatins [18], and 64TRP (64Truncated Rhipicephalus Protein) [19]. Although various strategies have been evaluated, no effective vaccine is available for tick-borne diseases (TBDs), except against tick-borne encephalitis virus [20].
In VBDs, the skin is an essential organ where the arthropod co-inoculates the pathogens and its saliva [21]. Therefore, a vaccine that would induce an immune response in the vertebrate host to neutralize the pathogen in the skin should be effective. We thus developed a new strategy to identify vaccine candidates present in the skin of the vertebrate host. We evaluated proteomics on skin biopsies of Borrelia-infected mice to identify such vaccine candidates [22]. In Lyme borreliosis, bacteria are co-inoculated with tick saliva in the dermis where an intense Borrelia multiplication occurs, followed by dissemination to the target organs: the heart, the nervous system, and the joints [23]. However, some bacteria persist in the skin, pointing out the major role of the skin as an immune-tolerant organ. In this study, we used a well-established C3H/HeN mouse model of Lyme borreliosis to select Borrelia proteins by non-targeted mass spectrometry that could be further tested as candidates in a Lyme vaccine.
2. Material and Methods
2.1. Mouse Infection and Bacterial Strains
We selected three Borrelia burgdorferi sensu stricto (ss) strains: 297 (AC Steere, Boston, MA, USA, isolated from cerebrospinal fluid samples), IBS 19 (B Jaulhac, Strasbourg, France, isolated from an erythema migrans), and BL 642 (CS Pavia, New-York, NY, USA, isolated from blood samples). All strains were cultured in BSK-H complete medium (Sigma-Aldrich, Saint Quentin Fallavier, France) at 33 °C and used at low passage (<7) for mouse infection. Spirochetes were counted and viability was checked using dark-field microscopy.
Three- to four-week old C3H/HeN mice were inoculated with 103 spirochetes in 0.1 mL BSK by intradermal injection in the dorsal thoracic area. The infection kinetics was assessed, and skin samples of mice were collected at various time points after Borrelia inoculation: day 3, 5, 7, and 15. An area of approximately 1 cm of mouse skin was collected at the inoculation site and stored at −80 °C.
For infection via tick bites, Ixodes ricinus nymphs infected with Borrelia afzelii strain NE4049 were generated according to a previously described protocol [14]. Briefly, each mouse was infected with 10 infected nymphs until blood meal completion. The infection rate of nymphs infected with Borrelia was 60–80%. For infection with field-collected ticks, three I. ricinus females from an endemic area for Lyme borreliosis [24] were fed per mouse. On day 3, the females were removed from the mice, and after 7 days, samples of potentially infected skin were collected. Borrelia detection and quantification were performed by PCR.
2.2. Quantification of B. burgdorferi DNA by PCR in Mouse Skin
DNA was extracted from skin samples of each individual mouse on a MagNA Pure system (Roche Diagnostics, Meylan, France), and the load of spirochetes in the skin was subsequently estimated by quantitative PCR targeting the flagellin gene (flaB) as previously described [23]. Briefly, quantification of Borrelia-specific flaB gene was performed on a LightCycler system (Roche Diagnostics, Meylan, France). Quantification of the mouse-specific glyceraldehyde-3-phosphate dehydrogenase (gapdh) gene was performed on an ABI Prism 7500 instrument (ThermoFisher, Illkirch-Graffenstaden, France), using a commercial kit (TaqMan rodent GAPDH control reagent; ThermoFisher, Illkirch-Graffenstaden,, France). The number of B. burgdorferi spirochetes in the mouse skin was standardized to 10,000 gapdh gene copies.
2.3. Proteomics
Samples (5 mg) of mouse skin biopsies were manually extracted by Laemmli buffer (200 µL), and proteins (50 µg) were pre-fractionated onto 12% SDS-PAGE as previously described [22]. Gel bands (10 ± 1 bands) of 2 mm were manually excised. After reduction, alkylation, and trypsin digestion, peptides were extracted using 60% acetonitrile and 0.1% HCOOH for 1 hour at room temperature.
For biopsies infected via syringe inoculation, we suspended peptides in 200 µL of 0.1% HCOOH after evaporation, and nanoLC–MS/MS analyses (1 µL injected) were performed on a nano-ACQUITY UPLC (UltraPerformance Liquid Chromatography) system (Waters, Milford, MA, USA) hyphenated to a quadrupole time-of-flight MaXis4G (Bruker Daltonics, Bremen, Germany) as previously described [25] with the following modifications: elution gradient of 6–35% solvent B over 28 min at 60 °C and external mass calibration in the mass range of 100–2200 m/z (scan speed 25 Hz). For tandem MS experiments, acquisition was performed by sequentially selecting the maximum of precursors for a cycle time of 3.5 s, with a preference for multiply charged ions and strict exclusion of monocharged ions. Acquisition speed in MS/MS was adjusted according to the precursor intensity. Selected ions were excluded for 1 min and optionally reconsidered if the measured intensity was three times higher than the previously measured intensity. Mass data analysis was performed as previously described [22,26], except for a 25 ppm parent mass tolerance, a 0.05 Da fragment mass tolerance, and a maximum of one missed cleavage.
For biopsies infected via tick bites, we suspended peptides in 50 µL of 0.1% HCOOH after evaporation, and nanoLC–MS/MS analyses (3 µL injected) were performed on a nano-ACQUITY UPLC system (Waters, Milford, MA, USA) hyphenated to a Q-Exactive Plus mass spectrometer (Thermo Scientific, Bremen, Germany) as previously described [26], except for an elution gradient of 5–35% solvent B over 60 min, then 35–90% solvent B over 1 min, and capillary voltage set to 1.6 kV at 250 °C. Mass data analysis was performed as previously described [26], and searches were performed against an inhouse-generated database containing all protein sequences of B. afzelii Pko (10 October 2016, 2186 entries) and mouse (10 October 2016, 16,806 entries) (extracted from NCBInr and UniProtKB/Swiss-Prot, respectively).
For all biopsies, a target-decoy database search allowed us to control the false-positive identification rate, which was set at 1% with a minimum of one peptide per protein. All results were loaded into Scaffold software (Proteome Software, Portland, OR, USA) to validate peptide identifications.
2.4. In Silico Evaluation of Protein Polymorphism Among Borrelia burgdorferi Coding DNA Sequences
We performed the analysis of protein variability in the coding DNA sequences (CDs) of proteins selected at the early cutaneous phase described in B. burgdorferi either using translated sequences of the validated subset of UniProtKB-Swiss-Prot or annotated genes in UniprotKB-TrEMBL database, among the 15 publicly available B. burgdorferi genomes curated on the website borreliabase.org [27]. These Borrelia genomes have a taxonomic identification at the species level confirmed by phylogenomics. Sequence comparisons with reference protein sequences were performed using bidirectional protein-protein BLAST (BlastP) sequence comparison of translated open reading frames. Proteins with amino acid sequence similarities ≥65% and E-value ≤10−10 were considered homologous [28].
2.5. Ethics
The protocols carried out in this study were approved by and complied with the requirements of the CREMEAS Committee on the Ethics of Animal Experiments of the University of Strasbourg (Comité Régional d’Ethique en Matière d’Expérimentation Animale Strasbourg—Ministry Permit Number: APAFIS 2015062414395551) in the animal facilities D-67-482-34.
2.6. Statistical Analyses
Statistical analyses between the various groups were made with Fisher’s exact test to compare proportions, a non-parametric Kruskal–Wallis test, or a Mann–Whitney test when there were only two groups to compare. A p-value of 0.05 was considered statistically significant. These statistical analyses were performed with Prism 7 software v7.0a (Graphpad, La Jolla, CA, USA). Exact confidence interval of a frequency, i.e., binomial confidence interval, was determined for each percentage according to the Clopper–Pearson method using the web server http://statpages.info/confint.html.
3. Results
3.1. Efficacy of Infection and Kinetics of Bacterial Multiplication in the Mouse Skin for the Three B. burgdorferi ss Strains
We first evaluated the efficacy of Borrelia transmission in mice by detection of B. burgdorferi DNA (flaB gene) in the skin at the inoculation site after 3, 5, 7, or 15 days post-infection (Table 1). No significant difference was observed in the rate of positivity between the three studied strains at each time point (Fisher’s exact test, p-value ≥ 0.1). Maximum positive detections in the skin samples of mice were observed on day 7 post-inoculation for the three B. burgdorferi strains, reaching 100% of tested animals. These results also indicate that infection was effective with a syringe inoculation of 103 B. burgdorferi spirochetes in the skin of mice.

Table 1.
Detection of Borrelia burgdorferi sensu stricto (ss) DNA in the skin of infected mice 3, 5, 7, or 15 days post-inoculation (dpi). This table presents the results of all the mice included in the study. Abbreviations: CI 95%, 95% confidence interval; PCR, polymerase chain reaction.
We then quantified Borrelia at the inoculation site at the various time points of the kinetics study by real-time PCR targeting flagellin gene with a standard curve. The number of flaB copies in samples was standardized to 10,000 copies of the housekeeping murine gene gapdh (Figure 1). Three- and 15-days post-inoculation (dpi), spirochetes were near the limit of quantification for all three B. burgdorferi strains (0.05 flagellin copies per 10,000 murine gapdh). On day 5, for some skin samples, bacterial load was detected but it did not exceed 200 flagellin copies per 10,000 murine gapdh. The bacterial load reached maximum density for the three strains on day 7 (p-values < 0.05) with median values of 219 (min. 9; max. 515), 65 (min. 7; max. 111), and 138 (min. 60; max. 268) flagellin copies per 10,000 murine gapdh for strains 297, IBS 19, and BL 642, respectively. Altogether, the kinetics of bacterial load suggested that the spirochetal multiplication peak in mouse skin occurs approximately 7 days after inoculation under such experimental conditions. Values of bacterial loads were not statistically different between strains at each time point, except for strain IBS 19, which exhibited a lower median value of spirochetal load on day 7 (p-value < 0.01), but its kinetic profile and the scale of values were comparable to strains 297 and BL 642 (see the median values indicated above).
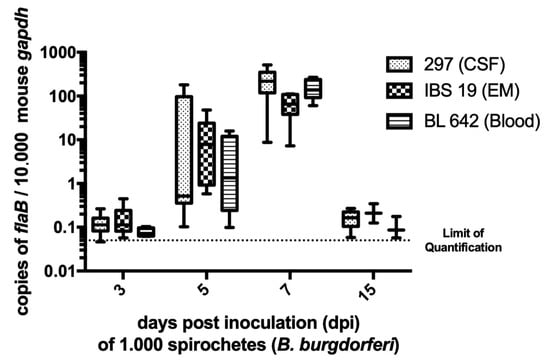
Figure 1.
Borrelia load in the mouse skin. The spirochetal burden in the skin at the inoculation site was measured by quantitative PCR for the Borrelia flagellin (flaB) gene and normalized to copies of mouse glyceraldehyde-3-phosphate dehydrogenase (gapdh). Values presented in box plot correspond to relative quantification in each positive mouse skin (all the mice referred to as positive in Table 1). In each box plot, the box embraces values between the first and third quartile, the median is a line, the higher error bar corresponds to the maximum value, and the lower error bar corresponds to the minimum value. Limit of quantification was around 0.05 flagellin copies per 10.000 murine gapdh. Medians of bacterial load were statistically compared [1] within strains at the same day and [2] for a same strain at each time post-inoculation. The bacterial load reached maximum density for the three strains on day 7 (p-values < 0.05). Values of bacterial load were not statistically different between strains at each time point, except for strain IBS 19 that exhibited a lower median value of spirochetal load on day 7 (p-value < 0.01). Abbreviations: CSF, cerebrospinal fluid; EM, erythema migrans.
3.2. Detection of Borrelia Proteins on Day 7 by Proteomics
Using non-targeted mass spectrometry on mice infected via syringe inoculation, we selected samples of infected skin on day 7—i.e., the peak of Borrelia multiplication in the skin—to increase the probability of finding a significant amount of bacterial proteins. We identified between two and nine proteins of Borrelia burgdorferi ss among an average of 1620 mouse proteins with flagellin, OspC, and GAPDH, the most frequently detected proteins (Table 2). According to Borrelia strains, we detected additional proteins such as chaperonin (GroEL), enolase, lipoprotein BbA36, and p66.

Table 2.
Identification of bacterial proteins in mouse skin infected by intradermal inoculation of different Borrelia burgdorferi strains, or via infectious tick bite. Proteins were identified using Mascot algorithm. Numbers of identified peptides are given.
Interestingly, when mice were infected by ticks (either with the ones infected under laboratory conditions or collected in the field), we also detected OspC and flagellin of B. afzelii. This is the predominant species circulating in ticks and isolated in patients in Europe.
Thanks to the Q-Orbitrap mass spectrometer, we identified additional proteins (average of 5920 mouse proteins) when mice were infected through an infectious tick bite, and we also detected 12 additional proteins of Borrelia (Table 2). Given the difference in the two types of equipment, the detection of additional bacterial proteins cannot be strictly assigned to the inoculation route, but also (if not exclusively) to the enhanced sensitivity of Q-Orbitrap mass spectrometer.
3.3. Peptidic Polymorphism of the Selected Coding DNA Sequences Among B. burgdorferi ss Genomes
We evaluated peptide variability of the coding DNA sequences (CDSs) for the six Borrelia proteins identified by proteomics (major flagellar protein FlaB, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 60 kDa chaperonin (GroEL), enolase, lipoprotein BbA36, and outer surface protein C (OspC). To this end, we performed an in silico comparison between the coding DNA peptide sequences from 15 B. burgdorferi ss genomes on the webserver borreliabase.org. The location of genes encoding for flagellin, GAPDH, GroEL, and enolase proteins is chromosomal, whereas lipoprotein BbA36 and OspC are plasmid-encoded in the lp54 and cp26 plasmids, respectively. The most conserved peptide sequences corresponded to the flagellin, GAPDH, GroEL, and enolase proteins, with homologs detected in all 15 genomes and excellent features of identity (≥99%) and similarity (100%) between strains, with the exception of the enolase CDS in strain 29805 that appeared truncated (Table 3). Homologs of lipoprotein BbA36 were detected in 13 B. burgdorferi ss strains and not in two strains for which the lp54 plasmid had not been sequenced. Identity and similarity features were high (≥97%), and we observed minor differences from the reference sequence including a supplementary amino acid detected in six strains and differences in the first eight amino acids that were specific to strain B31. OspC peptide sequences were significantly more polymorphic with 82 to 88% identity and 74 to 81% similarity for the homologous sequences detected in the 12 genomes harboring cp26 and other than the reference genome (Table 3).

Table 3.
Peptide variability of six protein-coding sequences in the species B. burgdorferi sensu stricto. The 15 B. burgdorferi sensu stricto strains studied were B31, CA382, 64b, ZS7, BOL26, WI91-23, 29805, N40, JD1, 156a, 94a, CA-11.2A, 72a, 118a, and CA8. Abbreviations: aa, amino acids; CDS, coding DNA sequence; Id., identity; S., similarity; WGS, whole genome sequences.
4. Discussion
Although various antigens have been identified as vaccine candidates for Lyme borreliosis, none have been particularly effective in protecting animals and humans [29]. It is well-established that the antibody response can control the bacterial infection, but it is not long-lasting. The complexity of this immune response against Borrelia is linked to high surface antigen variation, especially with up- and downregulation of VlsE (Variable major protein-like sequence, Expressed) [30], and inhibition of complement lysis pathway [31]. Interestingly, recent data show that Borrelia infection actually induces a temporary immunosuppression, which explains the lack of long-term immunity for this disease [32]. Presently, several vaccines have been developed and marketed in veterinary medicine for dogs. They are based on whole-lysate antigens or recombinant OspC or OspA protein. In humans, clinical trials are underway in Europe using a modified OspA antigen [33]. The vaccine is based on six different OspA serotypes of different Borrelia species occurring in the United States and Europe and formulated with aluminum hydroxide [34]. This vaccine (VLA15), previously tested in a mouse model, induced a good protection after three immunizations against a challenge of Borrelia inoculated using a syringe or via infected tick bites [35].
Up to now, this unsuccessful strategy to get an effective vaccine against Lyme borreliosis could be either due to inefficient vaccine candidates, or failure in the delivery system of the vaccine, or both, in terms of the ability to elicit a protective immune response. As the skin is an essential interface in VBDs, we found the development of a new strategy to identify relevant Borrelia proteins expressed in the skin during the early process of transmission to be particularly interesting. Indeed, antibodies directed against these proteins would neutralize bacteria before they further disseminate in deep organs: the heart, the joints, or the central nervous system. We decided to use the discovery proteomics approach, Ge-LC–MS/MS, to identify Borrelia proteins in the skin of infected mice after syringe inoculation or after an infectious tick bite. The C3H mouse model of Lyme borreliosis has been shown to be very effective in analyzing mechanisms of Borrelia transmission and persistence [26,36]. In this study, we showed that non-targeted mass spectrometry allows for the identification of at least 30 Borrelia proteins. Some of them have already been described as potential vaccine candidates, such as OspC, enolase, and DbpA.
As Borreliae are highly diverse [37], some bacterial proteins were only detected in some Borrelia strains, as shown, for example, for Borrelia glycosaminoglycan binding protein (Bgp), which is present only in the strain BL642, or GAPDH present in strains 297c4 and BL642, or GroEl protein detected in B. burgdorferi ss strain 297 and in B. afzelii from mice infected through the bite of field-collected ticks.
The proteins OspC and FlaB (p41) are immunodominant antigens of Borrelia that are implicated in the initial human immune response against spirochetes [38]. Mainly composed of the non-glycosylated major flagellar protein FlaB, the periplasmic flagellar filaments are essential in the motility and mammal infectivity of B. burgdorferi [39]. Proteomic assays highlighted that FlaB was abundant in the skin at the inoculation site, with a higher load 7 days after inoculation, i.e., at the time of the putative multiplication of spirochetes, as confirmed by quantification of the FlaB transcript. This relative abundance during the early cutaneous phase of the infection and the remarkable peptide sequence homogeneity between strains that we reported (no polymorphism from 15 B. burgdorferi ss genomes) may suggest that FlaB represents a relevant vaccine candidate as purified recombinant protein. However, Fikrig et al. [40] showed that FlaB had no role in the protective immunity of C3H mice against spirochetal infection by B. burgdorferi, unlike OspA and OspB. However, bacterial flagellin has recently been studied as a potential adjuvant in the development of novel vaccines [41]. As a vaccine candidate, flagellin has been tested as a protein target (e.g., in the Campylobacter jejuni vaccination of poultry [42]) or as an adjuvant (e.g., in the Clostridium difficile vaccination [43]). In this context, a thorough study of FlaB immunogenicity in Lyme borreliosis deserves to be performed using the protein as adjuvant and/or in combination with other Borrelia antigens.
Outer surface protein OspC is a well-known lipoprotein of B. burgdorferi sensu lato (sl) induced during nymphal tick feeding. It is essential for the tick-to-mammal transmission of the bacteria [44]. It is therefore not surprising to detect it in the skin by proteomics. However, the high rate of peptide polymorphism observed between strains obtained in our study confirmed the hypervariability of this protein reported by previous works [29,45]. Due to this high strain specificity, OspC cannot be a good vaccine candidate.
Enolase is an enzyme involved in glycolysis. It constitutes a plasminogen receptor and mediates the activation of plasmin and extracellular matrix degradation [46]. As a protein expressed on cancer cells to facilitate their dissemination, this protein is very well studied and has been proposed as a potential vaccine candidate in cancer [47]. It is also a ubiquitous protein in animals. Borrelia is known to bind to plasminogen via this protein to facilitate its dissemination through host tissues. Experiments of mouse immunization with recombinant enolase did not induce protective immunity against subsequent B. burgdorferi infection. However, mice immunized with this protein reduced pathogen survival within feeding ticks [48]. In our study, the enolase peptides were weakly detected in the various Borrelia strains and species.
GroEL is a 60-kDa heat shock protein that prevents misfolding and promotes the refolding and proper assembly of unfolded polypeptides generated under stress conditions. It mainly represents a cytosolic protein with a few proteins inserted in inner membrane [49]. As a ubiquitous protein in both prokaryotes and eukaryotes and as a highly conserved protein, it has been used to genotype various B. burgdorferi sensu lato strains [50]. Interestingly, it has been detected in the skin of mice infected with B. burgdorferi ss and B. afzelii, irrespective of the inoculation mode.
GAPDH (glyceraldehyde-3-phosphate dehydrogenase) has an enzymatic role in the glycolytic pathway. It has been detected by 2D gel (2-DE) immunoblotting with matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). Antigens of B. garinii were analyzed for their reactivity with sera from patients at the early and late stage of Lyme borreliosis. Among the 20 antigens identified, GAPDH was detected [51]. In our study, this protein was only detected after syringe inoculation of Borrelia.
Elongation factor Tu is a cytosolic protein, just like enolase, and can be localized on the surface of bacteria. It has been shown to be very immunogenic and to induce antibodies in patients. However, this protein has not proved protective after immunization in mice [52]. Interestingly, in our study, we found this protein in the mouse skin after syringe inoculation of Borrelia burgdorferi ss, but also in mice naturally infected with B. afzelii-infected ticks.
Vaccines are the most effective way of preventing VBDs [53]. Presently, only a few vaccines are available. They target viral pathogens such as the vaccines against tick-borne encephalitis, yellow fever, and Japanese encephalitis. It points out to weaknesses in the design of such vaccines, especially against bacteria and parasites. Pitfalls have to be identified and could be due to an incorrect identification/selection of vaccine candidates, an inefficient adjuvant, and/or a poor delivery system of vaccination. Borrelia antigens detected in the skin by mass spectrometry might constitute a reliable approach to identify vaccine candidates and to elaborate cocktails of antigens that are immunogenic and protective. We selected the skin samples of infected mice on day 7 as we know that Borrelia multiplies very actively on day 7 [23]. In the design of vaccine for vector-borne diseases, the adjuvant and the delivery system also play a major role to elicit strong immunity. Indeed, the skin is a complex immune organ and it constitutes a persistent site for several vector-borne pathogens [54,55]. The skin is also a key interface of specific immune response during inoculation of these pathogens [21]. It constitutes the best interface to address the specific interactions of the host immune system and the pathogen proteome in VBDs. Besides the technique of mass spectrometry to identify vaccine candidates in the skin, data bank searches are also essential to overcome the problems of gene variability for these vaccine candidates.
New concepts have emerged in the development of vaccines with the advances of “omics” and reverse vaccinology [56,57]. In the field of proteomics, various techniques have been suggested to identify potential vaccine candidates. Some studied the surface proteome of Borrelia [58]. However, the three studied Borrelia species were issued from in vitro culture. Interactome studies have also been performed [59], but they once again relied on in vitro study interaction. It is now well-documented that Borrelia adapts to its various hosts during its enzootic cycle through modifications of its transcriptome [60,61]. It is therefore essential to work in vivo with animal models, mimicking as closely as possible the natural environment of pathogens. The skin thus constitutes an excellent organ to identify Borrelia proteins essential for the early transmission of this TBD. Thanks to its specific immunity, temperature, and microbiome, the skin also represents a peculiar environment [62] where some pathogens can persist for months by inducing immune tolerance [26].
5. Conclusions
The identification of good vaccine candidates is key to the development of a vaccine, and so is the choice of the adjuvants and antigen delivery system in order to induce protective immunity. Besides aluminum hydroxide, which is largely used in vaccines, new adjuvants are being developed to improve the strength and quality of the immune response such as AS01 adjuvant [63]. Innovative techniques have also been developed lately to better deliver antigens in the outermost layer of the skin by microneedles [64]. This has already been tested in human clinical trials for immunization against influenza virus, with promising results [65].
Facing all the unsuccessful strategies developed in the field of vaccines against VBDs, it is essential to reevaluate the various techniques used to identify vaccine candidates. It is also essential to better understand skin immunity against vector-borne pathogens that are inoculated, which multiply intensively and persist in the skin. In this context, a broader multidisciplinary approach in vaccinology instead of the approach used up until now should open up new avenues to control VBDs.
Author Contributions
Conceptualization, N.B., B.J., and L.E.-S.; methodology, M.A.R., C.B., B.W., G.S., and P.C.; validation, E.T.-R. and L.E.-S.; formal analysis, L.E.-S., E.T.-R., and N.B.; investigation, M.A.R., P.C., B.W., G.S., and E.T.-R.; writing—original draft: E.T.-R. and N.B.; preparation, N.B. and L.E.-S.; writing—review and editing, N.B. and L.E.-S.; visualization, N.B.; supervision, N.B., L.E.-S., and B.J.; project administration, N.B.; funding acquisition, N.B. and L.E.-S. All authors have read and agreed to the published version of the manuscript.
Funding
We are grateful to the SATT Conectus for the financial support of the vaccine project, Virbac for the financial support of the student M. Raess, and the French Proteomics Infrastructure (ProFI; ANR-10-INSB-08-03).
Acknowledgments
Elody Collin and CNR Borrelia for technical support. We thank Marthe Moren for editing the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Steere, A.; Strle, F.; Wormser, G.; Hu, L.; Branda, J.; Hovius, J.; Li, X.; Mead, P. Lyme borreliosis. Nat. Rev. Dis. Prim. 2016, 2, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Solecki, M.; Arnaboldi, P.M.; Backenson, P.B.; Benach, J.L.; Cooper, C.L.; Dattwyler, R.J.; Diuk-Wasser, M.; Fikrig, E.; Hovius, J.W.; Laegreid, W.; et al. Protective immunity and new vaccines for lyme disease. Clin. Infect. Dis. 2019. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, J.; Piesman, J.; de Silva, A. Antigenic and genetic heterogeneity of Borrelia burgdorferi populations transmitted by ticks. Proc. Nati. Acad. Sci. USA 2001, 98, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Tilly, K.; Bestor, A.; Jewett, M.W.; Rosa, P. Rapid clearance of Lyme disease spirochetes lacking OspC from skin. Infect. Immun. 2007, 75, 1517–1519. [Google Scholar] [CrossRef] [PubMed]
- Earnhart, C.; Marconi, R. An octavalent lyme disease vaccine induces antibodies that recognize all incorporated OspC type-specific sequences. Hum. Vaccin. 2007, 3, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Pal, U.; Li, X.; Wang, T.; Montgomery, R.; Ramamoorthi, N.; Desilva, A.; Bao, F.; Yang, X.; Pypaert, M.; Pradhan, D.; et al. TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell 2004, 119, 457–468. [Google Scholar] [CrossRef]
- Schaible, U.; Wallich, R.; Kramer, M.; Gern, L.; Anderson, J.F.; Museteanu, C.; Simon, M. Immune sera to individual Borrelia burgdorferi isolates or recombinant OspA thereof protect SCID mice against infection with homologous strains but only partially or not at all against those of different OspA/OspB genotype. Vaccine 1993, 11, 1049–1054. [Google Scholar] [CrossRef]
- Abbott, A. Lyme disease: Uphill struggle. Nature 2006, 439, 524–525. [Google Scholar] [CrossRef]
- Wressnigg, N.; Barrett, P.; Pöllabauer, E.; O’Rourke, M.; Portsmouth, D.; Schwendinger, M.; Crowe, B.; Livey, I.; Dvorak, T.; Schmitt, B.; et al. A novel multivalent OspA vaccine against Lyme borreliosis is safe and immunogenic in an adult population previously infected with Borrelia burgdorferi sensu lato. Clin. Vaccine Immunol. 2014, 21, 1490–1499. [Google Scholar] [CrossRef]
- Hagman, K.; Yang, X.; Wikel, S.; Schoeler, G.; Caimano, M.; Radolf, J.; Norgard, M. Decorin-binding protein A (DbpA) of Borrelia burgdorferi is not protective when immunized mice are challenged via tick infestation and correlates with the lack of DbpA expression by B. burgdorferi in ticks. Infect. Immun. 2000, 68, 4759–4764. [Google Scholar] [CrossRef]
- Brown, E.L.; Kim, J.H.; Reisenbichler, E.S.; Höök, M. Multicomponent Lyme vaccine: Three is not a crowd. Vaccine 2005, 23, 3687–3696. [Google Scholar] [CrossRef] [PubMed]
- Tsao, J.I.; Wootton, J.T.; Bunikis, J.; Luna, M.G.; Fish, D.; Barbour, A.G. Elimination of Borrelia burgdorferi from vector ticks feeding on OspA-immunized mice. Proc. Nati. Acad. Sci. USA 2004, 89, 5418–5421. [Google Scholar] [CrossRef]
- Gomes-Solecki, M.J.C.; Brisson, D.R.; Dattwyler, R.J. Oral vaccine that breaks the transmission cycle of the Lyme disease spirochete can be delivered via bait. Vaccine 2006, 24, 4440–4449. [Google Scholar] [CrossRef] [PubMed]
- Merino, O.; Almazan, C.; Canales, M.; Villar, M.; Moreno-Cid, J.A.; Galindo, R.C.; de la Fuente, J. Targeting the tick protective antigen subolesin reduces vector infestations and pathogen infection by Anaplasma marginale and Babesia bigemina. Vaccine 2011, 29, 8575–8579. [Google Scholar] [CrossRef] [PubMed]
- Schuijt, T.J.; Hovius, J.W.R.; van Burgel, N.D.; Ramamoorthi, N.; Fikrig, E.; van Dam, A.P. The tick salivary protein Salp15 inhibits the killing of serum-sensitive Borrelia burgdorferi sensu lato isolates. Infect. Immun. 2008, 76, 2888–2894. [Google Scholar] [CrossRef]
- Valenzuela, J.G.; Charlab, R.; Mather, T.N.; Ribeiro, J.M. Purification, cloning, and expression of a novel salivary anticomplement protein from the tick, Ixodes scapularis. JBC 2000, 275, 18717–18723. [Google Scholar] [CrossRef]
- Francischetti, I.M.B.; Valenzuela, J.G.; Andersen, J.F.; Mather, T.N.; Ribeiro, J.M.C. Ixolaris, a novel recombinant tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick, Ixodes scapularis: Identification of factor X and factor Xa as scaffolds for the inhibition of factor VIIa/tissue factor complex. Blood 2002, 99, 3602–3612. [Google Scholar] [CrossRef]
- Kotsyfakis, M.; Sá-Nunes, A.; Francischetti, I.M.B.; Mather, T.N.; Andersen, J.F.; Ribeiro, J.M.C. Antiinflammatory and immunosuppressive activity of sialostatin L, a salivary cystatin from the tick ixodes scapularis. J. Biol. Chem. 2006, 281, 26298–26307. [Google Scholar] [CrossRef]
- Labuda, M.; Trimnell, A.R.; Licková, M.; Kazimírová, M.; Davies, G.M.; Lissina, O.; Hails, R.S.; Nuttall, P. An antivector vaccine protects against a lethal vector-borne pathogen. PLoS Pathog. 2006, 2, e27. [Google Scholar] [CrossRef]
- Kunz, C. TBE vaccination and the Austrian experience. Vaccine 2003, 21, S50–S55. [Google Scholar] [CrossRef]
- Bernard, Q.; Jaulhac, B.; Boulanger, N. Smuggling across the border: How arthropod-borne pathogens evade and exploit the host defense system of the skin. J. Investif. Dermatol. 2014, 134, 1211–1219. [Google Scholar] [CrossRef]
- Schnell, G.; Boeuf, A.; Westermann, B.; Jaulhac, B.; Carapito, C.; Boulanger, N.; Ehret-Sabatier, L. Discovery and targeted proteomics on cutaneous biopsies: A promising work toward an early diagnosis of Lyme disease. Mol. Cell. Proteom. 2015, 14, 1254–1264. [Google Scholar] [CrossRef] [PubMed]
- Kern, A.; Schnell, G.; Bernard, Q.; Bœuf, A.; Jaulhac, B.; Collin, E.; Barthel, C.; Ehret-Sabatier, L.; Boulanger, N. Heterogeneity of borrelia burgdorferi sensu stricto population and its involvement in borrelia pathogenicity: Study on murine model with specific emphasis on the skin interface. PLoS ONE 2015, 10, e0133195. [Google Scholar] [CrossRef] [PubMed]
- Nebbak, A.; Dahmana, H.; Almeras, L.; Raoult, D.; Boulanger, N.; Jaulhac, B.; Mediannikov, O.; Parola, P. Co-infection of bacteria and protozoan parasites in Ixodes ricinus nymphs collected in the Alsace region, France. Ticks Tick. Borne. Dis. 2019, 10, 101241. [Google Scholar] [CrossRef] [PubMed]
- Gissot, M.; Hovasse, A.; Chaloin, L.; Schaeffer-Reiss, C.; Van Dorsselaer, A.; Tomavo, S. An evolutionary conserved zinc finger protein is involved in Toxoplasma gondii mRNA nuclear export. Cell. Microbiol. 2017, 19, e12644. [Google Scholar] [CrossRef] [PubMed]
- Grillon, A.; Westermann, B.; Cantero, P.; Jaulhac, B.; Voordouw, M.; Kapps, D.; Collin, E.; Barthel, C.; Ehret-Sabatier, L.; Boulanger, N. Identification of Borrelia protein candidates in mouse skin for potential diagnosis of disseminated Lyme borreliosis. Sci. Rep. 2017, 7, 16719. [Google Scholar] [CrossRef]
- Di, L.; Pagan, P.E.; Packer, D.; Martin, C.L.; Akther, S.; Ramrattan, G.; Mongodin, E.F.; Fraser, C.M.; Schutzer, S.E.; Luft, B.J.; et al. BorreliaBase: A phylogeny-centered browser of Borrelia genomes. BMC Bioinform. 2014, 15, 233. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Embers, M.E.; Narasimhan, S. Vaccination against Lyme disease: Past, present, and future. Front. Cell. Infect. Microbiol. 2013, 3. [Google Scholar] [CrossRef]
- Rogovskyy, A.S.; Bankhead, T. Variable VlsE is critical for host reinfection by the lyme disease spirochete. PLoS ONE 2013, 8, e61226. [Google Scholar] [CrossRef]
- Kraiczy, P.; Stevenson, B. Complement regulator-acquiring surface proteins of Borrelia burgdorferi: Structure, function and regulation of gene expression. Ticks Tick. Borne. Dis. 2013, 4, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Elsner, R.A.; Hastey, C.J.; Olsen, K.J.; Baumgarth, N. Suppression of long-lived humoral immunity following borrelia burgdorferi infection. PLoS Pathog. 2015, 11, e1004976. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, L. Inner workings: Lyme disease vaccines face familiar challenges, both societal and scientific. Proc. Natl. Acad. Sci. USA 2019, 116, 19214–19217. [Google Scholar] [CrossRef]
- Comstedt, P.; Hanner, M.; Schüler, W.; Meinke, A.; Lundberg, U. Design and development of a novel vaccine for protection against Lyme borreliosis. PLoS ONE 2014, 9, e113294. [Google Scholar] [CrossRef] [PubMed]
- Comstedt, P.; Schüler, W.; Meinke, A.; Lundberg, U. The novel Lyme borreliosis vaccine VLA15 shows broad protection against Borrelia species expressing six different OspA serotypes. PLoS ONE 2017, 12, e0184357. [Google Scholar] [CrossRef] [PubMed]
- Barthold, S.; de Souza, M.; Janotka, J.; Smith, A.; Persing, D. Chronic Lyme borreliosis in the laboratory mouse. Am. J. Pathol. 1993, 143, 959–971. [Google Scholar]
- Margos, G.; Vollmer, S.A.; Ogden, N.H.; Fish, D. Population genetics, taxonomy, phylogeny and evolution of Borrelia burgdorferi sensu lato. Infect. Genet. Evol. 2011, 11, 1545–1563. [Google Scholar] [CrossRef]
- Aguero-Rosenfeld, M.E.; Nowakowski, J.; McKenna, D.F.; Carbonaro, C.A.; Wormser, G.P. Serodiagnosis in early Lyme disease. J. Clin. Microbiol. 1993, 31, 3090–3095. [Google Scholar] [CrossRef]
- Sultan, S.Z.; Manne, A.; Stewart, P.E.; Bestor, A.; Rosa, P.A.; Charon, N.W.; Motaleb, M.A. Motility is crucial for the infectious life cycle of borrelia burgdorferi. Infect. Immun. 2013, 81, 2012–2021. [Google Scholar] [CrossRef]
- Fikrig, E.; Barthold, S.W.; Marcantonio, N.; Deponte, K.; Kantor, F.S.; Flavell, R.A. Roles of OspA, OspB, and flagellin in protective immunity to Lyme borreliosis in laboratory mice. Infect. Immun. 1992, 60, 657–661. [Google Scholar] [CrossRef]
- Cui, B.; Liu, X.; Fang, Y.; Zhou, P.; Zhang, Y.; Wang, Y. Flagellin as a vaccine adjuvant. Expert Rev. Vaccines 2018, 17, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Meunier, M.; Guyard-Nicodème, M.; Vigouroux, E.; Poezevara, T.; Béven, V.; Quesne, S.; Amelot, M.; Parra, A.; Chemaly, M.; Dory, D. A DNA prime/protein boost vaccine protocol developed against Campylobacter jejuni for poultry. Vaccine 2018, 36, 2119–2125. [Google Scholar] [CrossRef] [PubMed]
- Bruxelle, J.-F.; Mizrahi, A.; Hoÿs, S.; Collignon, A.; Janoir, C.; Péchiné, S. Clostridium difficile flagellin FliC: Evaluation as adjuvant and use in a mucosal vaccine against Clostridium difficile. PLoS ONE 2017, 12, e0187212. [Google Scholar] [CrossRef] [PubMed]
- Tilly, K.; Krum, J.G.; Bestor, A.; Jewett, M.W.; Grimm, D.; Bueschel, D.; Byram, R.; Dorward, D.; Vanraden, M.J.; Stewart, P.; et al. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 2006, 74, 3554–3564. [Google Scholar] [CrossRef]
- Brisson, D.; Baxamusa, N.; Schwartz, I.; Wormser, G.P. Biodiversity of Borrelia burgdorferi strains in tissues of Lyme disease patients. PLoS ONE 2011, 6, e22926. [Google Scholar] [CrossRef]
- Toledo, A.; Coleman, J.; Kuhlow, C.; Crowley, J.; Benach, J. The enolase of Borrelia burgdorferi is a plasminogen receptor released in outer membrane vesicles. Infect Immun. 2012, 80, 359–368. [Google Scholar] [CrossRef]
- Cappello, P.; Principe, M.; Bulfamante, S.; Novelli, F. Alpha-Enolase (ENO1), a potential target in novel immunotherapies. Front. Biosci. 2017, 22, 944–959. [Google Scholar] [CrossRef]
- Nogueira, S.V.; Smith, A.A.; Qin, J.-H.; Pal, U. A surface enolase participates in Borrelia burgdorferi-plasminogen interaction and contributes to pathogen survival within feeding ticks. Infect. Immun. 2012, 80, 82–90. [Google Scholar] [CrossRef]
- Nowalk, A.J.; Nolder, C.; Clifton, D.R.; Carroll, J.A. Comparative proteome analysis of subcellular fractions from Borrelia burgdorferi by NEPHGE and IPG. Proteomics 2006, 6, 2121–2134. [Google Scholar] [CrossRef]
- Lee, S.-H.; Lee, J.-H.; Park, H.-S.; Jang, W.-J.; Koh, S.-E.; Yang, Y.-M.; Kim, B.-J.; Kook, Y.-H.; Park, K.-H. Differentiation of Borrelia burgdorferi sensu lato through groEL gene analysis. FEMS Microbiol. Lett. 2003, 222, 51–57. [Google Scholar] [CrossRef]
- Jungblut, P.; Grabher, G.; Stöffler, G. Comprehensive detection of immunorelevant Borrelia garinii antigens by two-dimensional electrophoresis. Electrophoresis 1999, 20, 3611–3622. [Google Scholar] [CrossRef]
- Carrasco, S.; Yang, Y.; Troxell, B.; Yang, X.; Pal, U.; Yang, X. Borrelia burgdorferi elongation factor EF-Tu is an immunogenic protein during Lyme borreliosis. Emerg. Microbes Infect. 2015, 4, e54. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, J.; Kocan, K.; Almazan, C.; Blouin, E. Targeting the tick-pathogen interface for novel control strategies. Front Biosci. 2008, 13, 6947–6956. [Google Scholar] [CrossRef] [PubMed]
- Caljon, G.; Van Reet, N.; De Trez, C.; Vermeersch, M.; Pérez-Morga, D.; Van Den Abbeele, J. The Dermis as a delivery site of trypanosoma brucei for tsetse flies. PLoS Pathog. 2016, 12, e1005. [Google Scholar] [CrossRef] [PubMed]
- Ménard, R.; Tavares, J.; Cockburn, I.; Markus, M.; Zavala, F.; Amino, R. Looking under the skin: The first steps in malarial infection and immunity. Nat. Rev. Microbiol. 2013, 11, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Kuleš, J.; Horvatić, A.; Guillemin, N.; Galan, A.; Mrljak, V.; Bhide, M. New approaches and omics tools for mining of vaccine candidates against vector-borne diseases. Mol. Biosyst. 2016, 12, 2680–2694. [Google Scholar]
- Moxon, R.; Reche, P.A.; Rappuoli, R. Editorial: Reverse vaccinology. Front. Immunol. 2019, 10, 2776. [Google Scholar] [CrossRef]
- Gesslbauer, B.; Poljak, A.; Handwerker, C.; Schüler, W.; Schwendenwein, D.; Weber, C.; Lundberg, U.; Meinke, A.; Kungl, A.J. Comparative membrane proteome analysis of three Borrelia species. Proteomics 2012, 12, 845–858. [Google Scholar] [CrossRef]
- Bencurova, E.; Gupta, S.K.; Oskoueian, E.; Bhide, M.; Dandekar, T. Omics and bioinformatics applied to vaccine development against Borrelia. Mol. Omi. 2018, 14, 330–340. [Google Scholar] [CrossRef]
- Iyer, R.; Caimano, M.J.; Luthra, A.; Axline, D.; Corona, A.; Iacobas, D.A.; Radolf, J.D.; Schwartz, I. Stage-specific global alterations in the transcriptomes of Lyme disease spirochetes during tick feeding and following mammalian host adaptation. Mol. Microbiol. 2015, 95. [Google Scholar] [CrossRef]
- Caimano, M.J.; Groshong, A.M.; Belperron, A.; Mao, J.; Hawley, K.L.; Luthra, A.; Graham, D.E.; Earnhart, C.G.; Marconi, R.T.; Bockenstedt, L.K.; et al. The RpoS gatekeeper in Borrelia burgdorferi: An invariant regulatory scheme that promotes spirochete persistence in reservoir hosts and niche diversity. Front. Microbiol. 2019, 10, 1923. [Google Scholar] [CrossRef]
- Chen, Y.E.; Fischbach, M.A.; Belkaid, Y. Skin microbiota-host interactions. Nature 2018, 553, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Buckley, P.R.; Alden, K.; Coccia, M.; Chalon, A.; Collignon, C.; Temmerman, S.T.; Didierlaurent, A.M.; van der Most, R.; Timmis, J.; Andersen, C.A.; et al. Application of Modeling Approaches to Explore Vaccine Adjuvant Mode-of-Action. Front. Immunol. 2019, 10, 2150. [Google Scholar] [CrossRef] [PubMed]
- Donadei, A.; Kraan, H.; Ophorst, O.; Flynn, O.; O’Mahony, C.; Soema, P.C.; Moore, A.C. Skin delivery of trivalent Sabin inactivated poliovirus vaccine using dissolvable microneedle patches induces neutralizing antibodies. J. Control. Release 2019, 311–312, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Rouphael, N.G.; Paine, M.; Mosley, R.; Henry, S.; McAllister, D.V.; Kalluri, H.; Pewin, W.; Frew, P.M.; Yu, T.; Thornburg, N.J.; et al. The safety, immunogenicity, and acceptability of inactivated influenza vaccine delivered by microneedle patch (TIV-MNP 2015): A randomised, partly blinded, placebo-controlled, phase 1 trial. Lancet 2017, 390, 649–658. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).