Abstract
Vaccines are one of the efficient means available so far for preventing and controlling the infection rate of COVID-19. Several researchers have focused on the whole virus’s (SARS-CoV-2) inactivated vaccines which are economically efficient to produce. In Pakistan, multiple variants of SARS-CoV-2 have been reported since the start of the pandemic in February 2020. Due to the continuous evolution of the virus and economic recessions, the present study was designed to develop an indigenous inactivated SARS-CoV-2 vaccine that might help not only to prevent the COVID-19 in Pakistan, it will also save the country’s economic resources. The SARS-CoV-2 were isolated and characterized using the Vero-E6 cell culture system. The seed selection was carried out using cross-neutralization assay and phylogenetic analysis. The selected isolate of SARS-CoV-2 (hCoV-19/Pakistan/UHSPK3-UVAS268/2021) was inactivated using beta-propiolactone followed by vaccine formulation using Alum adjuvant, keeping the S protein concentration as 5 μg/dose. The vaccine efficacy was evaluated by in vivo immunogenicity testing in laboratory animals and in in vitro microneutralization test. The phylogenetic analysis revealed that all the SARS-CoV-2 isolates reported from Pakistan nested into different clades, representing multiple introductions of the virus into Pakistan. The antisera raised against various isolates from different waves in Pakistan showed a varied level of neutralization titers. However, the antisera produced against a variant (hCoV-19/Pakistan/UHSPK3-UVAS268/2021; fourth wave) efficiently neutralized (1:64–1:512) all the tested SARS-CoV-2 isolates. The inactivated whole virus vaccine of SARS-CoV-2 was safe and it also elicited a protective immune response in rabbits and rhesus macaques on the 35th-day post-vaccination. The activity of neutralizing antibodies of vaccinated animals was found at 1:256–1:1024 at 35 days post-vaccination, indicating the effectiveness of the double-dose regime of the indigenous SARS-CoV-2 vaccine.
1. Introduction
The first human infection reported in China in 2019 with the severe acute respiratory syndrome (SARS) was identified as a novel β-coronavirus; however, its zoonotic origin is yet to be defined [1].The index case of SARS-CoV-2 in Pakistan was reported in Karachi on 26 February 2020, followed by the country-wide spread of the disease. More than 6200 people contracted the infection, and 111 deaths were reported during the first seven weeks of the commencement of the epidemic of COVID-19 in Pakistan [2].
The vaccines proved to be the only functional defense against the COVID-19 [3]. Moreover, it is also important to develop an indigenous immunologically efficient vaccine by analyzing the phylogeny and viral pathogenicity of prevalent strains of the virus in a country. Globally, four COVID-19 vaccine categories were produced and tested including whole virus inactivated vaccines, purified protein-based vaccines, mRNA, and vector-based vaccines [4]. Among the available vaccine categories, the inactivated cell culture-based vaccines are easy to develop [5]. Regardless of the type, vaccines produce adaptive (humoral and/or cellular) immune responses against the putative virus/bacterium/fungus which, in turn, prevent propagation of the pathogens in the host. Immune response generated by the vaccines, prior to the viral infection, needs to cease after some time for restoration of the individual’s health. Traditional, inactivated or live attenuated vaccines do not interfere with this balance, while mRNA vaccines that linger for more than four weeks may disturb this balance and could compromise the health of the recipient of the vaccine [6,7]. Moreover, the modern day expression vaccines are not efficient in reducing the risk of disease (<2%) [8]. It is important to note that the activated T-cells against one SARS-CoV-2 variant will efficiently cross react with any other SARS-CoV-2 variant [9]. Thus, the humoral responses should be used as a readout system of the effectivity to activate a meaningful adaptive immune response than thinking to be able to judge an in vivo effectiveness of the vaccine against relevant viral respiratory tract infection. The respiratory tract’s immune defense is chiefly based upon the protection rendered by innate immune system while the adaptive one has very limited access to contain respiratory infections [9,10,11].
Since February 2020, multiple lineages have been identified as the variant of concern (VOC), including B.1.1.7 (Alpha), B.1.351 (Beta), B.1.1.28.1 (Gamma), and B.1.617.2 (Delta). These variants have increased transmissibility to cause infection in the naive population and have potential to evade the body’s immune system. Due to the continuous evolution of the SARS-CoV-2 variants, it would be wise to develop vaccines from the most recent strains [12]. In Pakistan, 58.14% (132.48 million people have been vaccinated to date (covid.gov.pk), a considerable proportion of the population is still unvaccinated. Hence, the risk of re-emergence of SARS-CoV-2 is maintained. Therefore, it is the need of the hour to develop an indigenous SARS-CoV-2 vaccine in Pakistan.
The traditional vaccination approach with a whole inactivated virus is appreciable and feasible for a limited resource country such as Pakistan; therefore, the present study aimed to develop a whole virus vaccine against the indigenously circulating variant. The present study will also provide guidelines for developing countries for the inactivated vaccine production for future epidemics.
2. Materials and Methods
2.1. Ethical Statement
All the protocols used under the current experimental study were evaluated and approved by the Institutional Ethical Review Committee (DR/459) of the University of Veterinary and Animal Sciences (UVAS), Lahore. The experimental animals were handled according to the local and international ARRIVE guidelines.
2.2. Isolation of Indigenous SARS-CoV-2 Variant/s from the Clinical Samples
For the isolation of SARS-CoV-2, nasopharyngeal swabs (n = 780)s were collected from different areas of Punjab Province from March, 2020 to November, 2021 in a viral transport medium (VTM) kit (Copan Diagnostics, Murrieta, CA, USA). The samples were tested using real-time PCR (Thermo Fisher Scientific, Waltham, MA, USA) at the Biosafety Level 3 (BSL-3) facility at the Institute of Microbiology University of Veterinary and Animal Sciences, Lahore. A total of 20 nasopharyngeal swab samples with Ct < 25 were selected and processed for the isolation of SARS-CoV-2.
The Vero-E6 cell line (African monkey green kidney cells (ATCC CRL-1586)) was procured from the Defense Science and Technology Organization (DESTO) Pakistan and cultured in Dulbeco’s Modified Eagle Medium (DMEM) (Sigma–Aldrich Company, Burlington, MA, USA). The medium was supplemented with 10% fetal bovine serum (FBS) (Gibco, Waltham, MA, USA). Penicillin (100 IU/mL), streptomycin (100 µg/mL) and amphotericin B (250 μg/mL) (Biological Industries, Kibbutz Beit-Haemek, Israel) were added to the media to avoid contamination.
The representative samples were diluted and inoculated on a Vero-E6 cell line having 80% confluent monolayer and agitated gently for 1 h at 37 °C for adsorption. The DMEM media was added and kept at 37 °C in the presence of 5% CO2. The flask was observed for cytopathic effects (CPEs) daily.
2.3. Selection of Vaccine Candidates Based on Phylogeny and Cross Neutralization
Thirteen SARS-CoV-2 isolates were sequenced via next-generation sequencing [13] (The Quadram Institute, Norwich, UK). All sequences were uploaded to NCBI and GISAID databases and accession numbers (MW031799 to MW031803, EPIISL548942-EPIISL548948, EPIISL5063601) were obtained. The sequences of SARS-CoV-2 isolates were compared with publicly available SARS-CoV-2 sequence databases (GISAID and NCBI) for phylogenetic analysis. Large dataset phylogenies of newly sequenced SARS-CoV-2 isolates, and maximum likelihood phylogenetic tree was constructed using FastTree in Geneious v7.1.9 (Biomatters Ltd., Auckland, New Zealand). The dataset were then sub-sampled to 3395 and compared with the 13 sequenced isolates of SARS-CoV-2 of the present study.
For assessing the cross-neutralization, hyper-immune sera were raised for each of the variants in COVID-19 negative-laboratory animals (rabbits) at the BSL-3 animal containment facility [14].
2.4. Virus Titration
The tissue culture infective dose 50% (TCID50) of the indigenous SARS-CoV-2 vaccine candidate virus was calculated following the Reed and Muench method [15]. Briefly, the propagated virus was diluted 10-fold in DMEM (Gibco), and 100 μL from each dilution was seeded to each well of a microtiter plate with 80% confluency. The plate was incubated at 37 °C with 5% CO2, and cytopathic effects were observed after 72 h. As 100 uL of the virus was used to perform the TCID50, after obtaining the results, the final value was multiplied by 10 to assume the TCID50 per mL of virus.
2.5. Production of Inactivated SARS-CoV-2 Vaccine
The SARS-CoV-2 strain hCoV-19/Pakistan/UHSPK3-UVAS268/2021 was propagated with the multiplicity of infection (MOI: 0.01) in serum-free medium and incubated at 37 °C in a CO2 incubator. After CPEs (80–90%) were manifested, the virus was harvested by three freeze–thaw cycles and stored at −80 °C until further use.
For inactivation, the harvested SARS-CoV-2 virus was centrifuged at 2000 RCF for 10 min to separate the cellular debris. The supernatant was harvested and used for virus inactivation using β-propiolactone (BPL) (Solarbio® Life Sciences, Beijing, China) with a final concentration of 1:1600 [16]. The mixture was incubated at 4 °C for 10 h with continuous stirring at 100 rpm. The BPL was hydrolyzed by incubating the tubes at 37 °C. Viral inactivation was confirmed on a cell culture system by seven consecutive blind serial passages with a lack of development of CPEs.
The virus was purified by Pierce™ Protein Concentrator PES, 100 K MWC0 (Thermo Fisher Scientific, Waltham, MA, USA). Briefly, the inactivated virus fluid was centrifuged through a 100 K MWC0 filter. The retentate was retained for further processing. For vaccine formulation, different antigen concentrations were adjusted to 5 µg per dose as described previously [17]. The Imject™ Alum (Thermo Fisher Scientific, Waltham, MA, USA) was used as an adjuvant (250 μg per dose) for the immunogenicity studies in rabbits.
2.6. Protein Profile of Viral Antigen
The stability of the receptor binding domain of the S-Protein of the virus before and after inactivation was confirmed and quantified using the RayBio® COVID-19 S-Protein (S1RBD) ELISA kit following the manufacturer’s instructions.
2.7. Immunogenicity Studies in Mice, Rabbits, and Rhesus Macaques
The immunogenicity of the indigenous inactivated COVID-19 vaccine was evaluated for its capability to elicit neutralizing antibodies in mice, rabbits, and rhesus macaques. A total of 40 male mice (BALB/c) with weight 18–22 g and age of 8 weeks were procured from Animal Reproduction Department, UVAS. The mice were randomly divided into two groups: group I had 30 mice, and group II had ten mice. The mice in group I were immunized through the intramuscular route, and the booster dose was administered at 14 days post-vaccination. While group II was kept as a negative control and administered with an equal volume of sterile phosphate-buffered saline (PBS). Similarly, rabbits (male, n = 20) were divided into two groups viz group I (n = 15, vaccinated) and group II (n = 5, mock). The non-human primates (rhesus macaques: male, n = 6) were also divided into two groups viz group I (n = 5, vaccinated) and group II (n = 1, mock). Group I of rabbits and non-human primates were vaccinated by an intramuscular route at day 0, followed by a booster at 28 days post-vaccination. While group II of rabbits and non-human primates were administered with an equal volume of sterile PBS. Blood samples from experimental animals were collected on days 0, 7, 14, 21, 28, and 35 post priming and sera were separated and stored at −20 °C for immunological investigations [17].
2.8. SARS-CoV-2 Nucleoprotein-Based ELISA
The collected serum samples were tested for anti- SARS-CoV-2 antibodies using ID Screen® SARS-CoV-2 Double Antigen Multi-species ELISA kit (ID.vet, Grabels, France) as per manufacturer’s instructions [18]. Briefly, serum samples, positive and negative controls (25 uL each) were pipetted into respective wells of antigen-coated 96-well ELISA plates. The plates were then incubated at 37 °C for 45 min. The wells were washed thrice with a wash buffer and tapped over a paper towel to drain the excess buffer. Afterward, 100 uL of the freshly prepared conjugate was added into each well and incubated for 30 min at room temperature. The wells were again washed as described earlier. After that, 100 uL of HRP substrate was added into each well and incubated for 20 min at room temperature in a dark place. At the final stage, 100 uL of stop solution was added into each well, and the plates were read immediately at 450 nm. All the samples and controls were processed in triplicate to get the most accurate readings.
2.9. Micro Neutralization Assay of Mice, Rabbits, and Rhesus Macaques Serum
The collected sera were also checked for neutralizing antibodies against the SARS-CoV-2 virus. Briefly, flat bottom 96-well microtiter plates were seeded with (3 × 105 cells/mL) and incubated until 80–90% confluency was achieved. The serum samples were preheated at 56 °C for 30 min prior to use. Sera were two-fold diluted in a separate microtiter plate. Afterwards, the diluted sera (1:2 to 1:1024) were mixed with SARS-CoV-2 virus (100 TCID50) in equal volume and incubated for one hour at 37 °C and transferred to 96-well microtiter plate containing Vero E6 cells. The plate was incubated for 72 h at 37 °C in a CO2 incubator and observed daily for CPEs. The highest dilution of each serum sample which inhibited CPEs in 50% of the tested wells was identified as the TCID50 titer of that serum sample [19].
2.10. Toxicity Test in Rabbits and Mice
To determine the single-dose toxicity of the developed vaccine, 25 rabbits were procured and randomly divided into three groups (10 rabbits in groups A and B and five rabbits in group C). Groups A and B were immunized with a standard clinical dose (1000 SU/dose) and a high dose (2000 SU/dose) of inactivated COVID-19 vaccine, respectively, via intramuscular (I/M) route, while Group C was inoculated intramuscularly with normal saline. A similar procedure was adopted for 25 BALB/c mice injected with a total volume of 0.3 mL/dose through the I/M route. After each injection, a local tolerance test for 72 h was assessed [17].
The possible toxicity reaction of the target organ was assessed after administration of SARS-CoV-2 vaccine. A total of 20 rabbits with similar bodyweights were randomly divided into two groups (A and B). Groups A and B served as the treatment and control groups, respectively. The rabbits in groups A and B were administered with 0.5 mL/1000 SU/dose and 0.5 mL/dose of normal saline, respectively. The vaccine safety was assessed using intramuscular injections on days 1, 8, and 15 till four weeks post-last-administration. All animals were monitored for clinical signs and mortality throughout the dose toxicity test. Furthermore, all experimental animals were weighed periodically, and feed consumption was recorded. Samples for clinical biochemistry and hematology were taken on days 2, 21, and 28, as described earlier [17].
2.11. Statistical Analysis
For analysis of the data, GraphPad Prism version 8.4.3 software was used [20]. The statistical significance was assessed using the Kruskal–Wallis between the control and the vaccinated groups.
3. Results
3.1. Isolation of Indigenous SARS-CoV-2 Variant/s from the Clinical Samples
A total of 20 nasopharyngeal swab samples with Ct < 25 were selected from 780 samples and processed for the isolation of SARS-CoV-2. A total of 13 samples showed the cytopathic effect on the Vero E6 cell line, as shown in Table 1 and Figure 1.

Table 1.
Details of the clinical samples used for virus adaptation.
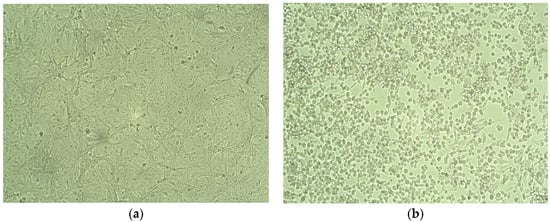
Figure 1.
Isolation of SARS-CoV-2. (a) Normal Vero Cells (b) Cytopathic effect of SARS-CoV-2 after 72 h of infection on vero cells showing plaques and disrupted monolayer.
3.2. Selection of Vaccine Candidates Based on Phylogeny and Cross Neutralization
A maximum likelihood phylogenetic tree was constructed for different waves separately. Our phylogenetic analysis revealed that multiple variants have been circulating in Pakistan since February 2020. The first wave of the SARS-CoV-2 infection was observed from February to May 2020. The results showed two major clades which were circulating during the first wave of COVID-19 in Pakistan. The second wave was observed from October to December 2020, and all reported sequences nested in a separate clade (Nextstrain clade classification). From May 2021 to June 2021, the Mu variant was predominantly circulating and nested in a separate clade, whereas from June to October 2021, the Delta variant was mainly found circulating and that was nested in separate clade (Figure 2).
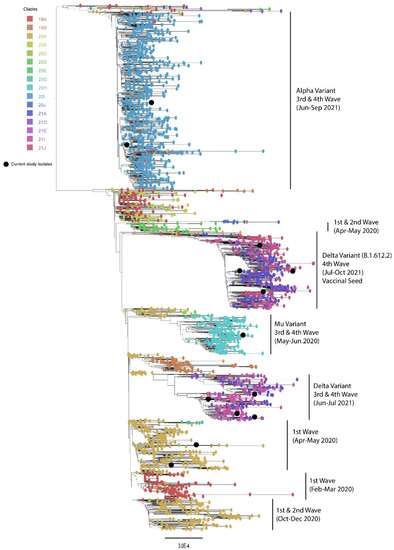
Figure 2.
Phylogenetic analysis of circulating SARS-CoV-2 virus strains in Pakistan (black balls) during the 1st–4th wave.
In addition to the genetic characterization of the SARS-CoV-2 isolates, anti-SARS-CoV-2 polyclonal sera were raised against the representative isolates in rabbits and cross neutralization results are shown in Table 2.

Table 2.
Cross-neutralization of different serum samples with viruses.
The hCoV-19/Pakistan/UHSPK3-UVAS268/2021 (EPIISL5063601) virus was selected for vaccine preparation. The virus was re-confirmed through real-time PCR and subsequently processed for virus inactivation.
3.3. TCID50 of the Vaccine Candidate Virus
The tissue culture infective dose 50% (TCID50) of the indigenous SARS-CoV-2 vaccine candidate virus was calculated as 107.5 virus particles.
3.4. SARS-CoV-2 Inactivated Vaccine Preparation
Different concentrations of the BPL (1:1000, 1:1300, 1:1600, and 1:2000) were used for the virus inactivation, and the 1:1600 concentration showed the complete inactivation of the virus having no CPEs in the seven blind passages. The virus was purified by Pierce™ Protein Concentrator PES, 100 K MWC0 (Thermo Fisher Scientific, Waltham, MA, USA). After concentrating the inactivated virus, the final concentration was found to be 1074 μg/mL, which was adjusted to 5 μg/dose of the vaccine.
3.5. Protein Profile of Viral Antigen
The quantification of the intact RBD of the S protein, immunologically determinant of SARS-CoV-2 virus was 1989.6 pg/mL.
3.6. SARS-CoV-2 Nucleoprotein-Based ELISA
ELISA results showed that the formulation having 5 μg/dose produced anti-SARS-CoV-2 antibodies in all animal models (mice, rabbits, and rhesus macaques).
3.7. Anti-SARS-CoV-2 Antibody Response in Animal Models
No adverse effects or clinical signs were observed during the experimentation in all the animal models (mice, rabbits, and rhesus macaques). The collected serum samples were processed for determination of presence of anti-SARS-CoV-2 immunoglobulin (IgG) antibodies. The results revealed that the inactivated SARS-CoV-2 vaccine developed in this study produced detectable IgG antibodies after three weeks. There was an increased level of antibodies IgG after 35 days of vaccination in all three animal models. The serum samples collected from all animals of the negative control group had no anti-SARS-CoV-2 IgG antibody, as shown in Figure 3.
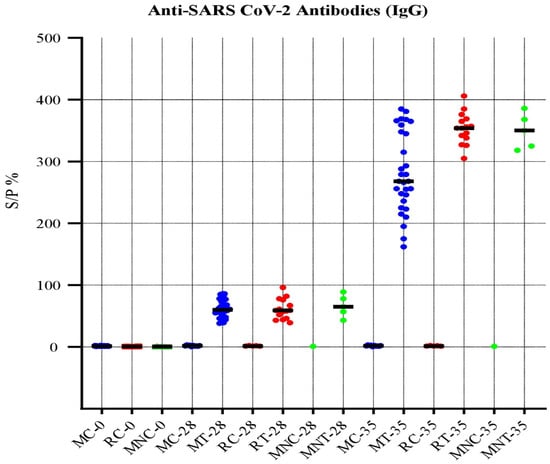
Figure 3.
Anti-SARS-CoV-2 antibody response of serum samples.
3.8. Micro Neutralization Assay
The results of microneutralization assay revealed that the serum samples of all vaccinated animals inhibited the development of CPEs by SARS-CoV-2 in Vero-E6 cells. The level of neutralizing antibodies on day 28 was 1:64 to 1:256, while on day 35, it was 1:256 to 1:1024 in all vaccinated animals. In comparison, serum samples from all the animals in control groups never showed any ability to neutralize the virus as found by the observation of CPEs.
3.9. Toxicity Test in Rabbits and Mice
The absence of clinical aberrant response and death was used as a measure of toxicity in the mice and rabbits. Our findings revealed no adverse effect on the animals’ bodyweight and feed intake. The primary organs and tissues of animals in each group were free of any gross abnormalities. Even after the administration of the double dose of vaccine, the results showed no abnormal drug administration effects, indicating that the maximum tolerated dose (MTD) was equivalent to or higher than 2000 SU/dose.
In order to determine the toxicity of SARS-CoV-2 vaccine after three administrations, no adverse reaction was found in clinical observations in rabbits of groups (A and B).
3.10. Post-Vaccination Blood Profile of Non-Human Primates
Results of the blood profile of non-human primates (rhesus macaques) on different days after the immunization revealed no difference in blood parameters among vaccinated and control animals as shown in Table 3 and Table 4.

Table 3.
Blood profile (post COVID-19 vaccine) of non-human primates (rhesus macaques).

Table 4.
LFT’s and RFT’s results (post COVID-19 vaccine) of non-human primates (rhesus macaques).
4. Discussion
Globally, several vaccines have been developed and granted permission for emergency use to combat the spread of SARS-CoV-2, including viral vectored vaccines (ChAdOx1 nCoV-19 and Ad-26.COV.2.S), the mRNA vaccine (mRNA-1273), the DNA vaccine (INO-4800), and inactivated vaccines (PiCoVacc and BBIBPCorV). However, clinical trials (phase III clinical trials) for the (mRNA-1273) and BNT162b2 (BioNTech/Pfizer) have been completed so far [17]. Due to rigorous cold-chain requirements of mRNA-based vaccines and their high prices, the worldwide access to these vaccines is limited, particularly in lower-income and lower-middle income countries such as Pakistan [21]. Therefore, more SARS-CoV-2 vaccines are required to fulfill the worldwide demand and to achieve herd immunity. Most vaccines developed are based on the SARS-CoV-2 spike protein only [22]. The advantage of inactivated whole virus vaccine over the other vaccines is that the immune system response to the inactivated whole virus vaccine would target all of the virus’s proteins, not only the spike protein. Moreover, the whole inactivated virus vaccine provides complex antigenicity and better adaptive or humoral immune response for better cross-reactivity [23]. Traditional vaccines such as the inactivated vaccines have been better explored and the risks associated with their use are better defined, whereas the risk/benefit ratio of mRNA vaccines are not well defined. Some of the recent publications have indicated that the risk of getting sick or having unwanted effects after using the mRNA-based vaccines is higher [24]. It is also supported from the previous publications that expression of spike protein in mRNA vaccine is uncontrollable and possibly disturbs the adaptive immune response due to the presence of artificially stabilized mRNA of the vaccine in the germinal center of secondary lymph organs [6,7].
Previously reported literature has mentioned that the vaccines developed using indigenous strains of SARS-CoV-2 are safe and effective in producing protective antibodies [25]. The current study demonstrated that indigenous inactivated whole virion (SARS-CoV-2) vaccine efficiently elicit immune responses in three animal models (mice, rabbits, and rhesus macaques) and produce neutralizing antibodies against the SARS-CoV-2 virus. Here, our results showed that the efficient immune response among experimental animals, including mice, rabbits, and rhesus macaques, was observed after a two-dose immunization regimen of the indigenous inactivated COVID-19 vaccine.
The selection of indigenous SARS-CoV-2 was carried out by phylogenetic analysis. The results revealed the circulation of several variants, which nested well in separate clades during the same period. This shows the diversification of circulating SARS-CoV-2, which might indicate the direct introduction of SARS-CoV-2 into Pakistan through international trade/travelers or globalization. The phylogenetic analysis of this study is in accordance with the previous reports which have reported the circulation of seven clades of SARS-CoV-2 in Pakistan during the first three waves [26]. These different variants of SARS-CoV-2 originated mainly due to the mutations in the spike protein, which ultimately leads to concerns of reduced vaccine efficacy [27,28].
The vaccine was formulated with aluminum hydroxide, which is considered as the safest and most widely used adjuvant [29,30]. In present study, the safety evaluation of the vaccine formulated with aluminum hydroxide showed no toxic effect in all the experimental animals (mice, rabbits, and rhesus macaques). In general, aluminum hydroxide adjuvant formulations produced significant titers of neutralizing antibodies (NAbs). Neutralizing antibodies are essential in protecting against SARS-CoV-2 infection and have been used to assess vaccination effectiveness as an immunological correlate of protection [31]. In preclinical trials, inactivated SARS-CoV-2 vaccine candidates were demonstrated to generate substantial levels of antigen binding and NAb titers [32,33].
Preclinical studies of the vaccine are critical to evaluate the immunogenicity of the vaccine. A mouse model and non-human primates have been widely used in different studies to evaluate the immunogenicity and effectiveness of the SARS-CoV-2 vaccines [34,35,36]. The immunogenicity of indigenous inactivated whole virion SARS-CoV-2 vaccine in all three animal models (mice, rabbit, monkey) showed a detectable level of antibodies IgG at 28 days of vaccination. In comparison, many studies showed a detectable level of antibodies at 21 days of vaccination [34,36,37,38,39]. This difference may be due to using different ELISA kits for the SARS-CoV-2 nucleoprotein IgG in the tested animal models. In other SARS-CoV-2 inactivated vaccine candidates such as PiCoVacc and BBIBP-CorV, studied in a non-human primate model, the NAb were observed from the first and second week, respectively, with a period of detection lasting for five weeks [36,40].
For the viral vaccine evaluation, neutralization ability is an important indicator. In the study, we also presented the in vitro evaluation of the vaccine using a microneutralization assay. For the subject matter, serum samples of vaccinated animals were used to neutralize SARS-CoV-2 and all the samples neutralized the virus and did not present CPEs on Vero-E6 cells. The antibody titer of all experimental animals vaccinated with SARS-CoV-2 inactivated vaccine was found to be 1:64 to 1:256 at the 28th day of vaccination while this titer was increased at 35th day of vaccination (1:256 to 1:1024). A previous study also reported that the booster immunization produces a high titer of neutralizing antibodies against SARS-CoV in mice (BALB/c) [41]. Without an effective antiviral drug against SARS-CoV-2, vaccines with good potency and safety are needed to achieve herd immunity [42].
The present study also had some limitations including the unavailability of data on T helper cells, which can also contribute towards the humoral response. To overcome this limitation, we determined the antibodies in response to the SARS-CoV-2 vaccine in three different animal models. Other limitations included the use of a small number of rhesus macaques for in vivo studies, unavailability of data on cross-neutralizing ability of this NAb with other related viruses such as SARS-CoV. However, previous studies have reported the cross-neutralizing ability of two SARS-CoV NAb to neutralize SARS-CoV-2 [42].
In conclusion, this study presents molecular characterization and selection of an indigenous SARS-CoV-2 variant to develop whole virion-based inactivated vaccine against SARS-CoV-2 in Pakistan. The study also reported the preclinical immunogenicity evaluation of this SARS-CoV-2 vaccine formulated in aluminum hydroxide adjuvant in three animal models including (mice, rabbits, and rhesus macaques). The present study revealed that inactivated SARS-CoV-2 vaccine with a two-dose regimen can elicit a robust immune response and may protect the animals challenged with SARS-CoV-2 infection, which requires further investigations. Developing indigenous SARS-CoV-2 vaccines may also save the budget allocated to import the SARS-CoV-2 vaccine in Pakistan. The present study will also provide a platform for the development of such viral vaccines in the country in future.
Author Contributions
M.N. and T.Y. designed the study; M.W.A., M.A., M.F.S. and M.A.A. collected data and samples; M.W.A., M.F.S., M.A. and M.A.A. performed the experiments; M.F.S., M.A., M.A.B.S. and N.M. conducted the formal analysis; T.Y., A.A.A. and M.H.M. provided overall supervision of the project; M.W.A. and M.A.B.S. were responsible for writing the original draft; M.W.A., M.A. and M.A.B.S. were responsible for reviewing and editing the drafts. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The animal study protocols were approved by the Institutional Review Board (or Ethics Committee) of UVAS (DR/459).
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data are available.
Acknowledgments
We are thankful to all the laboratories that submitted their sequences to the GISAID platforms. We are also grateful to the GISAID team for managing this platform.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| Severe acute respiratory syndrome coronavirus 2 | SARS-CoV-2 |
| Coronavirus infectious disease 19 | COVID-19 |
| Human coronavirus 19 | hCoV-19 |
| University of Health Sciences Pakistan | UHSPK3 |
| University of Veterinary and Animal Sciences | UVAS |
| Spike | S |
| Severe acute respiratory syndrome | SARS |
| Gross domestic product | GDP |
| World Health Organization | WHO |
| Nucleoprotein | N |
| Variant of Concern | VOC |
| Animal research reporting of in vitro experiments | ARRIVE |
| African green monkey kidney cells | VERO-E6 |
| American-type culture collection | ATCC CRL |
| Defense science and technology organization | DESTO |
| Dulbeco’s modified eagles media | DMEM |
| Fetal bovine serum | FBS |
| Micro gram per mililitre | μg/mL |
| United States of America | USA |
| Viral transport media | VTM |
| Polymerase chain reaction | PCR |
| Biosafety level 3 | BSL-3 |
| Institute of Microbiology | IOM |
| Threshold cycle | CT |
| Carbon dioxide | CO2 |
| National Center for Biotechnology Information | NCBI |
| Global Initiative on Sharing All Influenza Data | GISAID |
| Mean tissue culture infective dose 50 | TCID50 |
| Multiplicity of infection | MOI |
| Cytopathic effects | CPE’s |
| Revolution per minute | RCF |
| Beta propiolactone | BPL |
| Polyethersulfone | PES |
| Molecular weight cutoff 0 | MWC0 |
| Enzyme-linked immune sorbent assay | ELISA |
| Albino laboratory-bred strain of house mouse | BALB/c |
| Phosphate buffered saline | PBS |
| Microlitre | ul |
| Horseradish peroxidase | HRP |
| Standard unit per dose | SU/dose |
| Receptor binding domain | RBD |
| Immunoglobulin G | Ig-G |
| Neutralizing antibodies | Nab |
| Microneutralization test | MNT |
| Maximum tolerated dose | MTD |
| Day post-vaccination | DPV |
| Liver function test | LFT |
| Renal function test | RFT |
| Novel coronavirus 19 | nCov-19 |
| Massanger RNA | mRNA |
| Gamma-glutamyltransferase | GGT |
| Alanine transaminase | ALT |
| Aspartate transaminase | AST |
| Serum glutamic pyruvic transaminase | SGPT |
| Serum glutamic oxaloacetic transaminase | SGOT |
| Albumin/globulin | A/G |
| Inhaled nitric oxide | INO |
| Beijing institute of biological sciences corona vaccine | BBIBP-CorV |
| Intradermal units per milliliter | IU/ml |
| Sample to positive ratio | S/P |
| White blood cell | WBC |
| Lymphocyte | Lym. |
| Monocyte | Mon. |
| Granulucyte | Gra. |
| Red blood cell | RBC |
| Mean corpuscular volume | MCV |
| Hematocrit | Hct |
| Mean corpuscular haemoglobin | MCH |
| Mean corpuscular hemoglobin concentration | MCHC |
| Red cell distribution width | RDW |
| Hemoglobin | Hb |
| Thrombochytocrit | THR |
| Mean platelet volume | MPV |
| Procalcitonin | Pct |
| Platelet distribution width | PDW |
| Day post vaccine | DPV |
References
- Hossain, M.F.; Hasana, S.; Mamun, A.A.; Uddin, M.S.; Wahed, M.I.I.; Sarker, S.; Behl, T.; Ullah, I.; Begum, Y.; Bulbul, I.J. COVID-19 outbreak: Pathogenesis, current therapies, and potentials for future management. Front. Pharmacol. 2020, 11, 563478. [Google Scholar] [CrossRef] [PubMed]
- Mehla, R.; Kokate, P.; Bhosale, S.R.; Vaidya, V.; Narayanan, S.; Shandil, R.; Singh, M.; Rudramurthy, G.R.; Naveenkumar, C.N.; Bharathkumar, K. A Live Attenuated COVID-19 Candidate Vaccine for Children: Protection against SARS-CoV-2 Challenge in Hamsters. Vaccines 2023, 11, 255. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, N.; Azuine, R.E.; Tamiz, A. COVID-19 pandemic in Pakistan. Int. J. Transl. Med. Res. Public Health 2020, 4, 37–49. [Google Scholar] [CrossRef]
- Smith, K.A. Edward Jenner and the small pox vaccine. Front. Immunol. 2011, 2, 21. [Google Scholar] [CrossRef]
- Costanzo, M.; De Giglio, M.A.; Roviello, G.N. Anti-coronavirus vaccines: Past investigations on SARS-CoV-1 and MERS-CoV, the approved vaccines from BioNTech/Pfizer, Moderna, Oxford/AstraZeneca and others under Development Against SARSCoV-2 Infection. Curr. Med. Chem. 2022, 29, 4–18. [Google Scholar] [CrossRef]
- Gallo–Ramírez, L.E.; Nikolay, A.; Genzel, Y.; Reichl, U. Bioreactor concepts for cell culture-based viral vaccine production. Expert Rev. Vaccines 2015, 14, 1181–1195. [Google Scholar] [CrossRef]
- Röltgen, K.; Nielsen, S.C.; Silva, O.; Younes, S.F.; Zaslavsky, M.; Costales, C.; Yang, F.; Wirz, O.F.; Solis, D.; Hoh, R.A. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell 2022, 185, 1025–1040.e14. [Google Scholar] [CrossRef]
- Lederer, K.; Bettini, E.; Parvathaneni, K.; Painter, M.M.; Agarwal, D.; Lundgreen, K.A.; Weirick, M.; Muralidharan, K.; Castaño, D.; Goel, R.R. Germinal center responses to SARS-CoV-2 mRNA vaccines in healthy and immunocompromised individuals. Cell 2022, 185, 1008–1024.e15. [Google Scholar] [CrossRef]
- Olliaro, P.; Torreele, E.; Vaillant, M. COVID-19 vaccine efficacy and effectiveness—The elephant (not) in the room. Lancet Microbe 2021, 2, e279–e280. [Google Scholar] [CrossRef]
- Jordan, S.C.; Shin, B.-H.; Gadsden, T.-A.M.; Chu, M.; Petrosyan, A.; Le, C.N.; Zabner, R.; Oft, J.; Pedraza, I.; Cheng, S. T cell immune responses to SARS-CoV-2 and variants of concern (Alpha and Delta) in infected and vaccinated individuals. Cell. Mol. Immunol. 2021, 18, 2554–2556. [Google Scholar] [CrossRef]
- Farrag, M.A.; Almajhdi, F.N. Human respiratory syncytial virus: Role of innate immunity in clearance and disease progression. Viral Immunol. 2016, 29, 11–26. [Google Scholar] [CrossRef]
- Hartl, D.; Tirouvanziam, R.; Laval, J.; Greene, C.M.; Habiel, D.; Sharma, L.; Yildirim, A.Ö.; Cruz, C.S.D.; Hogaboam, C.M. Innate immunity of the lung: From basic mechanisms to translational medicine. J. Innate Immun. 2018, 10, 487–501. [Google Scholar] [CrossRef]
- Khandia, R.; Singhal, S.; Alqahtani, T.; Kamal, M.A.; Nahed, A.; Nainu, F.; Desingu, P.A.; Dhama, K. Emergence of SARS-CoV-2 Omicron (B. 1.1. 529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ. Res. 2022, 209, 112816. [Google Scholar] [CrossRef]
- Lei, S.; Gao, X.; Sun, Y.; Yu, X.; Zhao, L. Gas chromatography-mass spectrometry method for determination of β-propiolactone in human inactivated rabies vaccine and its hydrolysis analysis. J. Pharm. Anal. 2018, 8, 373–377. [Google Scholar] [CrossRef]
- Perera, R.A.; Ko, R.; Tsang, O.T.; Hui, D.S.; Kwan, M.Y.; Brackman, C.J.; To, E.M.; Yen, H.-l.; Leung, K.; Cheng, S.M. Evaluation of a SARS-CoV-2 surrogate virus neutralization test for detection of antibody in human, canine, cat, and hamster sera. J. Clin. Microbiol. 2021, 59, e02504-20. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating 50% endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- Zhao, F.; Liu, L.; Xu, M.; Shu, X.; Zheng, L.; Wei, Z. Assessments of different inactivating reagents in formulating transmissible gastroenteritis virus vaccine. Virol. J. 2020, 17, 163. [Google Scholar] [CrossRef]
- Pavel, S.T.I.; Yetiskin, H.; Uygut, M.A.; Aslan, A.F.; Aydın, G.; İnan, Ö.; Kaplan, B.; Ozdarendeli, A. Development of an inactivated vaccine against SARS-CoV-2. Vaccines 2021, 9, 1266. [Google Scholar] [CrossRef]
- Wernike, K.; Aebischer, A.; Michelitsch, A.; Hoffmann, D.; Freuling, C.; Balkema-Buschmann, A.; Graaf, A.; Müller, T.; Osterrieder, N.; Rissmann, M. Multi-species ELISA for the detection of antibodies against SARS-CoV-2 in animals. Transbound. Emerg. Dis. 2021, 68, 1779–1785. [Google Scholar] [CrossRef]
- Bennett, R.S.; Postnikova, E.N.; Liang, J.; Gross, R.; Mazur, S.; Dixit, S.; Kocher, G.; Yu, S.; Georgia-Clark, S.; Gerhardt, D. Scalable, Micro-Neutralization Assay for Assessment of SARS-CoV-2 (COVID-19) Virus-Neutralizing Antibodies in Human Clinical Samples. Viruses 2021, 13, 893. [Google Scholar] [CrossRef]
- Swift, M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Richman, D.D. COVID-19 vaccines: Implementation, limitations and opportunities. Glob. Health Med. 2021, 3, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Forni, G.; Mantovani, A. COVID-19 vaccines: Where we stand and challenges ahead. Cell Death Differ. 2021, 28, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, L.; Liang, Z.; Wang, N.; Liu, S.; Li, T.; Yu, Y.; Cui, Q.; Wu, X.; Nie, J. Cross-reactivity of eight SARS-CoV-2 variants rationally predicts immunogenicity clustering in sarbecoviruses. Signal Transduct. Target. Ther. 2022, 7, 256. [Google Scholar] [CrossRef] [PubMed]
- Fraiman, J.; Erviti, J.; Jones, M.; Greenland, S.; Whelan, P.; Kaplan, R.M.; Doshi, P. Serious adverse events of special interest following mRNA COVID-19 vaccination in randomized trials in adults. Vaccine 2022, 40, 5798–5805. [Google Scholar] [CrossRef]
- Basheer, A.; Zahoor, I. Genomic epidemiology of SARS-CoV-2 divulge B. 1, B. 1.36, and B. 1.1. 7 as the most dominant lineages in first, second, and third wave of SARS-CoV-2 infections in Pakistan. Microorganisms 2021, 9, 2609. [Google Scholar] [CrossRef]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021, 27, 717–726. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D. Antibody resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef]
- Sanders, B.; Koldijk, M.; Schuitemaker, H. Inactivated Viral Vaccines. Vaccine Analysis: Strategies, Principles, and Control; Springer: Berlin/Heidelberg, Germany, 2015; pp. 45–80. [Google Scholar]
- He, P.; Zou, Y.; Hu, Z. Advances in aluminum hydroxide-based adjuvant research and its mechanism. Hum. Vaccines Immunother. 2015, 11, 477–488. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.C.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020, 584, 437–442. [Google Scholar] [CrossRef]
- Yao, Y.-F.; Wang, Z.-J.; Jiang, R.-D.; Hu, X.; Zhang, H.-J.; Zhou, Y.-W.; Gao, G.; Chen, Y.; Peng, Y.; Liu, M.-Q. Protective efficacy of inactivated vaccine against SARS-CoV-2 infection in mice and non-human primates. Virol. Sin. 2021, 36, 879–889. [Google Scholar] [CrossRef]
- Ganneru, B.; Jogdand, H.; Daram, V.K.; Das, D.; Molugu, N.R.; Prasad, S.D.; Kannappa, S.V.; Ella, K.M.; Ravikrishnan, R.; Awasthi, A. Th1 skewed immune response of whole virion inactivated SARS-CoV-2 vaccine and its safety evaluation. Iscience 2021, 24, 102298. [Google Scholar] [CrossRef]
- Mohandas, S.; Yadav, P.D.; Shete-Aich, A.; Abraham, P.; Vadrevu, K.M.; Sapkal, G.; Mote, C.; Nyayanit, D.; Gupta, N.; Srinivas, V.K. Immunogenicity and protective efficacy of BBV152, whole virion inactivated SARS-CoV-2 vaccine candidates in the Syrian hamster model. Iscience 2021, 24, 102054. [Google Scholar] [CrossRef]
- Ganneru, B.; Jogdand, H.; Dharam, V.K.; Molugu, N.R.; Prasad, S.D.; Vellimudu, S.; Ella, K.M.; Ravikrishnan, R.; Awasthi, A.; Jose, J.; et al. Evaluation of safety and immunogenicity of an adjuvanted, TH-1 skewed, whole virion Inactivated SARS-CoV-2 vaccine-BBV152. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Huang, B.; Deng, W.; Quan, Y.; Wang, W.; Xu, W.; Zhao, Y.; Li, N.; Zhang, J. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell 2020, 182, 713–721.e9. [Google Scholar] [CrossRef]
- Muñoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.; Gsell, P.-S.; Riveros-Balta, A.X.; Albrecht, R.A.; Andersen, H.; Baric, R.S.; Carroll, M.W.; Cavaleri, M. Animal models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef]
- Yadav, P.D.; Ella, R.; Kumar, S.; Patil, D.R.; Mohandas, S.; Shete, A.M.; Vadrevu, K.M.; Bhati, G.; Sapkal, G.; Kaushal, H. Immunogenicity and protective efficacy of inactivated SARS-CoV-2 vaccine candidate, BBV152 in rhesus macaques. Nat. Commun. 2021, 12, 1386. [Google Scholar] [CrossRef]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef]
- Zhang, C.-h.; Lu, J.-h.; Wang, Y.-f.; Zheng, H.-y.; Xiong, S.; Zhang, M.-y.; Liu, X.-j.; Li, J.-x.; Wan, Z.-y.; Yan, X.-g. Immune responses in Balb/c mice induced by a candidate SARS-CoV inactivated vaccine prepared from F69 strain. Vaccine 2005, 23, 3196–3201. [Google Scholar] [CrossRef]
- Charitos, I.A.; Ballini, A.; Lovero, R.; Castellaneta, F.; Colella, M.; Scacco, S.; Cantore, S.; Arrigoni, R.; Mastrangelo, F.; Dioguardi, M. Update on COVID-19 and Effectiveness of a Vaccination Campaign in a Global Context. Int. J. Environ. Res. Public Health 2022, 19, 10712. [Google Scholar] [CrossRef]
- Lv, Z.; Deng, Y.-Q.; Ye, Q.; Cao, L.; Sun, C.-Y.; Fan, C.; Huang, W.; Sun, S.; Sun, Y.; Zhu, L. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 2020, 369, 1505–1509. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).