Abstract
Salivary gland neoplasms are a heterogeneous neoplasm group, including mucoepidermoid carcinoma (MECa), adenoid cystic carcinoma (AdCC), and many others. Objective: We aimed to identify new critical genes of MECa and AdCC using bioinformatics analysis. Methods: Gene expression profile of GSE153283 was analyzed by the GEO2R online tool to use the DAVID software for their subsequent enrichment. Protein–protein interactions (PPI) were visualized using String. Cytoscape with MCODE plugin followed by Kaplan–Meier online for overall survival analysis were performed. Results: 97 upregulated genes were identified for MECa and 86 for AdCC. PPI analysis revealed 22 genes for MECa and 63 for AdCC that were validated by Kaplan–Meier that showed FN1 and SPP1 for MECa, and EGF and ERBB2 for AdCC as more significant candidate genes for each neoplasm. Conclusion: With bioinformatics methods, we identify upregulated genes in MECa and AdCC. The resulting candidate genes as possible therapeutic targets were FN1, SPP1, EGF, and ERBB2, and all those genes had been tested as a target in other neoplasm kinds but not salivary gland neoplasm. The bioinformatic evidence is a solid strategy to select them for more extensive research with clinical impact.
1. Introduction
Salivary gland neoplasms can originate from structures that comprise the parenchymal or glandular stromal, constituting a group of morphologically diverse tumors [1]. Its incidence is 2.5 cases per 100,000 persons per year [2]. The biomolecular mechanisms involved in their etiology are unknown. Recent studies focused on investigating mutations in genes that may be associated with the origin of these tumors. According to the 2017 WHO classification, these neoplasms are categorized according to their biological behavior in 11 benign and 20 malignant entities, of which mucoepidermoid carcinoma (MECa) and adenoid cystic carcinoma (AdCC) are the most frequent malignant neoplasms, representing approximately 35% and 21.9% of cases, respectively [3,4].
MECa appears in wide age distribution, with a peak incidence in the second decade of life. It affects both major and minor salivary glands, with a predilection for the female sex [3]. Clinically and histopathologically, it shows various degrees of malignancy, ranging from non-aggressive or low-grade neoplasms and intermediate-grade neoplasms to aggressive or high-grade neoplasms. Cellular diversity and histological patterns of salivary gland neoplasms are frequently misdiagnosed as lymphomatous papillary cystadenoma, adenoid cystic carcinoma, and squamous cell carcinoma [5,6].
The etiopathogenesis of MECa remains uncertain, and recent research aimed to elucidate their development. Researchers have suggested changes in the DNA sequence, such as alterations in TP53, mutations in the transcription factor POU6F2 involved in cell differentiation, and the translocation t (11; 19) (q21; p13), which gives rise to the MECT1-MAML2 fusion oncoprotein [7,8,9,10].
Concerning AdCC, the mean age of patients diagnosed was 57 years old, without gender predilection. It could affect both the major and minor salivary glands [11]. Clinically and histopathologically, it shows three degrees of malignancy described in 1984 by Szanto et al. [12]. Low-grade neoplasms are those with a tubular pattern, intermediate-grade neoplasms are those with a cribriform pattern, and high-grade neoplasms are those with a solid component. Less than 30% of AdCC have a great affinity for perineural invasion and presenting distant metastases, which is why they are considered aggressive neoplasms. Their five-year survival represents 55% to 89%, which decreases at 15 years from 20 to 40% [11]. Their etiopathogenesis has not been fully elucidated. However, in more than 60% of cases, the AdCC has been associated with the chromosomal translocation t (6; 9) (q22–23; p23–24), which gives rise to the MYB-NFIB fusion oncoprotein [13].
Although these alterations continue to be studied, they cannot explain the development of MECa and AdCC. This study aimed to identify new target genes using bioinformatics analysis for the prognosis of both malignant salivary gland neoplasms.
2. Materials and Methods
2.1. Microarray Data Information
NCBI-GEO is regarded as a free public microarray/gene profile database, and we obtained the gene expression profile of GSE153283 in primary salivary gland carcinomas and normal salivary gland tissues. Microarray data were all on account of the Nano Stringn Counter human PanCancer Pathways Panel, which included 14 MEC and 11 AdCC, each with their respective normal tissue.
2.2. Data Processing of Differential Expressed Genes (DEGs)
Candidate differentially expressed genes (DEGs) for MECa and AdCC with their normal specimens were identified via the GEO2R online tool with [logFC] > 1, and adjusted p-value < 0.05. DEGs with log FC > 1 were considered as an upregulated gene.
2.3. Functional Enrichment Analyses
The >1 FC gene list was submitted to DAVID 6.8 (available online: https://david.ncifcrf.gov, accessed on 31 May 2022) [14] to analyze the functions of the DEGs by Gene Ontology (GO) with the biological process, molecular function and cellular component; and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. Statistical significance was set at p < 0.05. A Venn diagram was made for GO and KEGG of both neoplasms and determined which functions were similar for both.
2.4. Protein–Protein Interaction (PPI)
The Search Tool for the Retrieval of Interacting Genes (STRING; version 11.0, available online: https://string-db.org/, accessed on 31 May 2022) [15] database was employed to predict the protein–protein interaction (PPI) networks of the DEGs obtained. Next, the Cytoscape software was used to analyze the interaction with a combined score of >0.4 (Cytoscape; version 3.8.2, available online: http://cytoscape.org, accessed on 31 May 2022) [16]. The plugin molecular complex detection (MCODE) was used to detect the most significant module in the PPI networks with the MCODE score of degree cutoff = 2, node score cutoff = 0.2, k-core = 2, and max depth = 100.
2.5. Selection and Analyses of Hub Genes
For the selection of the hub genes, those clustered with MCODE score ≥ 6 were selected. Then, the effect of the hub genes on overall survival and disease-free survival was analyzed using the Kaplan–Meier plotter (KM plotter, available online: http://kmplot.com/analysis, accessed on 31 May 2022) by adjusting the follow-up threshold to 60 months [17]. To mimic the behavior of both neoplasm to the maximum extent concerning the candidate genes, both analyses were adjusted for head-neck squamous cell carcinoma (HNSCC).
3. Results
We identified 97 and 86 upregulated genes for MECa for AdCC, respectively, from an analysis of 32,000 gene sets. Both gene lists were analyzed by DAVID software, and the results of GO analysis for MECs indicated that the biological process was particularly enriched in extracellular matrix organization, collagen fibril organization, and collagen catabolic process; the molecular function was enriched in extracellular matrix structural constituent, identical protein binding, and integrin binding; and the cellular component was significantly enriched in extracellular region, space, and exosome. The AdCC showed that biological process was enriched in the positive regulation of the transcription from RNA polymerase II promoter, signal transduction, and positive regulation of transcription of the DNA template; molecular function was enriched in growth factor activity, protein binding, and Ras guanyl-nucleotide exchange factor activity; and the cellular component was significantly enriched in the plasma membrane, extracellular region, and membrane raft (Table 1). Five terms were matched when the common functions were analyzed—three cellular components (extracellular region, extracellular space, and extracellular exosome) and two molecular functions (Wnt-protein binding and growth factor activity).

Table 1.
GO analysis of differentially expressed genes associated with Mucoepidermoid and Adenoid cystic carcinoma.
KEGG analysis data (Table 2) showed that the upregulated genes were particularly enriched in ECM-receptor interaction, PI3K-Akt signaling pathway, and Focal adhesion for mucoepidermoid carcinoma; for adenoid cystic carcinoma, we observed enriched in pathways in cancer, the Ras signaling pathway, and transcriptional misregulation in cancer. The matched pathways observed for both neoplasms were the PI3K-Akt signaling pathway, pathways in cancer, and the Wnt signaling pathway; however, the genes displayed differed for each neoplasm.

Table 2.
KEGG pathway analysis of differentially expressed genes associated with mucoepidermoid and adenoid cystic carcinoma.
3.1. PPI Network and Modular Analysis
A total of 22 DEGs, including 21 nodes and 70 edges for MECa, and 63 DEGs, with 25 nodes and 23 edges for AdCC, were imported into the PPI network complex. We then applied Cytoscape MCODE for further analysis. The results revealed 11 nodes and 45 edges for mucoepidermoid carcinoma and 8 nodes and 13 edges for adenoid cystic carcinoma, showing 10 clustered genes for mucoepidermoid and 6 clustered genes for adenoid cystic carcinoma (Figure 1 and Table 3).
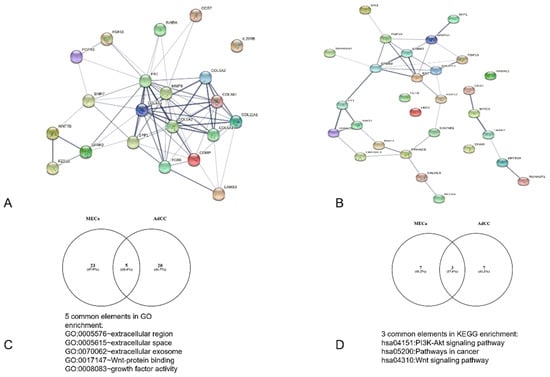
Figure 1.
Analysis of the PPI interaction and sharedGO and KEGG interactions between MECa and AdCC. (A) PPI of MECa with central interactions with FN1, (B) PPI of ACC with principal interaction for EGF, (C,D) analysis of common shared elements of bioinformatic analysis.

Table 3.
Prognostic information of mucoepidermoid and adenoid cystic carcinomas hub candidate genes analyzed by KM plotter.
3.2. Analysis of Core Genes Using a Kaplan–Meier Plotter
A Kaplan–Meier plotter was used to identify the survival data for the clustered genes. Only FN1 and SPP1 for MECa and EGF and ERBB2 for AdCC were significantly associated with poor survival (Figure 2).
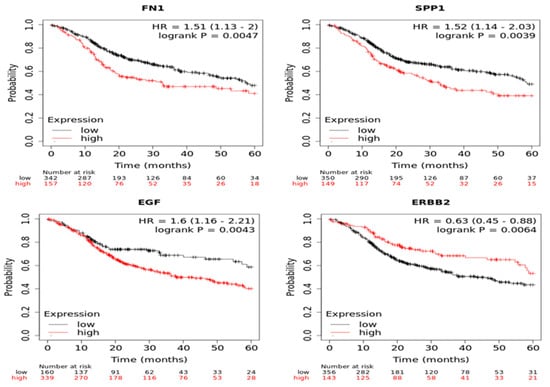
Figure 2.
Analysis of core genes using Kaplan-Meier plotter FN1 and SPP1 for MECa and EGF and ERBB2 for AdCC.
4. Discussion
Cancer is a multistep process involving alterations or changes in the transcriptional activity of genes associated with many cellular processes for tumor development, including proliferation, senescence, and metastasis. Salivary gland neoplasms represent a significant challenge because their biological diversity leads to unpredictable treatment responses. Nowadays, it is not possible to improve worldwide the five-year survival rates of these lesions, which are generally reported between 60–80%, but can decrease to 37% in high-grade tumors [18,19,20].
MECa is the most common malignant salivary gland tumor, followed by AdCC. Tumor stage and grade have been used as survival predictors. It has been considered that low-grade MECa arises more often in minor salivary glands, and high-grade MEC arises more often in major salivary glands (frequently in the parotid gland) [21].
The chromosomal aberration due to CRTC1/3-MAML2 fusion is the most reported genetic alteration, whose fusion protein activates the transcription of cAMP target genes; however, this translocation act as a potential primary driver mutation. Germinal and somatic mutation, copy number variations, and gain or loss of different genes have been associated with CRTC1/3-MAML2 [22]. The AdCC is a slow-growing but mortal salivary gland neoplasm. Similar to MECa, the main genomic alteration is gene fusion (MYB-NFIB fusion is present in 29% to 86% of neoplasms). Applying bioinformatics methods on microarray profile datasets represents an important strategy to identify more useful therapeutic or prognostic biomarkers of salivary glands carcinomas. We obtained 97 and 86 upregulated genes for MECa and AdCC, respectively. The GO function and KEGG pathway analysis showed a heterogeneous expression pattern, with similarities in gene functions and pathways; however, the genes displayed in them were different. For this reason, the PPI network was constructed individually for each neoplasm. The genes that resulted from KM plotting validation were FN1 and SPP1 for MECa and EGF and ERBB2 for AdCC.
In recent years, high-throughput molecular biology techniques, such as high-throughput sequencing, rna-seq, gene microarray, or proteome analysis, have been used to search new markers to determine prognosis, histological lineage, or therapeutic targets. The information obtained from thousands of candidate genes or biomarkers has required the use of different bioinformatic tools, which can handle this large amount of information to be able to classify it, understand it, and give it clinical utility. Bioinformatic studies and their verification through molecular assays, such as PCR, Western blot, immunohistochemistry, or other bioinformatic databases, are a prerequisite to estimating the robustness and give us evidence or possible applications of differentially expressed candidate genes [23,24,25,26]. In this study, we use our analysis algorithm based on the genome-proteome-clinical utility premise; that is, we first explore results through GO and KEGG, and then look for their validated protein interaction, selecting only the genes with the highest correlation (clustered). Finally, we estimate its clinical usefulness based on one of the most important parameters, the five-year survival.
The FN1 is a glycoprotein that usually exists as a dimer. It has plasma FN, synthesized by liver hepatocytes, or cellular FN1, produced by fibroblasts, chondrocytes, myocytes, and synovial cells, that could assemble into insoluble fibrils. The FN interacts with other extracellular matrix (ECM) proteins, growth factors, glycosaminoglycans, cell surface receptors, and other molecules. These interactions are fundamental to inducing specific cell functions, such as differentiation and epithelial-mesenchymal transition. Fibril assembly is often upregulated during cancer [27]. In cancer, FN1 is expressed by cancer-associated fibroblasts and by the cancer cells. Leivo I et al. showed that salivary glands, including MECa, present overexpression of several genes, including FN1. Their analysis suggests that FN1 overexpression could be related to cell adhesion or shape. However, it is essential to consider that histopathological diversity and severity degree could be related to clinical behavior differences [28]. Many studies have postulated that ECM targeting is a possible alternative to cancer treatment. Inhibition of ECM components, remodeling enzymes, blockers for cell surface receptors as integrins that bind FN1, or targeting cancer-associated fibroblasts are other alternatives for cancer treatment [29]. These targets could be explored for MECa treatment.
Osteopontin (OPN) is a phosphoglycoprotein with many reported functions expressed by osteoclasts, osteoblasts, neurons, epithelial cells, T, B, NK, NK T, myeloid, and innate lymphoid cells. Similar to FN1, in cancer, OPN is overexpressed by tumoral parenchymal and stromal cells, and it has been implicated in invasion, metastasis, and treatment resistance. Its expression is associated with poor prognosis in glioma, melanoma, hepatocellular, prostate, lung, breast, colorectal, ovarian, and head and neck cancer [30]. It has been shown that OPN acts as a negative regulator of T cell activation and promotes the recruitment of macrophages. It has been proposed that these macrophages promote tumor growth by COX-2 through the promotion of angiogenesis and migration of cancer cells. Recently, immunotherapy to OPN in cancer has been proposed to be an alternative by neutralizing mAbs [31]. It has been reported that in salivary gland tumors, particularly in MECa, OPN preserves their overexpressed pattern compared with normal salivary gland tissue [32,33]. However, Fok et al. reported that OPN expression correlated with histological grade, which is an essential feature, since it reinforces that OPN can be used as a therapeutic target.
Epidermal growth factor (EGF) is secreted by ectodermic cells, monocytes, kidneys, and duodenal glands. It stimulates epithelial cell growth by binding their transmembrane receptor kinases to promote cell proliferation, survival, adhesion, migration, and differentiation. The EGF receptor (EGFR) family consists of four transmembrane receptors, including EGFR (HER1/erbB-1), HER2 (erbB-2/neu), HER3 (erbB-3), and HER4 (erbB-4). HER1 and HER2 are overexpressed in breast, non–small-cell lung, head and neck, and colon cancers, which is related to poor prognosis. The activation of ligand-receptor EGF-EGFR complex has been related to EMT and MMP9 in head and neck carcinoma and salivary adenoid cystic carcinoma [34,35,36]. Considering that this complex is a more accessible target, employing monoclonal antibodies is a recurrent strategy. Huang et al. reported that nimotuzumab, a humanized neutralizing G1 monoclonal antibody, can cause cell cycle arrest by suppressing in vitro proliferation of AdCC cells, which represents an important feature because the inhibition of these neoplasm cells could be translated to reduce or eliminate some clinical features of AdCC as metastasizes to lungs, bone, liver or brain, or recurrence. Trastuzumab and Pertuzumab are monoclonal antibodies against HER2, another member of the EGFR receptors family. HER2 (erbB-2/neu) is a tyrosine kinase that activates MAPK and PI3K pathways to cell growth and differentiation. In tumors, it is possible to observe gene amplification or overexpression that could be related to more aggressive and metastatic behavior, decreasing the survival expectation of a patient with many carcinomas, including AdCC [37].
An important variable to consider is activating one erbb member pathway could generate resistance in another [38]. Therefore, inhibiting the receptors may not be sufficient, and inhibiting the ligand is required. Considering that EGF is one of the essential ligands for EGFR, their concentration in human serum is variable. A high EGF amount plus EGFR overexpression could create conditions for tumor growth, even without specific EGFR mutations. Crombet et al. have proposed that targeting EGF is necessary for a complete treatment [39]. It has been reported that the most common genetic alteration in MECa and ACC is the presence of gene fusions [23,24] and that these fusions can even be used to subclassify the neoplasm, as well as from a molecular point of view be responsible for common gene alterations in carcinogenesis of salivary gland neoplasms. Although the bioinformatic approach can give greater order and understanding to a complex and uncertain panorama such as MECa and ACC, it is necessary to try to validate these results to achieve better results or applications. Chen Z et al. [24], through their bioinformatic analysis, have reported that their candidate genes may be the target of combined therapy against EGFR through drugs already approved by the FDA. These data, which coincide with our findings on EGF and erbb2, fulfill this therapeutic possibility, which provides a significant advance on the possible path that can be taken in the treatment of MECa and ACC.
5. Conclusions
Our bioinformatic analysis identified two hub genes (FN1 and SPP1) for MECa and two (EGF and ERBB2) for AdCC. The results supported in bioinformatic platforms showed that these genes play critical roles in the pathogenesis, progression, and prognosis of both salivary gland neoplasms. It is necessary to consider them as therapeutic targets to identify how we can affect MECa and AdCC in a specific way as tumor growth or metastasis; however, these results provide new and useful information for testing the new biomarkers.
Author Contributions
Proposal, Conceptualization, Study design development, methodology C.M.R.-M., L.F.J.-A., J.P.-R. and E.R.L.-H.; software, validation, validation, formal analysis, data curation C.M.R.-M. and L.F.J.-A.; writing—original draft and preparation, writing—review and editing, C.M.R.-M., L.P.C.-H., L.F.J.-A. and E.R.L.-H. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by PAPIIT IN226720, UNAM.
Institutional Review Board Statement
This research was based on a simulation of biomarkers using information available in GEO (Gene Expression Omnibus), functional genomics open access data repository. This research does not require approval by the ethics and research committee because no humans or animals were involved.
Informed Consent Statement
Not applicable.
Data Availability Statement
The supporting data for reported results can be found in Targeted RNA quantification of 770 genes in primary salivary gland carcinomas, DataSets for GSE153283, on line available in: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153283 (accessed on 31 May 2022).
Acknowledgments
We would like to thank the Oral Medicine and Pathology Department, Postgraduate and Research Division, Dentistry School, National Autonomous University of Mexico for facilitating our research project.
Conflicts of Interest
The authors have no relevant financial or non-financial interest to disclose.
References
- Speight, P.M.; Barret, A.W. Salivary Gland Tumors. Oral Dis. 2002, 8, 229–240. [Google Scholar] [CrossRef]
- Eveson, J.W.; Cawson, R. Tumours of the minor (oropharyngeal) salivary glands: A demographic study of 336 cases. J. Oral Pathol. 1985, 14, 500. [Google Scholar] [CrossRef]
- El-Naggar, A.; Chan, J.; Grandis, J.; Takata, T.; Slootweg, P. IARC WHO Classification of Head and Neck Tumours; WHO: Geneva, Switzerland, 2017; Volume 9. [Google Scholar]
- Eveson, J.W.; Auclair, P.; Gnepp, D.R. Tumours of the salivary glands. In Pathology & Genetics Head and Neck Tumours; IARC Press: Lyon, France, 2005; pp. 209–281. [Google Scholar]
- Mullin, M.H.; Brierley, D.J.; Speight, P.M. Second opinion reporting in head and neck pathology: The pattern of referrals and impact on final diagnosis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2015, 119, 656–660. [Google Scholar] [CrossRef]
- Chenevert, J.; Barnes, L.E.; Chiosea, S.I. Mucoepidermoid carcinoma: A five-decade journey. Virchows. Arch. 2011, 458, 133–140. [Google Scholar] [CrossRef]
- Kang, H.; Tan, M.; Bishop, J.A.; Jones, S.; Sausen, M.; Ha, P.K.; Agrawal, N. Whole-Exome Sequencing of Salivary Gland Mucoepidermoid Carcinoma. Clin. Cancer Res. 2017, 23, 283–288, clincanres-0720. [Google Scholar] [CrossRef]
- Tonon, G.; Modi, S.; Wu, L.; Kubo, A.; Coxon, A.B.; Komiya, T.; O’Neil, K.; Stover, K.; El-Naggar, A.; Griffin, J.D.; et al. t (11; 19)(q21; p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003, 33, 208–213. [Google Scholar] [CrossRef]
- Ilic-Dimitrijević, I.; Loening, T.; Falk, M.; Konstantinović, V.; Vukadinović, M.; Tepavčević, Z.; Tabaković, S.Z.; Pejčić, N.; Miličić, B.; Milašin, J. Incidence and clinical relevance of t (11; 19) translocation in salivary gland mucoepidermoid carcinoma. Genetika 2014, 46, 601–610. [Google Scholar] [CrossRef]
- Miyabe, S.; Okabe, M.; Nagatsuka, H.; Hasegawa, Y.; Inagaki, A.; Ijichi, K.; Nagai, N.; Eimoto, T.; Yokoi, M.; Shimozato, K.; et al. Prognostic Significance of p27Kip1, Ki-67, and CRTC1-MAML2 Fusion Transcript in Mucoepidermoid Carcinoma: A Molecular and Clinicopathologic Study of 101 Cases. J. Oral Maxillofac. Surg. 2009, 67, 1432–1441. [Google Scholar] [CrossRef]
- Van Weert, S.; Reinhard, R.; Bloemena, E.; Buter, J.; Witte, B.I.; Vergeer, M.R.; Leemans, C.R. Differences in patterns of survival in metastatic adenoid cystic carcinoma of the head and neck. Head Neck 2016, 39, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Szanto, P.A.; Luna, M.A.; Tortoledo, M.E.; White, R.A. Histologic grading of adenoid cystic carcinoma of the salivary glands. Cancer 1984, 54, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.S.; Kannan, K.; Roy, D.M.; Morris, L.G.T.; Ganly, I.; Katabi, N.; Ramaswami, D.; Walsh, L.; Eng, S.; Huse, J.T.; et al. The mutational landscape of adenoid cystic carcinoma. Nat. Genet. 2013, 45, 791–798. [Google Scholar] [CrossRef] [PubMed]
- DAVID Bioinformatics Resources 6.8 [Internet]. 2022. Available online: https://david.ncifcrf.gov/ (accessed on 31 May 2022).
- STRING [Internet]. 2022. Available online: https://string-db.org/ (accessed on 31 May 2022).
- Cytoscape [Internet) 2022. Available online: http://cytoscape.org (accessed on 31 May 2022).
- KM plotter [Internet]. 2022. Available online: http://kmplot.com/analysis (accessed on 31 May 2022).
- Terhaard, C.H.J.; Lubsen, H.; Van der Tweel, I.; Hilgers, F.; Eijkenboom, W.; Marres, H.; Tjho-Heslinga, R.; De Jong, J. On behalf of the Dutch Head and Neck Oncology Cooperative Group (NWHHT) Salivary gland carcinoma: Independent prognostic factors for locoregional control, distant metastases, and overall survival: Results of the Dutch head and neck oncology cooperative group. Head Neck 2004, 26, 681–693, discussion 692-3. [Google Scholar] [CrossRef]
- Fonseca, F.P.; Carvalho, M.D.V.; de Almeida, O.P.; Rangel, A.L.C.A.; Takizawa, M.C.H.; Bueno, A.G.; Vargas, P.A. Clinicopathologic analysis of 493 cases of salivary gland tumors in a Southern Brazilian population. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2012, 114, 230–239. [Google Scholar] [CrossRef]
- Bell, R.B.; Dierks, E.J.; Homer, L.; Potter, B.E. Management and Outcome of Patients with Malignant Salivary Gland Tumors. J. Oral Maxillofac. Surg. 2005, 63, 917–928. [Google Scholar] [CrossRef]
- Peraza, A.; Gómez, R.; Beltran, J.; Amarista, F. Mucoepidermoid carcinoma. An update and review of the literature. J. Stomatol. Oral Maxillofac. Surg. 2020, 121, 713–720. [Google Scholar] [CrossRef]
- Toper, M.H.; Sarioglu, S. Molecular Pathology of Salivary Gland Neoplasms: Diagnostic, Prognostic, and Predictive Perspective. Adv. Anat. Pathol. 2021, 28, 81–93. [Google Scholar] [CrossRef]
- Karpinets, T.V.; Mitani, Y.; Liu, B.; Zhang, J.; Pytynia, K.B.; Sellen, L.D.; Karagiannis, D.T.; Ferrarotto, R.; Futreal, A.P.; El-Naggar, A.K. Whole-Genome Sequencing of Common Salivary Gland Carcinomas: Subtype-Restricted and Shared Genetic Alterations. Clin. Cancer Res. 2021, 27, 3960–3969. [Google Scholar] [CrossRef]
- Chen, Z.; Ni, W.; Li, J.-L.; Lin, S.; Zhou, X.; Sun, Y.; Li, J.W.; Leon, M.E.; Hurtado, M.D.; Zolotukhin, S.; et al. The CRTC1-MAML2 fusion is the major oncogenic driver in mucoepidermoid carcinoma. JCI Insight 2021, 6, e139497. [Google Scholar] [CrossRef]
- Lu, H.; Han, N.; Xu, W.; Zhu, Y.; Liu, L.; Liu, S.; Yang, W. Screening and bioinformatics analysis of mRNA, long non-coding RNA and circular RNA expression profiles in mucoepidermoid carcinoma of salivary gland. Biochem. Biophys. Res. Commun. 2018, 508, 66–71. [Google Scholar] [CrossRef]
- Liu, Z.; Gao, J.; Yang, Y.; Zhao, H.; Ma, C.; Yu, T. Potential targets identified in adenoid cystic carcinoma point out new directions for further research. Am. J. Transl. Res. 2021, 13, 1085–1108. [Google Scholar]
- Dalton, C.J.; Lemmon, C.A. Fibronectin: Molecular Structure, Fibrillar Structure and Mechanochemical Signaling. Cells 2021, 10, 2443. [Google Scholar] [CrossRef]
- Leivo, I.; Jee, K.J.; Heikinheimo, K.; Laine, M.; Ollila, J.; Nagy, B.; Knuutila, S. Characterization of gene expression in major types of salivary gland carcinomas with epithelial differentiation. Cancer Genet. Cytogenet. 2005, 156, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zheng, X.; Zheng, Y.; Chen, Y.; Fei, W.; Wang, F.; Zheng, C. Extracellular Matrix: Emerging Roles and Potential Therapeutic Targets for Breast Cancer. Front. Oncol. 2021, 11, 650453. [Google Scholar] [CrossRef]
- Moorman, H.R.; Poschel, D.; Klement, J.D.; Lu, C.; Redd, P.S.; Liu, K. Osteopontin: A Key Regulator of Tumor Progression and Immunomodulation. Cancers 2020, 12, 3379. [Google Scholar] [CrossRef]
- Klement, J.D.; Paschall, A.V.; Redd, P.S.; Ibrahim, M.L.; Lu, C.; Yang, D.; Celis, E.; Abrams, S.I.; Ozato, K.; Liu, K. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J. Clin. Investig. 2018, 128, 5549–5560. [Google Scholar] [CrossRef]
- Bjørndal, K.; Larsen, S.R.; Godballe, C.; Krogdahl, A. Osteopontin expression in salivary gland carcinomas. J. Oral Pathol. Med. 2010, 40, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Fok, T.; Lapointe, H.; Tuck, A.; Chambers, A.; Jackson-Boeters, L.; Daley, T.; Darling, M. Expression and localization of osteopontin, homing cell adhesion molecule/CD44, and integrin αvβ3 in mucoepidermoid carcinoma and acinic cell adenocarcinoma of salivary gland origin. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2014, 118, 320–329. [Google Scholar] [CrossRef]
- Smith, A.; Teknos, T.N.; Pan, Q. Epithelial to mesenchymal transition in head and neck squamous cell carcinoma. Oral Oncol. 2013, 49, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yu, T.; Fu, X.; Chen, J.; Liu, Y.; Li, C.; Xia, Y.; Zhang, Z.; Li, L. EGFR inhibition prevents in vitro tumor growth of salivary adenoid cystic carcinoma. BMC Cell Biol. 2013, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, J.; Wang, Y.; Ye, W.; Zhang, X.; Ju, H.; Xu, D.; Liu, L.; Ye, D.; Zhang, L.; et al. EGFR activation induced Snail-dependent EMT and myc-dependent PD-L1 in human salivary adenoid cystic carcinoma cells. Cell Cycle 2018, 17, 1457–1470. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Ren, W.; Rao, G. erbB2 gene silencing and its effect on PTEN in SACC-83 salivary adenoid cystic carcinoma cells. Oncol. Rep. 2010, 24, 1291–1296. [Google Scholar] [CrossRef]
- ZZhang, L.; Castanaro, C.; Luan, B.; Yang, K.; Fan, L.; Fairhurst, J.L.; Rafique, A.; Potocky, T.B.; Shan, J.; Delfino, F.J.; et al. ERBB3/HER2 Signaling Promotes Resistance to EGFR Blockade in Head and Neck and Colorectal Cancer Models. Mol. Cancer Ther. 2014, 13, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- Ramos, T.C.; Morales, O.S.; Dy, G.K.; Monzón, K.L.; Dávila, A.L. The Position of EGF Deprivation in the Management of Advanced Non-Small Cell Lung Cancer. Front. Oncol. 2021, 11, 639745. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).