Pro-Inflammatory Cytokines and Interferon-Stimulated Gene Responses Induced by Seasonal Influenza A Virus with Varying Growth Capabilities in Human Lung Epithelial Cell Lines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Virus Infection of Cells
2.3. RNA Extraction and Polymerase Chain Reaction
2.4. Expression of Pro-Inflammatory Cytokines (TNF-α, IL-6, and IFN-β) and ISGs (ISG-15, IFIM1, and TRIM22)
2.5. IFN-β and ISG-15 Protein Production
2.6. JAK Inhibition
2.7. NS1 Sequencing
2.8. Statistical Analysis
3. Results
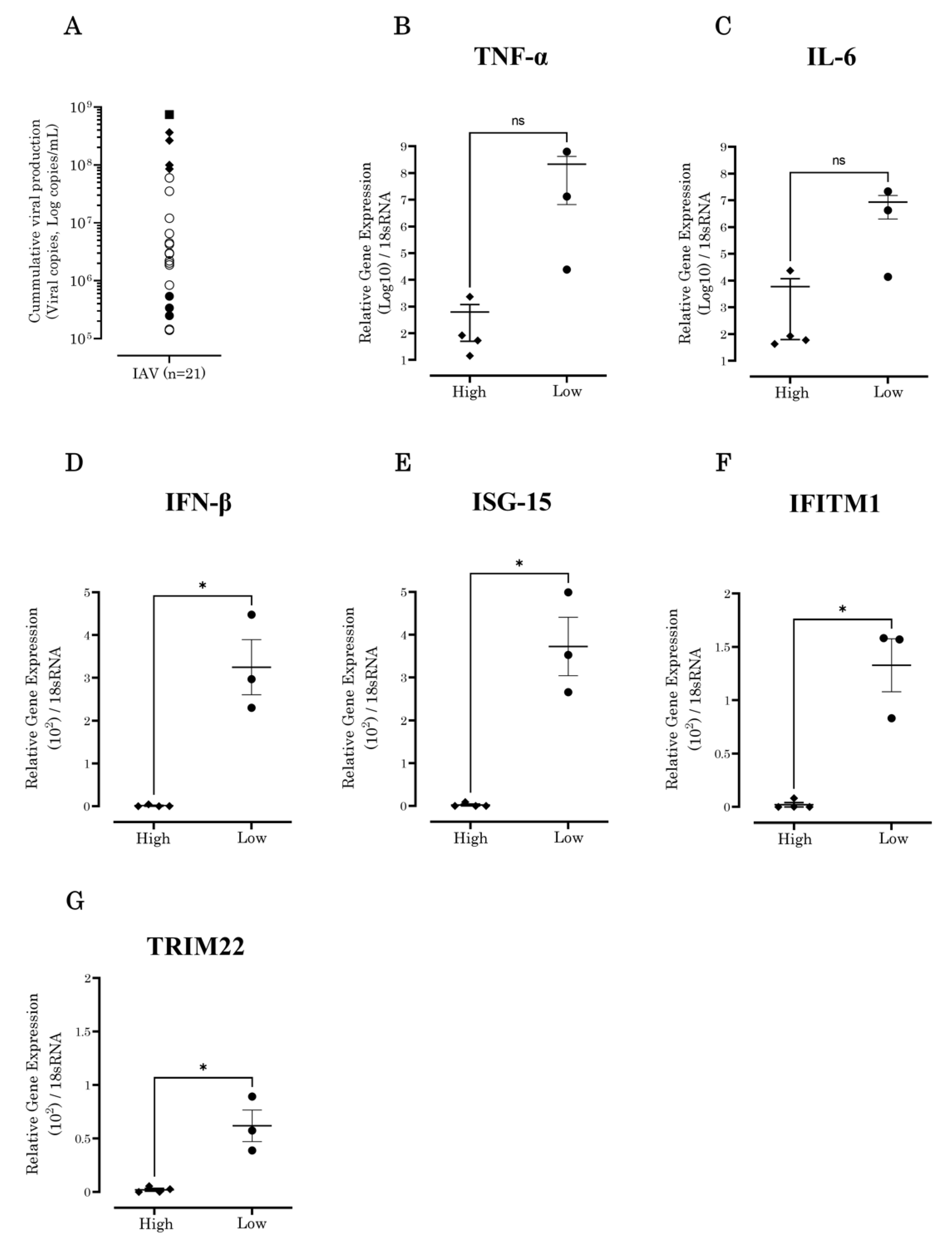
3.1. IAV Showed Diverse Growth Capabilities In Vitro
3.2. IFN-β and Antiviral ISG Expression Levels Increased in A549 Cells Inoculated with Low-Growth-Capability IAV Strains
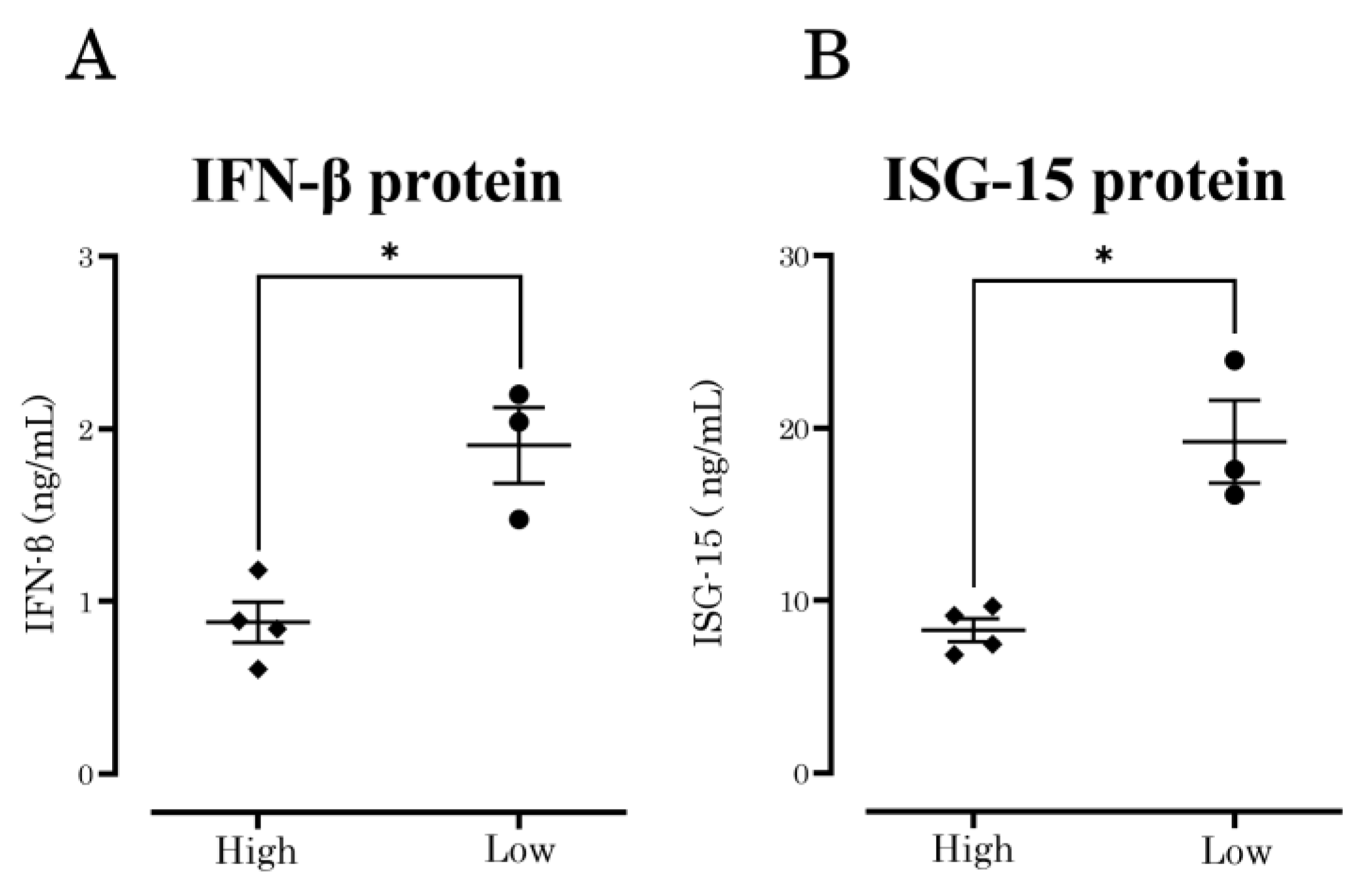
3.3. IFN-β and ISG-15 Protein Production
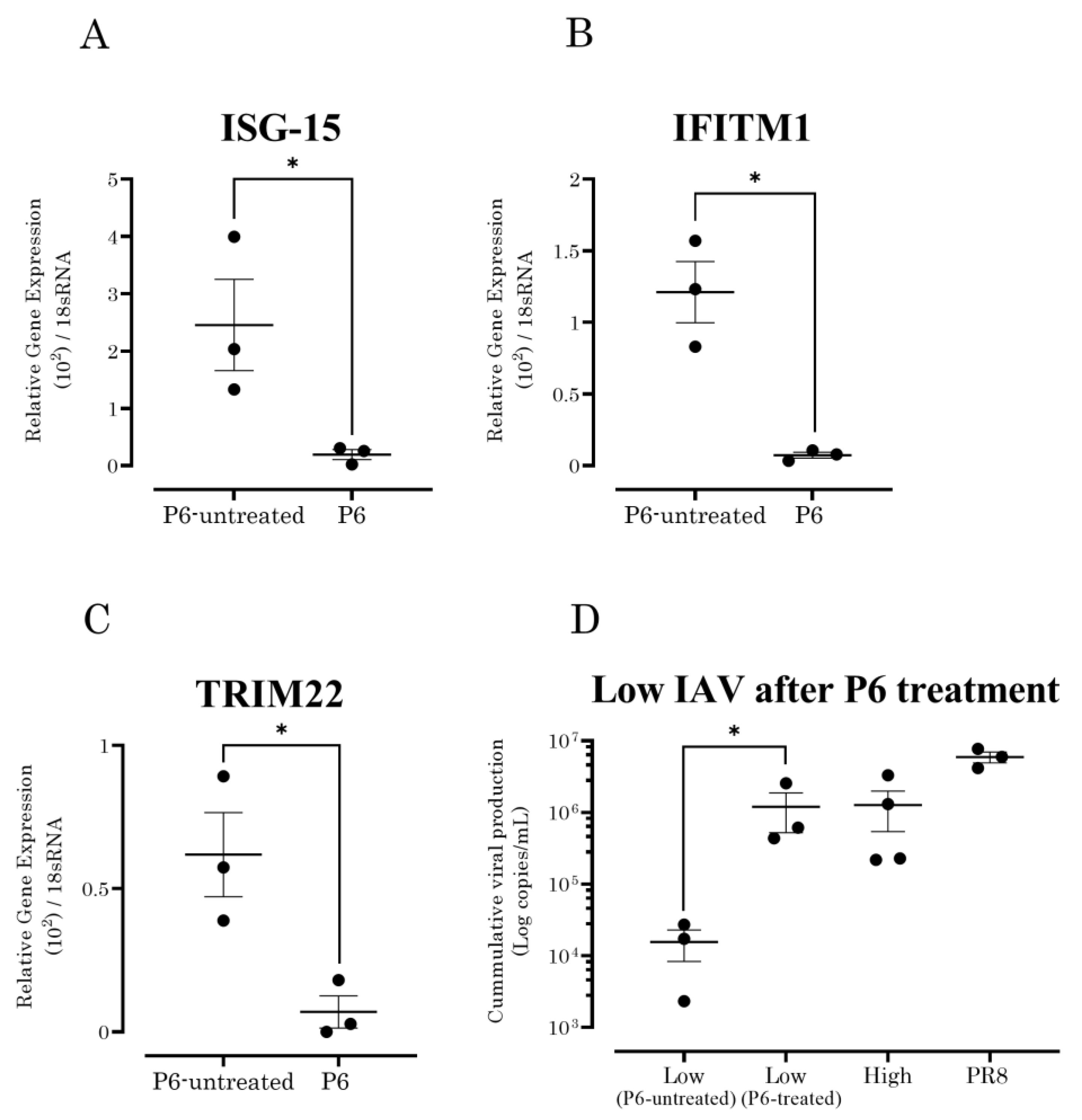
3.4. JAK/STAT Pathway Inhibition Increased the Susceptibility of A549 Cells to Seasonal IAV Strains with Low Growth Capability
3.5. NS1 Mutations Are Present in the Circulating Seasonal IAV Strains
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peteranderl, C.; Herold, S.; Schmoldt, C. Respiratory Viral Infections: Human Influenza Virus Infections. Semin Respir. Crit. Care Med. 2016, 37, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Herold, S.; Becker, C.; Ridge, K.M.; Budinger, G.S. Influenza virus-induced lung injury: Pathogenesis and implications for treatment. Eur. Respir. J. 2015, 45, 1463–1478. [Google Scholar] [CrossRef] [PubMed]
- Lalueza, A.; Folgueira, D.; Muñoz-Gallego, I.; Trujillo, H.; Laureiro, J.; Hernández-Jiménez, P.; Moral-Jiménez, N.; Castillo, C.; Ayuso, B.; Díaz-Pedroche, C.; et al. Influence of viral load in the outcome of hospitalized patients with influenza virus infection. Eur. J. Clin. Microbiol. 2019, 38, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Alves, V.R.G.; Perosa, A.H.; Luna, L.K.D.S.; Cruz, J.S.; Conte, D.D.; Bellei, N. Influenza A(H1N1)pdm09 infection and viral load analysis in patients with different clinical presentations. Mem. Inst. Oswaldo Cruz. 2020, 115, e200009. [Google Scholar] [CrossRef]
- Tsuneki-Tokunaga, A.; Kanai, K.; Itagaki, A.; Tsuchie, H.; Okada, T.; Kasagi, M.; Tanaka, K.; Aoki, M.; Hinay, A.J.A.; Kageyama, S. Growth capability of epidemic influenza viruses in Japan since the 2009 H1N1 pandemic. Arch. Virol. 2021, 166, 1193–1196. [Google Scholar] [CrossRef]
- Tsuneki, A.; Itagaki, A.; Tsuchie, H.; Tokuhara, M.; Okada, T.; Narai, S.; Kasagi, M.; Tanaka, K.; Kageyama, S. Reduced replication capacity of influenza A(H1N1)pdm09 virus during the 2010-2011 winter season in Tottori, Japan. J. Med. Virol. 2013, 85, 1871–1877. [Google Scholar] [CrossRef]
- Zohari, S.; Munir, M.; Metreveli, G.; Belák, S.; Berg, M. Differences in the ability to suppress interferon β production between allele A and allele B NS1 proteins from H10 influenza A viruses. Virol. J. 2010, 7, 376. [Google Scholar] [CrossRef]
- Samuel, C.E. Antiviral Actions of Interferons. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar] [CrossRef]
- Haller, O.; Kochs, G.; Weber, F. The interferon response circuit: Induction and suppression by pathogenic viruses. Virology 2005, 344, 119–130. [Google Scholar] [CrossRef]
- Wu, W.; Metcalf, J.P. The Role of Type I IFNs in Influenza: Antiviral Superheroes or Immunopathogenic Villains? J. Innate Immun. 2020, 12, 437–447. [Google Scholar] [CrossRef]
- Villalón-Letelier, F.; Brooks, A.G.; Saunders, P.M.; Londrigan, S.L.; Reading, P.C. Host Cell Restriction Factors that Limit Influenza A Infection. Viruses 2017, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Perng, Y.-C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Genet. 2018, 16, 423–439. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Hsiang, T.-Y.; Kuo, R.-L.; Krug, R.M. ISG15 conjugation system targets the viral NS1 protein in influenza A virus–infected cells. Proc. Natl. Acad. Sci. USA 2010, 107, 2253–2258. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, L.; Luo, J.; He, H. NP and NS1 proteins of H5N1 virus significantly upregulated IFITM1, IFITM2, and IFITM3 in A549 cells. Afr. Health Sci. 2019, 19, 1402–1410. [Google Scholar] [CrossRef] [PubMed]
- Lamotte, L.-A.; Tafforeau, L. How Influenza A Virus NS1 Deals with the Ubiquitin System to Evade Innate Immunity. Viruses 2021, 13, 2309. [Google Scholar] [CrossRef]
- Di Pietro, A.; Kajaste-Rudnitski, A.; Oteiza, A.; Nicora, L.; Towers, G.J.; Mechti, N.; Vicenzi, E. TRIM22 Inhibits Influenza A Virus Infection by Targeting the Viral Nucleoprotein for Degradation. J. Virol. 2013, 87, 4523–4533. [Google Scholar] [CrossRef]
- Le Goffic, R.; Bouguyon, E.; Chevalier, C.; Vidic, J.; Da Costa, B.; Leymarie, O.; Bourdieu, C.; Decamps, L.; Dhorne-Pollet, S.; Delmas, B. Influenza A Virus Protein PB1-F2 Exacerbates IFN-β Expression of Human Respiratory Epithelial Cells. J. Immunol. 2010, 185, 4812–4823. [Google Scholar] [CrossRef]
- Hale, B.G.; Randall, R.E.; Ortín, J.; Jackson, D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008, 89, 2359–2376. [Google Scholar] [CrossRef]
- Pauli, E.-K.; Schmolke, M.; Wolff, T.; Viemann, D.; Roth, J.; Bode, J.G.; Ludwig, S. Influenza A Virus Inhibits Type I IFN Signaling via NF-κB-Dependent Induction of SOCS-3 Expression. PLOS Pathog. 2008, 4, e1000196. [Google Scholar] [CrossRef]
- Murakami, S.; Horimoto, T.; Mai, L.Q.; Nidom, C.A.; Chen, H.; Muramoto, Y.; Yamada, S.; Iwasa, A.; Iwatsuki-Horimoto, K.; Shimojima, M.; et al. Growth Determinants for H5N1 Influenza Vaccine Seed Viruses in MDCK Cells. J. Virol. 2008, 82, 10502–10509. [Google Scholar] [CrossRef] [Green Version]
- Evseev, D.; Magor, K.E. Molecular Evolution of the Influenza A Virus Non-structural Protein 1 in Interspecies Transmission and Adaptation. Front. Microbiol. 2021, 12, 2853. [Google Scholar] [CrossRef] [PubMed]
- Kaewborisuth, C.; Zanin, M.; Häcker, H.; Webby, R.J.; Lekcharoensuk, P. G45R mutation in the nonstructural protein 1 of A/Puerto Rico/8/1934 (H1N1) enhances viral replication independent of dsRNA-binding activity and type I interferon biology. Virol. J. 2016, 13, 127. [Google Scholar] [CrossRef] [PubMed]
- DeDiego, M.L.; Nogales, A.; Lambert-Emo, K.; Martinez-Sobrido, L.; Topham, D.J. NS1 Protein Mutation I64T Affects Interferon Responses and Virulence of Circulating H3N2 Human Influenza A Viruses. J. Virol. 2016, 90, 9693–9711. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, L.; Zhang, R.; Chi, X.; Yang, Z.; Xie, Y.; Shu, S.; Liao, Y.; Chen, J.-L. Identification of two residues within the NS1 of H7N9 influenza A virus that critically affect the protein stability and function. Vet. Res. 2018, 49, 98. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Martinez-Sobrido, L.; Topham, D.J.; DeDiego, M.L. NS1 Protein Amino Acid Changes D189N and V194I Affect Interferon Responses, Thermosensitivity, and Virulence of Circulating H3N2 Human Influenza A Viruses. J. Virol. 2017, 91, e01930-16. [Google Scholar] [CrossRef]
- Kuchipudi, S.V.; Tellabati, M.; Nelli, R.K.; White, G.A.; Perez, B.B.; Sebastian, S.; Slomka, M.J.; Brookes, S.M.; Brown, I.H.; Dunham, S.P.; et al. 18S rRNAis a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virol. J. 2012, 9, 230. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Seng, L.-G.; Daly, J.; Chang, K.-C.; Kuchipudi, S.V. High Basal Expression of Interferon-Stimulated Genes in Human Bronchial Epithelial (BEAS-2B) Cells Contributes to Influenza A Virus Resistance. PLoS ONE 2014, 9, e109023. [Google Scholar] [CrossRef]
- Tsuneki-Tokunaga, A.; Kondo, T.; Kanai, K.; Itagaki, A.; Tsuchie, H.; Okada, T.; Kasagi, M.; Tanaka, K.; Hinay, A.J.A.; Kageyama, S. Local spread of influenza A (H1N1) viruses without a mutation for the maximum duration of an epidemic season in Japan. Arch. Virol. 2021, 167, 1–5. [Google Scholar] [CrossRef]
- Pedranzini, L.; Dechow, T.; Berishaj, M.; Comenzo, R.; Zhou, P.; Azare, J.; Bornmann, W.; Bromberg, J. Pyridone 6, A Pan-Janus–Activated Kinase Inhibitor, Induces Growth Inhibition of Multiple Myeloma Cells. Cancer Res. 2006, 66, 9714–9721. [Google Scholar] [CrossRef] [Green Version]
- Ortigoza, M.; Dibben, O.; Maamary, J.; Martinez-Gil, L.; Leyva-Grado, V.; Abreu, P.; Ayllon, J.; Palese, P.; Shaw, M.L. A Novel Small Molecule Inhibitor of Influenza A Viruses that Targets Polymerase Function and Indirectly Induces Interferon. PLoS Pathog. 2012, 8, e1002668. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Shen, S.-M.; Yin, J.; Zhang, P.-P.; Shi, Y. Influenza virus non-structural protein 1 inhibits the production of interferon β of alveolar epithelial cells upon the infection of influenza A H1N1. Mol. Med. Rep. 2017, 16, 4553–4560. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Yang, F.; Wang, Q.; Xu, N.; Xie, Y.; Chen, S.; Qin, T.; Peng, D. Influenza a virus antagonizes type I and type II interferon responses via SOCS1-dependent ubiquitination and degradation of JAK1. Virol. J. 2020, 17, 1–10. [Google Scholar] [CrossRef]
- Jia, D.; Rahbar, R.; Chan, R.W.Y.; Lee, S.M.Y.; Chan, M.C.W.; Wang, B.X.; Baker, D.P.; Sun, B.; Peiris, J.S.M.; Nicholls, J.M.; et al. Influenza Virus Non-Structural Protein 1 (NS1) Disrupts Interferon Signaling. PLoS ONE 2010, 5, e13927. [Google Scholar] [CrossRef] [PubMed]
- Kochs, G.; Garcia-Sastre, A.; Martinez-Sobrido, L. Multiple Anti-Interferon Actions of the Influenza A Virus NS1 Protein. J. Virol. 2007, 81, 7011–7021. [Google Scholar] [CrossRef]
- Park, E.-S.; Dezhbord, M.; Kim, A.R.L.; Kim, K.-H. The Roles of Ubiquitination in Pathogenesis of Influenza Virus Infection. Int. J. Mol. Sci. 2022, 23, 4593. [Google Scholar] [CrossRef]
- Killip, M.J.; Fodor, E.; Randall, R.E. Influenza virus activation of the interferon system. Virus Res. 2015, 209, 11–22. [Google Scholar] [CrossRef]
- Li, J.; Zhang, K.; Chen, Q.; Zhang, X.; Sun, Y.; Bi, Y.; Zhang, S.; Gu, J.; Li, J.; Liu, D.; et al. Three amino acid substitutions in the NS1 protein change the virus replication of H5N1 influenza virus in human cells. Virology 2018, 519, 64–73. [Google Scholar] [CrossRef]
- Elshina, E.; Velthuis, A.J.W.T. The influenza virus RNA polymerase as an innate immune agonist and antagonist. Experientia 2021, 78, 7237–7256. [Google Scholar] [CrossRef]
| Primers | Sequence (5′-3′) |
|---|---|
| Virus Quantification | |
| FLUA-MAT-F | CTTCTAACCGAGGTCGAAACGTA |
| FLUA-MAT-R | GGTGACAGGATTGGTCTTGTCTTTA |
| Gene Expression | |
| 18S rRNA-F | GGAGCCTGCGGCTTAATTTG |
| 18S rRNA-R | CCACCCACGGAATCGAGAAA |
| TNF-α-F | GCGACGTGGAACTGGCAGAAG |
| TNF-α-R | GGTACAACCCATCGGCTGGCA |
| IL-6-F | AGGATACCACTCCCAACAGACCT |
| IL-6-R | CAAGTGCATCATCGTTGTTCATAC |
| IFN-β-F | AACTGCAACCTTTCGAAGCC |
| IFN-β-R | TGTCGCCTACTACCTGTTGTGC |
| ISG-15-F | GAGAGGCAGCGAACTCATCT |
| ISG-15-R | CTTCAGCTCTGACACCGACA |
| IFITM1-F | ACTCCGTGAAGTCTAGGGACA |
| IFITM1-R | TGTCACAGAGCCGAATACCAG |
| TRIM22-F | GGGTGGACGTGATGCTGA |
| TRIM22-R | TCACTTGTCTCTGATCCACAGAAATA |
| NS1 Sequencing | |
| MBTuni-12 | ACGCGTGATCAGCAAAAGCAGG |
| MBTuni-13 | ACGCGTGATCAGTAGAAACAAGG |
| A-NS-M13-F | TGTAAAACGACGGCCAGTAGCAAAAGCAGGGTGACAAAGACA |
| A-NS-M13-R | CAGGAAACAGCTATGACCAGTAGAAACAAGGGTGTTTTTTAT |
| Influenza A Virus Subtypes | NS1 Mutations That May Influence the Viral Replication of IAV | |||
|---|---|---|---|---|
| Increase | Decrease | |||
| High Growth Capability | G45R | K66E | I64T | E55X a |
| A/Tottori/ST215/2009 (H1N1) | R | E | - | - |
| A/Tottori/ST1349/2014 (H3N2) | R | E | - | K |
| A/Tottori/ST488/2014 (H3N2) | R | E | - | K |
| A/Tottori/ST1350/2013 (H1N1) | R | E | - | K |
| LowGrowth Capability | ||||
| A/Tottori/TT039/2011 (H1N1) | R | - | T | N |
| A/Tottori/ST1705/2014 (H3N2) | - | - | T | H |
| A/Tottori/ST777/2011 (H3N2) | - | - | T | H |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hinay, A.A., Jr.; Kakee, S.; Kageyama, S.; Tsuneki-Tokunaga, A.; Perdana, W.Y.; Akena, Y.; Nishiyama, S.; Kanai, K. Pro-Inflammatory Cytokines and Interferon-Stimulated Gene Responses Induced by Seasonal Influenza A Virus with Varying Growth Capabilities in Human Lung Epithelial Cell Lines. Vaccines 2022, 10, 1507. https://doi.org/10.3390/vaccines10091507
Hinay AA Jr., Kakee S, Kageyama S, Tsuneki-Tokunaga A, Perdana WY, Akena Y, Nishiyama S, Kanai K. Pro-Inflammatory Cytokines and Interferon-Stimulated Gene Responses Induced by Seasonal Influenza A Virus with Varying Growth Capabilities in Human Lung Epithelial Cell Lines. Vaccines. 2022; 10(9):1507. https://doi.org/10.3390/vaccines10091507
Chicago/Turabian StyleHinay, Alfredo A., Jr., Sosuke Kakee, Seiji Kageyama, Akeno Tsuneki-Tokunaga, Waldy Y. Perdana, Yui Akena, Shota Nishiyama, and Kyosuke Kanai. 2022. "Pro-Inflammatory Cytokines and Interferon-Stimulated Gene Responses Induced by Seasonal Influenza A Virus with Varying Growth Capabilities in Human Lung Epithelial Cell Lines" Vaccines 10, no. 9: 1507. https://doi.org/10.3390/vaccines10091507
APA StyleHinay, A. A., Jr., Kakee, S., Kageyama, S., Tsuneki-Tokunaga, A., Perdana, W. Y., Akena, Y., Nishiyama, S., & Kanai, K. (2022). Pro-Inflammatory Cytokines and Interferon-Stimulated Gene Responses Induced by Seasonal Influenza A Virus with Varying Growth Capabilities in Human Lung Epithelial Cell Lines. Vaccines, 10(9), 1507. https://doi.org/10.3390/vaccines10091507






