Increased Vaccination Diversity Leads to Higher and Less-Variable Neutralization of TBE Viruses of the European Subtype
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Sera
2.2. Virus Strains
2.3. Virus Cultivation Quantification and Microserum Neutralization Tests
2.4. Phylogenetic Analysis of the TBEV-EU Genome Sequences
2.5. Statistical Analysis
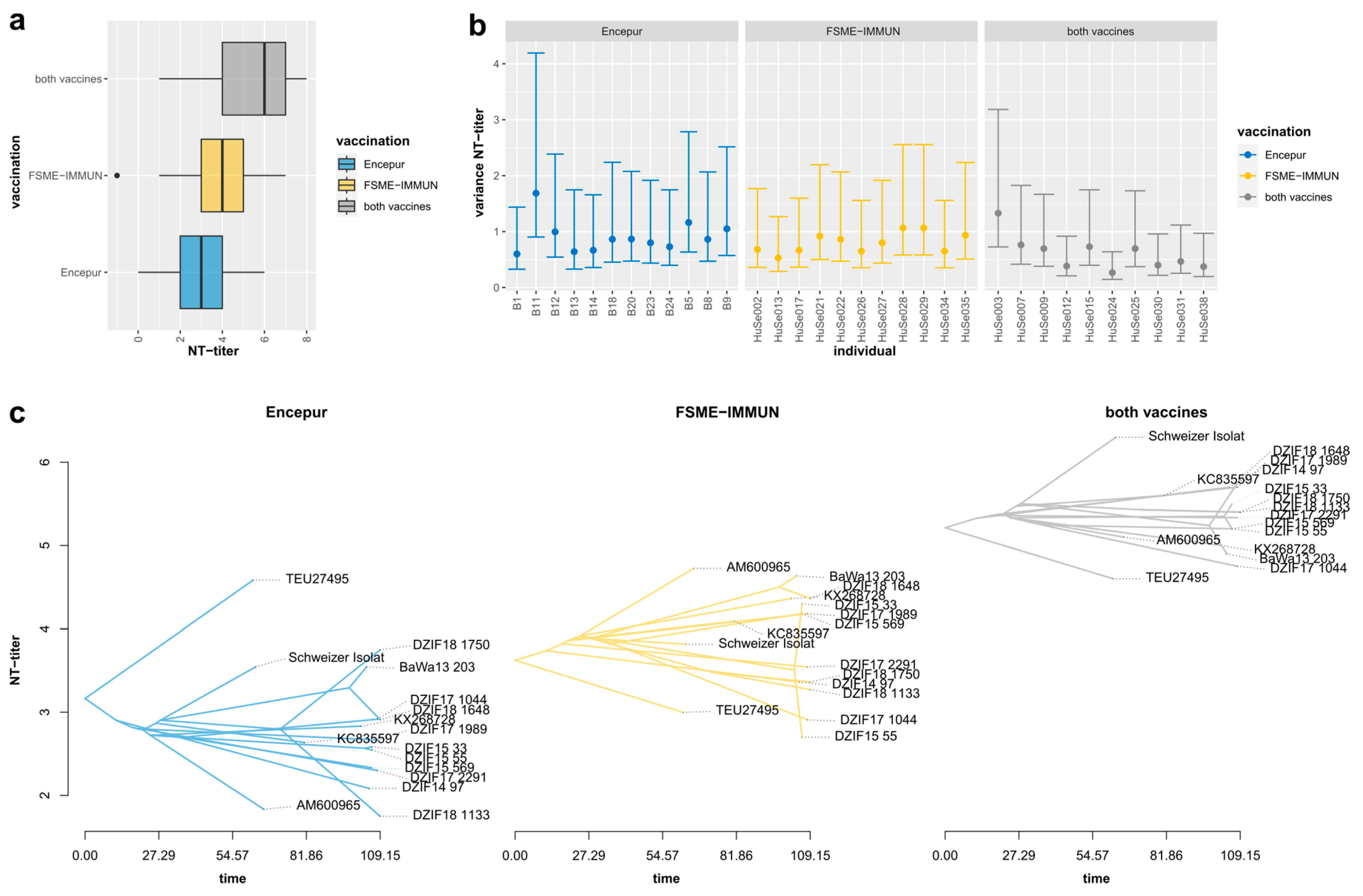
3. Results
3.1. High Inter-Serum Variation and Low Sampling Bias
3.2. Genotypes of the Diverse and Highly Divergent TBEV-EU Panel Were Neutralized Independent of Their Phylogenetic Position
3.3. Neutralization Titers Differed Significantly, Depending on Serum Immunization Profiles
3.4. Sequence Analysis of Possible Genetic Determinants for the NT-Titer Phenoytpes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dobler, G. Update Zur Frühsommer-Meningoenzephalitis. MMW-Fortschritte Med. 2020, 162, 40–43. [Google Scholar] [CrossRef]
- Boelke, M.; Bestehorn, M.; Marchwald, B.; Kubinski, M.; Liebig, K.; Glanz, J.; Schulz, C.; Dobler, G.; Monazahian, M.; Becker, S.C. First Isolation and Phylogenetic Analyses of Tick-Borne Encephalitis Virus in Lower Saxony, Germany. Viruses 2019, 11, 462. [Google Scholar] [CrossRef]
- Holding, M.; Dowall, S.D.; Medlock, J.M.; Carter, D.P.; Pullan, S.T.; Lewis, J.; Vipond, R.; Rocchi, M.S.; Baylis, M.; Hewson, R. Tick-Borne Encephalitis Virus, United Kingdom. Emerg. Infect. Dis. 2020, 26, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Camino, E.; Schmid, S.; Weber, F.; Pozo, P.; de Juan, L.; König, M.; Cruz-Lopez, F. Detection of Antibodies against Tick-Borne Encephalitis Flaviviruses in Breeding and Sport Horses from Spain. Ticks Tick-Borne Dis. 2020, 11, 101487. [Google Scholar] [CrossRef] [PubMed]
- Wallenhammar, A.; Lindqvist, R.; Asghar, N.; Gunaltay, S.; Fredlund, H.; Davidsson, Å.; Andersson, S.; Överby, A.K.; Johansson, M. Revealing New Tick-Borne Encephalitis Virus Foci by Screening Antibodies in Sheep Milk. Parasites Vectors 2020, 13, 185. [Google Scholar] [CrossRef] [PubMed]
- Brockmann, S.; Oehme, R.; Buckenmaier, T.; Beer, M.; Jeffery-Smith, A.; Spannenkrebs, M.; Haag-Milz, S.; Wagner-Wiening, C.; Schlegel, C.; Fritz, J.; et al. A Cluster of Two Human Cases of Tick-Borne Encephalitis (TBE) Transmitted by Unpasteurised Goat Milk and Cheese in Germany, May 2016. Eurosurveillance 2018, 23, 17-00336. [Google Scholar] [CrossRef]
- Holzmann, H.; Aberle, S.W.; Stiasny, K.; Werner, P.; Mischak, A.; Zainer, B.; Netzer, M.; Koppi, S.; Bechter, E.; Heinz, F.X. Tick-Borne Encephalitis from Eating Goat Cheese in a Mountain Region of Austria. Emerg. Infect. Dis. 2009, 15, 1671–1673. [Google Scholar] [CrossRef]
- Tick-Borne Encephalitis Outbreak Linked to Raw Milk Cheese in France. Available online: https://www.foodsafetynews.com/2020/06/tick-borne-encephalitis-outbreak-linked-to-raw-milk-cheese-in-france/ (accessed on 9 July 2020).
- Dai, X.; Shang, G.; Lu, S.; Yang, J.; Xu, J. A New Subtype of Eastern Tick-Borne Encephalitis Virus Discovered in Qinghai-Tibet Plateau, China. Emerg. Microbes Infect. 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Ecker, M.; Allison, S.L.; Meixner, T.; Heinz, F.X. Sequence Analysis and Genetic Classification of Tick-Borne Encephalitis Viruses from Europe and Asia. J. Gen. Virol. 1999, 80 Pt 1, 179–185. [Google Scholar] [CrossRef]
- Kovalev, S.; Mukhacheva, T.A. Reconsidering the Classification of Tick-Borne Encephalitis Virus within the Siberian Subtype Gives New Insights into Its Evolutionary History. Infect. Genet. Evol. 2017, 55, 159–165. [Google Scholar] [CrossRef]
- Bogovic, P.; Strle, F. Tick-Borne Encephalitis: A Review of Epidemiology, Clinical Characteristics, and Management. World J. Clin. Cases WJCC 2015, 3, 430–441. [Google Scholar] [CrossRef]
- Ruzek, D.; Avšič Županc, T.; Borde, J.; Chrdle, A.; Eyer, L.; Karganova, G.; Kholodilov, I.; Knap, N.; Kozlovskaya, L.; Matveev, A.; et al. Tick-Borne Encephalitis in Europe and Russia: Review of Pathogenesis, Clinical Features, Therapy, and Vaccines. Antiviral Res. 2019, 164, 23–51. [Google Scholar] [CrossRef]
- Dobler, G.; Gniel, D.; Petermann, R.; Pfeffer, M. Epidemiology and Distribution of Tick-Borne Encephalitis. Wien. Med. Wochenschr. 2012, 162, 230–238. [Google Scholar] [CrossRef]
- Kollaritsch, H.; Paulke-Korinek, M.; Holzmann, H.; Hombach, J.; Bjorvatn, B.; Barrett, A. Vaccines and Vaccination against Tick-Borne Encephalitis. Expert Rev. Vaccines 2012, 11, 1103–1119. [Google Scholar] [CrossRef]
- Gritsun, T.S.; Lashkevich, V.A.; Gould, E.A. Tick-Borne Encephalitis. Antivir. Res. 2003, 57, 129–146. [Google Scholar] [CrossRef]
- Mansfield, K.L.; Johnson, N.; Phipps, L.P.; Stephenson, J.R.; Fooks, A.R.; Solomon, T. Tick-Borne Encephalitis Virus—A Review of an Emerging Zoonosis. J. Gen. Virol. 2009, 90, 1781–1794. [Google Scholar] [CrossRef]
- Lang, D.; Chitimia-Dobler, L.; Bestehorn-Willmann, M.; Lindau, A.; Drehmann, M.; Stroppel, G.; Hengge, H.; Mackenstedt, U.; Kaier, K.; Dobler, G.; et al. The Emergence and Dynamics of Tick-Borne Encephalitis Virus in a New Endemic Region in Southern Germany. Microorganisms 2022, 10, 2125. [Google Scholar] [CrossRef]
- Kutschera, L.S.; Wolfinger, M.T. Evolutionary Traits of Tick-Borne Encephalitis Virus: Pervasive Non-Coding RNA Structure Conservation and Molecular Epidemiology. Virus Evol. 2022, 8, veac051. [Google Scholar] [CrossRef]
- WHO|Vaccine Position Papers. Available online: http://www.who.int/immunization/documents/positionpapers/en/ (accessed on 30 July 2020).
- Klockmann, U.; Krivanec, K.; Stephenson, J.; Hilfenhaus, J. Protection against European Isolates of Tick-Borne Encephalitis Virus after Vaccination with a New Tick-Borne Encephalitis Vaccine. Vaccine 1991, 9, 210–212. [Google Scholar] [CrossRef]
- Mandl, C.W.; Heinz, F.X.; Stöckl, E.; Kunz, C. Genome Sequence of Tick-Borne Encephalitis Virus (Western Subtype) and Comparative Analysis of Nonstructural Proteins with Other Flaviviruses. Virology 1989, 173, 291–301. [Google Scholar] [CrossRef]
- Wallner, G.; Mandl, C.W.; Kunz, C.; Heinz, F.X. The Flavivirus 3’-Noncoding Region: Extensive Size Heterogeneity Independent of Evolutionary Relationships among Strains of Tick-Borne Encephalitis Virus. Virology 1995, 213, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, K.L.; Horton, D.L.; Johnson, N.; Li, L.; Barrett, A.D.T.; Smith, D.J.; Galbraith, S.E.; Solomon, T.; Fooks, A.R. Flavivirus-Induced Antibody Cross-Reactivity. J. Gen. Virol. 2011, 92, 2821–2829. [Google Scholar] [CrossRef]
- Saron, W.A.A.; Rathore, A.P.S.; Ting, L.; Ooi, E.E.; Low, J.; Abraham, S.N.; John, A.L.S. Flavivirus Serocomplex Cross-Reactive Immunity Is Protective by Activating Heterologous Memory CD4 T Cells. Sci. Adv. 2018, 4, eaar4297. [Google Scholar] [CrossRef]
- Antigenic Variation among Members of the Tick-Borne Encephalitis Complex|Microbiology Society. Available online: https://www.microbiologyresearch.org/content/journal/jgv/10.1099/0022-1317-65-1-81;jsessionid=s_-q09J2Y6hqMFua3VCKkV2E.mbslive-10-240-10-152 (accessed on 1 February 2021).
- McAuley, A.J.; Sawatsky, B.; Ksiazek, T.; Torres, M.; Korva, M.; Lotrič-Furlan, S.; Avšič-Županc, T.; von Messling, V.; Holbrook, M.R.; Freiberg, A.N.; et al. Cross-Neutralisation of Viruses of the Tick-Borne Encephalitis Complex Following Tick-Borne Encephalitis Vaccination and/or Infection. NPJ Vaccines 2017, 2, 5. [Google Scholar] [CrossRef]
- Mandl, C.W.; Holzmann, H.; Meixner, T.; Rauscher, S.; Stadler, P.F.; Allison, S.L.; Heinz, F.X. Spontaneous and Engineered Deletions in the 3′ Noncoding Region of Tick-Borne Encephalitis Virus: Construction of Highly Attenuated Mutants of a Flavivirus. J. Virol. 1998, 72, 2132–2140. [Google Scholar] [CrossRef]
- Beck, Y.; Fritz, R.; Orlinger, K.; Kiermayr, S.; Ilk, R.; Portsmouth, D.; Pöllabauer, E.-M.; Löw-Baselli, A.; Hessel, A.; Kölch, D.; et al. Molecular Basis of the Divergent Immunogenicity of Two Pediatric Tick-Borne Encephalitis Virus Vaccines. J. Virol. 2016, 90, 1964–1972. [Google Scholar] [CrossRef]
- Rockstroh, A.; Moges, B.; Berneck, B.S.; Sattler, T.; Revilla-Fernández, S.; Schmoll, F.; Pacenti, M.; Sinigaglia, A.; Barzon, L.; Schmidt-Chanasit, J.; et al. Specific Detection and Differentiation of Tick-borne Encephalitis and West Nile Virus Induced IgG Antibodies in Humans and Horses. Transbound. Emerg. Dis. 2019, 66, 1701–1708. [Google Scholar] [CrossRef]
- Bradt, V.; Malafa, S.; von Braun, A.; Jarmer, J.; Tsouchnikas, G.; Medits, I.; Wanke, K.; Karrer, U.; Stiasny, K.; Heinz, F.X. Pre-Existing Yellow Fever Immunity Impairs and Modulates the Antibody Response to Tick-Borne Encephalitis Vaccination. NPJ Vaccines 2019, 4, 38. [Google Scholar] [CrossRef]
- Dobler, G.; Bestehorn, M.; Antwerpen, M.; Överby-Wernstedt, A. Complete Genome Sequence of a Low-Virulence Tick-Borne Encephalitis Virus Strain. Genome Announc. 2016, 4, e01145-16. [Google Scholar] [CrossRef]
- Weidmann, M.; Frey, S.; Freire, C.C.M.; Essbauer, S.; Růžek, D.; Klempa, B.; Zubrikova, D.; Vögerl, M.; Pfeffer, M.; Hufert, F.T.; et al. Molecular Phylogeography of Tick-Borne Encephalitis Virus in Central Europe. J. Gen. Virol. 2013, 94, 2129–2139. [Google Scholar] [CrossRef]
- Baer, A.; Kehn-Hall, K. Viral Concentration Determination Through Plaque Assays: Using Traditional and Novel Overlay Systems. J. Vis. Exp. JoVE 2014, 93, e52065. [Google Scholar] [CrossRef]
- Haut, M.; Girl, P.; Oswald, B.; Romig, T.; Obiegala, A.; Dobler, G.; Pfeffer, M. The Red Fox (Vulpes vulpes) as Sentinel for Tick-Borne Encephalitis Virus in Endemic and Non-Endemic Areas. Microorganisms 2020, 8, 1817. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Single-Cell Genomes and Mini-Metagenomes From Chimeric MDA Products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for Large-Scale Multiple Sequence Alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Sagulenko, P.; Puller, V.; Neher, R.A. TreeTime: Maximum-Likelihood Phylodynamic Analysis. Virus Evol. 2018, 4, vex042. [Google Scholar] [CrossRef]
- Revell, L.J. Phytools: An R Package for Phylogenetic Comparative Biology (and Other Things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Yu, G. Data Integration, Manipulation and Visualization of Phylogenetic Treess, 1st ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2022. [Google Scholar]
- Yu, G. Using Ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Wang, L.-G.; Lam, T.T.-Y.; Xu, S.; Dai, Z.; Zhou, L.; Feng, T.; Guo, P.; Dunn, C.W.; Jones, B.R.; Bradley, T.; et al. Treeio: An R Package for Phylogenetic Tree Input and Output with Richly Annotated and Associated Data. Mol. Biol. Evol. 2020, 37, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Kahle, D.; Wickham, H. Ggmap: Spatial Visualization with Ggplot2. R J. 2013, 5, 144–161. [Google Scholar] [CrossRef]
- Padgham, M.; Rudis, B.; Lovelace, R.; Salmon, M. Osmdata. J. Open Source Softw. 2017, 2, 1. [Google Scholar] [CrossRef]
- Blomberg, S.P.; Garland, T.; Ives, A.R. Testing for Phylogenetic Signal in Comparative Data: Behavioral Traits Are More Labile. Evol. Int. J. Org. Evol. 2003, 57, 717–745. [Google Scholar] [CrossRef]
- Pennell, M.W.; Eastman, J.M.; Slater, G.J.; Brown, J.W.; Uyeda, J.C.; FitzJohn, R.G.; Alfaro, M.E.; Harmon, L.J. Geiger v2.0: An Expanded Suite of Methods for Fitting Macroevolutionary Models to Phylogenetic Trees. Bioinformatics 2014, 30, 2216–2218. [Google Scholar] [CrossRef]
- Kayser, M.; Klein, H.; Paasch, I.; Pilaski, J.; Blenk, H.; Heeg, K. Human Antibody Response to Immunization with 17D Yellow Fever and Inactivated TBE Vaccine. J. Med. Virol. 1985, 17, 35–45. [Google Scholar] [CrossRef]
- Garner-Spitzer, E.; Poellabauer, E.-M.; Wagner, A.; Guzek, A.; Zwazl, I.; Seidl-Friedrich, C.; Binder, C.J.; Stiasny, K.; Kundi, M.; Wiedermann, U. Obesity and Sex Affect the Immune Responses to Tick-Borne Encephalitis Booster Vaccination. Front. Immunol. 2020, 11, 860. [Google Scholar] [CrossRef]
- Janik, M.; Płaczkowska, S.; Woźniak, M.; Bil-Lula, I. Analysis of Multiple Risk Factors for Seronegative Rate of Anti-Tick-Borne Encephalitis Virus Immunization in Human Serum. Medicina 2020, 56, 244. [Google Scholar] [CrossRef]
- Baldovin, T.; Mel, R.; Bertoncello, C.; Carpenè, G.; Soppelsa, F.; Giliberti, A.; Baldo, V. Persistence of Immunity to Tick-Borne Encephalitis after Vaccination and Natural Infection. J. Med. Virol. 2012, 84, 1274–1278. [Google Scholar] [CrossRef]
- Hansson, K.E.; Rosdahl, A.; Insulander, M.; Vene, S.; Lindquist, L.; Gredmark-Russ, S.; Askling, H.H. Tick-Borne Encephalitis Vaccine Failures: A 10-Year Retrospective Study Supporting the Rationale for Adding an Extra Priming Dose in Individuals Starting at Age 50 Years. Clin. Infect. Dis. 2020, 70, 245–251. [Google Scholar] [CrossRef]
- Stiasny, K.; Aberle, J.; Keller, M.; Grubeck-Loebenstein, B.; Heinz, F.X. Age Affects Quantity but Not Quality of Antibody Responses after Vaccination with an Inactivated Flavivirus Vaccine against Tick-Borne Encephalitis. PLoS ONE 2012, 7, e34145. [Google Scholar] [CrossRef] [PubMed]
- Nygren, T.M.; Pilic, A.; Böhmer, M.M.; Wagner-Wiening, C.; Wichmann, O.; Harder, T.; Hellenbrand, W. Tick-Borne Encephalitis Vaccine Effectiveness and Barriers to Vaccination in Germany. Sci. Rep. 2022, 12, 11706. [Google Scholar] [CrossRef] [PubMed]
- Heinz, F.X.; Holzmann, H.; Essl, A.; Kundi, M. Field Effectiveness of Vaccination against Tick-Borne Encephalitis. Vaccine 2007, 25, 7559–7567. [Google Scholar] [CrossRef]
- Erber, W.; Khan, F.; Zavadska, D.; Freimane, Z.; Dobler, G.; Böhmer, M.M.; Jodar, L.; Schmitt, H.-J. Effectiveness of TBE Vaccination in Southern Germany and Latvia. Vaccine 2022, 40, 819–825. [Google Scholar] [CrossRef]
- Askling, H.H.; Vene, S.; Rombo, L.; Lindquist, L. Immunogenicity of Delayed TBE-Vaccine Booster. Vaccine 2012, 30, 499–502. [Google Scholar] [CrossRef]
- Steffen, R. Tick-Borne Encephalitis (TBE) in Children in Europe: Epidemiology, Clinical Outcome and Comparison of Vaccination Recommendations. Ticks Tick-Borne Dis. 2019, 10, 100–110. [Google Scholar] [CrossRef]
- Salat, J.; Mikulasek, K.; Larralde, O.; Pokorna Formanova, P.; Chrdle, A.; Haviernik, J.; Elsterova, J.; Teislerova, D.; Palus, M.; Eyer, L.; et al. Tick-Borne Encephalitis Virus Vaccines Contain Non-Structural Protein 1 Antigen and May Elicit NS1-Specific Antibody Responses in Vaccinated Individuals. Vaccines 2020, 8, 81. [Google Scholar] [CrossRef]
- Loew-Baselli, A.; Konior, R.; Pavlova, B.G.; Fritsch, S.; Poellabauer, E.; Maritsch, F.; Harmacek, P.; Krammer, M.; Barrett, P.N.; Ehrlich, H.J. Safety and Immunogenicity of the Modified Adult Tick-Borne Encephalitis Vaccine FSME-IMMUN®: Results of Two Large Phase 3 Clinical Studies. Vaccine 2006, 24, 5256–5263. [Google Scholar] [CrossRef]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological Memory to SARS-CoV-2 Assessed for up to 8 Months after Infection. Science 2021, 371, eabf4063. [Google Scholar] [CrossRef]
- Bröker, M.; Schöndorf, I. Are Tick-Borne Encephalitis Vaccines Interchangeable? Expert Rev. Vaccines 2006, 5, 461–466. [Google Scholar] [CrossRef]

| Sera | Vaccination | Count | Age | Sex | YF Vaccination |
|---|---|---|---|---|---|
| HuSe002 | FSME-IMMUN® | 5 | 30–40 | Male | Vaccinated |
| HuSe013 | FSME-IMMUN® | 3 | 20–30 | Male | No vaccination |
| HuSe017 | FSME-IMMUN® | 3 | 40–60 | Female | No vaccination |
| HuSe021 | FSME-IMMUN® | 3 | 20–30 | Female | No vaccination |
| HuSe022 | FSME-IMMUN® | 4 | 40–60 | Female | Vaccinated |
| HuSe026 | FSME-IMMUN® | 5 | 30–40 | Male | Vaccinated |
| HuSe027 | FSME-IMMUN® | 6 | 30–40 | Female | No vaccination |
| HuSe028 | FSME-IMMUN® | 6 | 30–40 | Female | No vaccination |
| HuSe029 | FSME-IMMUN® | 5 | 20–30 | Male | Vaccinated |
| HuSe035 | FSME-IMMUN® | 3 | 20–30 | Female | No vaccination |
| HuSe034 | FSME-IMMUN® | 6 | 40–60 | Female | Vaccinated |
| HuSe003 | MIX | 5 | 40–60 | Male | Vaccinated |
| HuSe007 | MIX | 5 | 30–40 | Female | Vaccinated |
| HuSe009 | MIX | 6 | 30–40 | Female | Vaccinated |
| HuSe012 | MIX | 5 | 30–40 | Female | No vaccination |
| HuSe015 | MIX | 5 | 20–30 | Male | No vaccination |
| HuSe024 | MIX | 5 | 20–30 | Female | No vaccination |
| HuSe025 | MIX | 5 | 40–60 | Female | No vaccination |
| HuSe030 | MIX | 7 | 30–40 | Male | No vaccination |
| HuSe031 | MIX | 3 | 30–40 | Female | Vaccinated |
| HuSe038 | MIX | 4 | 20–30 | Male | No vaccination |
| B1 | Encepur® | 4 | 20–30 | Female | No vaccination |
| B5 | Encepur® | 3 | 20–30 | Female | No vaccination |
| B8 | Encepur® | 5 | 20–30 | Male | No vaccination |
| B9 | Encepur® | 4 | 20–30 | Female | No vaccination |
| B11 | Encepur® | 5 | 20–30 | Female | No vaccination |
| B12 | Encepur® | 4 | 20–30 | Female | No vaccination |
| B13 | Encepur® | 6 | 20–30 | Female | No vaccination |
| B14 | Encepur® | 4 | 20–30 | Male | No vaccination |
| B18 | Encepur® | 6 | 20–30 | Female | No vaccination |
| B20 | Encepur® | 4 | 20–30 | Male | No vaccination |
| B23 | Encepur® | 4 | 20–30 | Female | No vaccination |
| B24 | Encepur® | 5 | 40–60 | Female | No vaccination |
| Strain ID | Acc. | Country | Region/City | Year of Isolation | Isolation Source | Passage History | Reference | Sequencing Method |
|---|---|---|---|---|---|---|---|---|
| K2 (Karlsruhe); K23 | AM600965 | Germany | Karlsruhe | 1975 | Ixodes ricinus | 4. BMB 1. A549 | [21] provided by FX Heinz, Vienna, Austria | Amplicon sequencing with Illumina TruSeq |
| Neudörfl | TEU27495 | Austria | Neudoerfl | 1971 | Ixodes ricinus | Unknown | [22,23] | Amplicon sequencing with Illumina TruSeq |
| DZIF14 97 | Austria | Aschau/Zillertal | 2014 | Ixodes ricinus | A549 Passage 01 | Isolate of IMB | Amplicon sequencing with Illumina TruSeq | |
| DZIF15 33 | Germany/Austria | Eglsee/Kufstein | 2015 | Ixodes ricinus | A549 Passage 01 | Isolate of IMB | Amplicon sequencing with Illumina TruSeq | |
| DZIF15 569 | Austria | Wald/Pitztal | 2015 | Ixodes ricinus | A549 Passage 01 | Isolate of IMB | Amplicon sequencing with Illumina TruSeq | |
| DZIF17 1044 | Germany | Mühlau | 2017 | Ixodes ricinus | A549 Passage 01 | Isolate of IMB | Amplicon sequencing with Illumina TruSeq | |
| DZIF17 1989 | Germany | Petting | 2017 | Ixodes ricinus | A549 Passage 01 | Isolate of IMB | Amplicon sequencing with Illumina TruSeq | |
| DZIF18 1133 | Italy | Tres | 2018 | Ixodes ricinus | A549 Passage 01 | Isolate of IMB | Amplicon sequencing with Illumina TruSeq | |
| BaWa11 171 | KX268728 | Germany | Heselbach | 2011 | Ixodes ricinus | A549 Passage 01 | [32] | Sanger sequencing |
| Schweizer_Isolat (40) | Switzerland | Schaffhausen | 1972 | Ixodes ricinus | 1. BMB 1. A549 | provided by Franz X. Heinz | Amplicon sequencing with Illumina TruSeq | |
| BaWa13 203 | Germany | Haselmühl | 2013 | Ixodes ricinus | A549 Passage 01 | Isolate of IMB | Amplicon sequencing with Illumina TruSeq | |
| DZIF17 2291 | Germany | Nürnberg | 2017 | Ixodes ricinus | A549 Passage 01 | Isolate of IMB | Amplicon sequencing with Illumina TruSeq | |
| DZIF18 1750 | Germany | Gleissenbach | 2018 | Ixodes ricinus | A549 Passage 01 | Isolate of IMB | Amplicon sequencing with Illumina TruSeq | |
| DZIF18 1648 | Germany | Schnaittenbach | 2018 | Ixodes ricinus | A549 Passage 01 | Isolate of IMB | Amplicon sequencing with Illumina TruSeq | |
| CG/223_1990 | KC835597 | Slovakia | Zahorska Ves | 1990 | Myodes glareolus | 5. BMB 3. Vero 1. A549 | [33] | Sanger sequencing |
| DZIF15 55 | Germany/Austria | Hechtsee | 2015 | Ixodes ricinus | A549 Passage 01 | Isolate of IMB | Amplicon sequencing with Illumina TruSeq |
| Neutralization Titers (Mean ± SD)/10 LOG2 | |
|---|---|
| Sex of the sample donors | |
| Female | n = 22 (66%) 3.79 ± 1.85 |
| Male | n = 11 (33%) 4.17 ± 1.99 |
| Wilcoxon Test | p = 0.063 |
| Yellow fever vaccination | |
| YF+ | n = 9 (27%) 3.8 ± 1.41 |
| YF− | n = 24 (73%) 3.96 ± 2.06 |
| Wilcoxon Test | p = 0.79 |
| Number of Vaccinations (nV) | |
| 3 | n = 6 (18%) 3.83 ± 1.68 |
| 4 | n = 8 (24%) 3.37 ± 1.93 |
| 5 | n = 12 (36%) 4.19 ± 1.98 |
| 6 | n = 6 (18%) 3.81 ± 1.73 |
| 7 | n = 1 (4%) 6 |
| Kendall’s rank correlation test | R = 0.109, p = 0.0018 ** |
| Generalized linear model | nV-coefficient = 0.2393 p = 0.00212 ** |
| Immunization profile | |
| Encepur | n = 12 (36.3%) 2.8 ± 1.43 |
| FSME-IMMUN | n = 11 (33.3%) 3.8 ± 1.62 |
| MIX | n = 10 (30.3%) 5.35 ± 1.74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bestehorn-Willmann, M.; Girl, P.; Greiner, F.; Mackenstedt, U.; Dobler, G.; Lang, D. Increased Vaccination Diversity Leads to Higher and Less-Variable Neutralization of TBE Viruses of the European Subtype. Vaccines 2023, 11, 1044. https://doi.org/10.3390/vaccines11061044
Bestehorn-Willmann M, Girl P, Greiner F, Mackenstedt U, Dobler G, Lang D. Increased Vaccination Diversity Leads to Higher and Less-Variable Neutralization of TBE Viruses of the European Subtype. Vaccines. 2023; 11(6):1044. https://doi.org/10.3390/vaccines11061044
Chicago/Turabian StyleBestehorn-Willmann, Malena, Philipp Girl, Franziska Greiner, Ute Mackenstedt, Gerhard Dobler, and Daniel Lang. 2023. "Increased Vaccination Diversity Leads to Higher and Less-Variable Neutralization of TBE Viruses of the European Subtype" Vaccines 11, no. 6: 1044. https://doi.org/10.3390/vaccines11061044
APA StyleBestehorn-Willmann, M., Girl, P., Greiner, F., Mackenstedt, U., Dobler, G., & Lang, D. (2023). Increased Vaccination Diversity Leads to Higher and Less-Variable Neutralization of TBE Viruses of the European Subtype. Vaccines, 11(6), 1044. https://doi.org/10.3390/vaccines11061044






