The Immune Subtypes and Landscape of Advanced-Stage Ovarian Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Extraction and Preprocessing
2.2. Generation of the Immune-Related Gene List
2.3. Identification and Validation of the Immune Subtypes
2.4. Characteristics of TIICs of the Immune Subtypes
2.5. Prediction of the Efficacy of Immunotherapy and Chemotherapy
2.6. Assessment of the Immune Landscape
2.7. Construction of Co-Expression Modules of OC
2.8. Tissue Specimens
2.9. Total RNA Extraction, Quantitative Real-Time PCR and Western Blotting
2.10. Statistical Analysis
3. Results
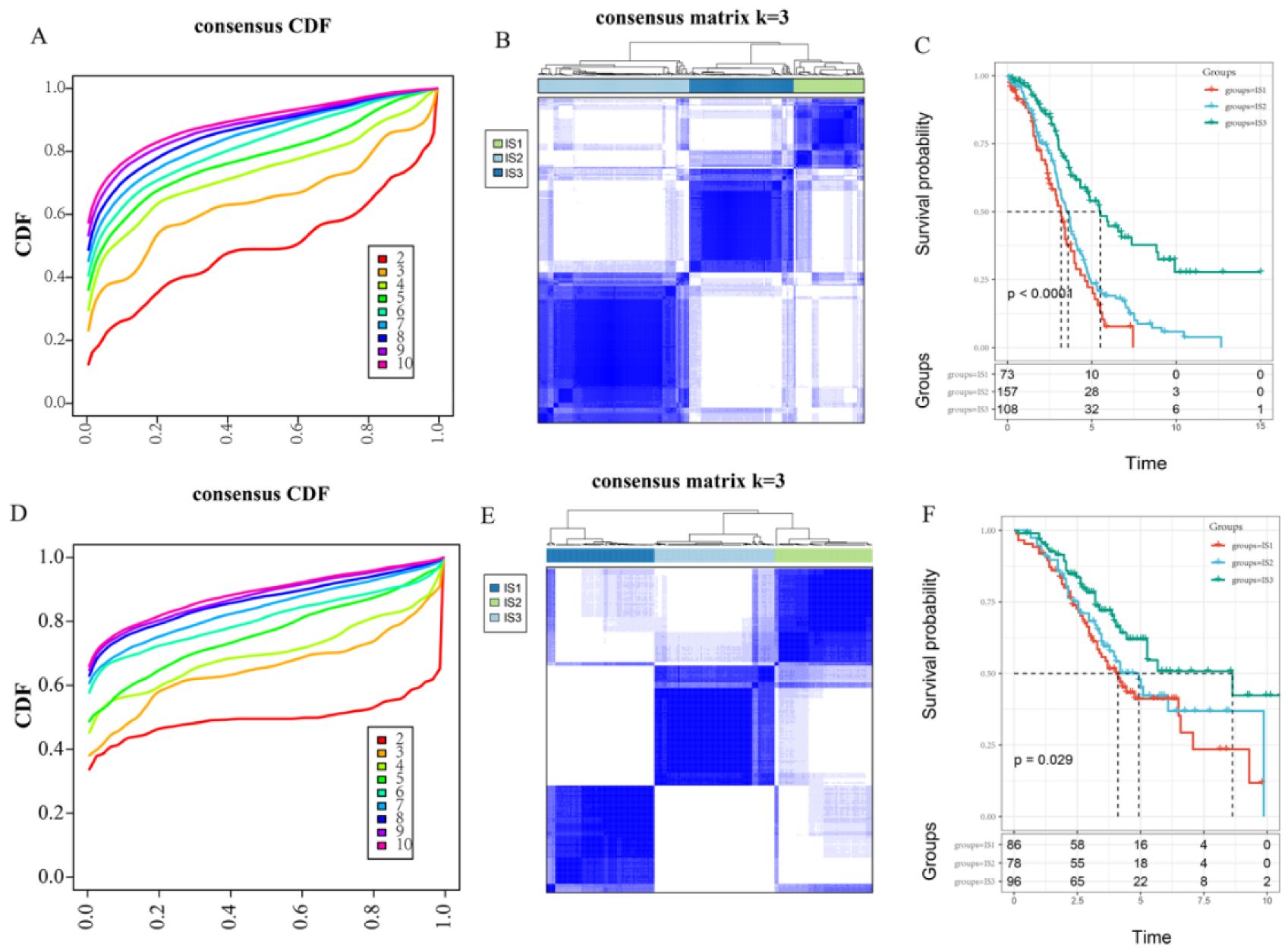
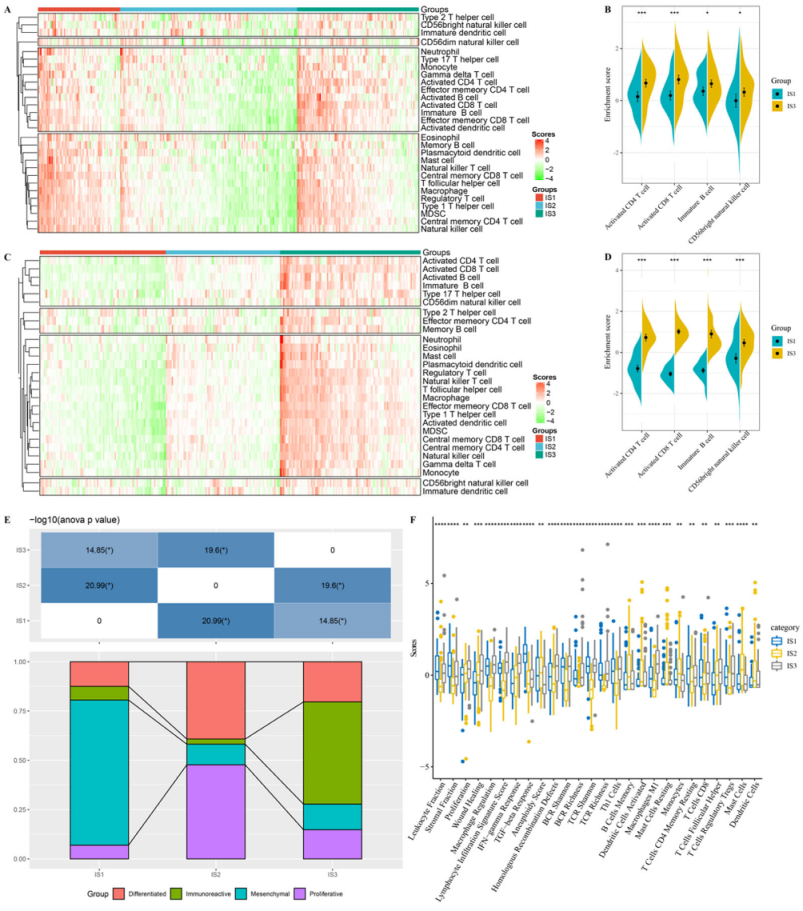
3.1. Identification and Validation of the Immune Subtypes
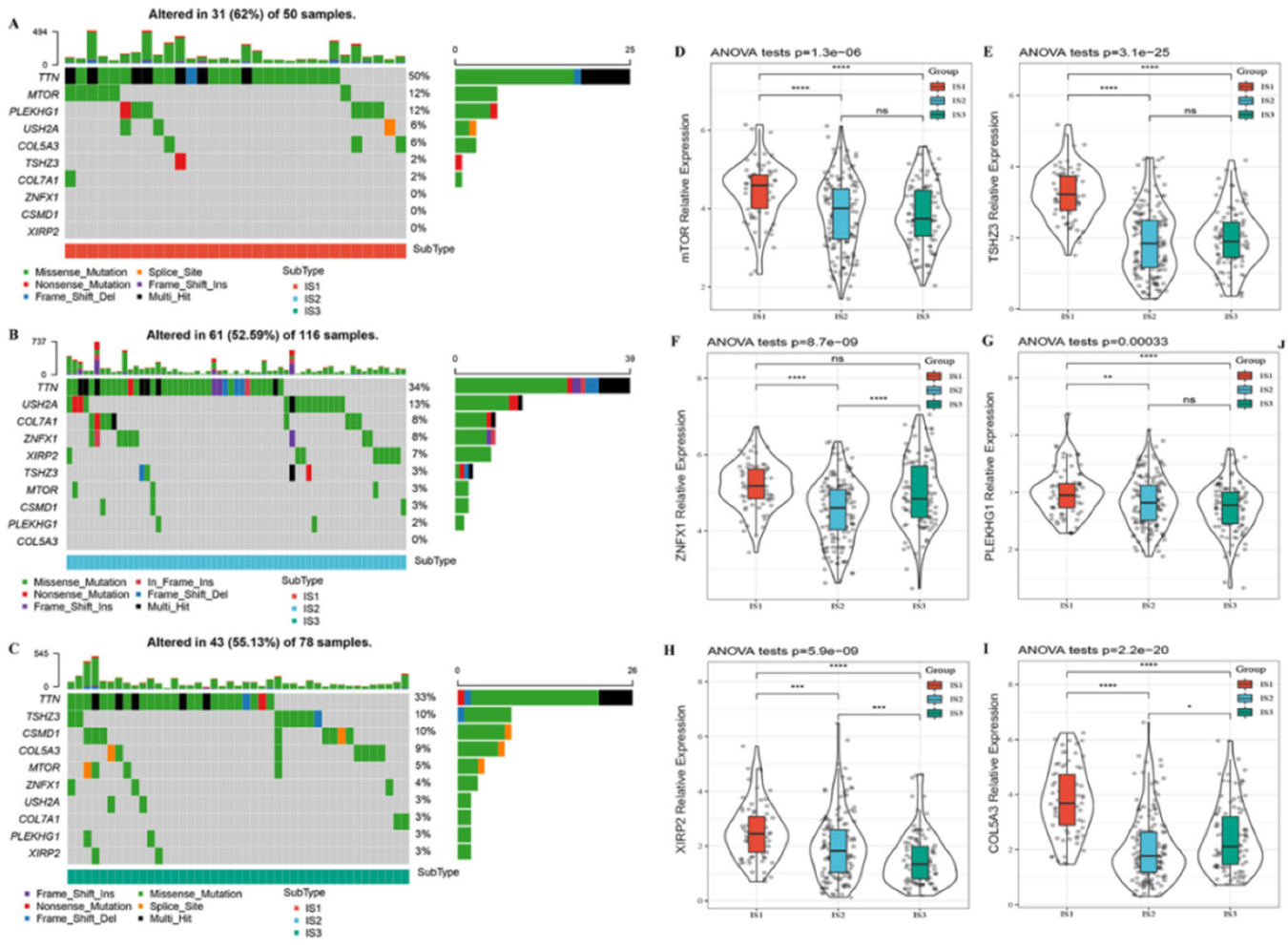
3.2. Hub Mutation Gene Features in Different Immune Subtypes
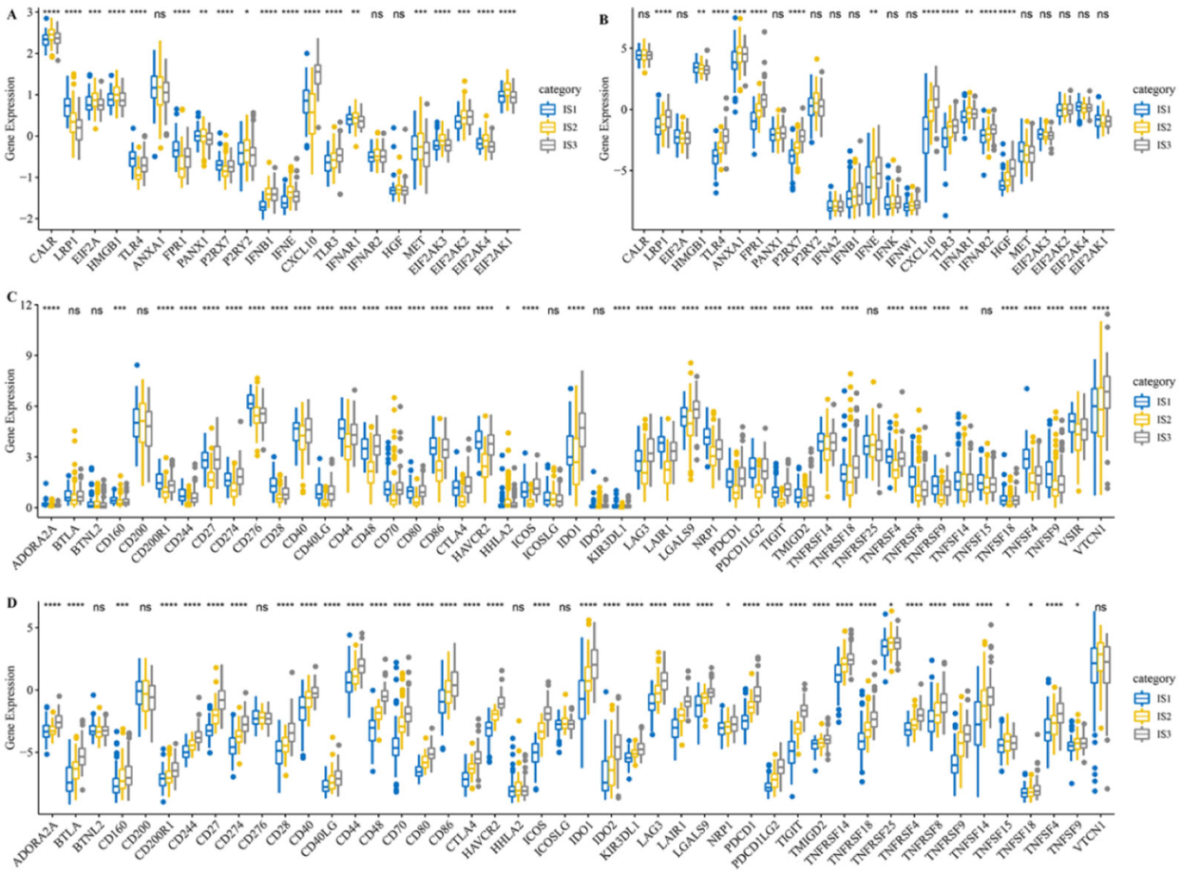
3.3. Different Expression of Immune Modulators among Three Immune Subtypes
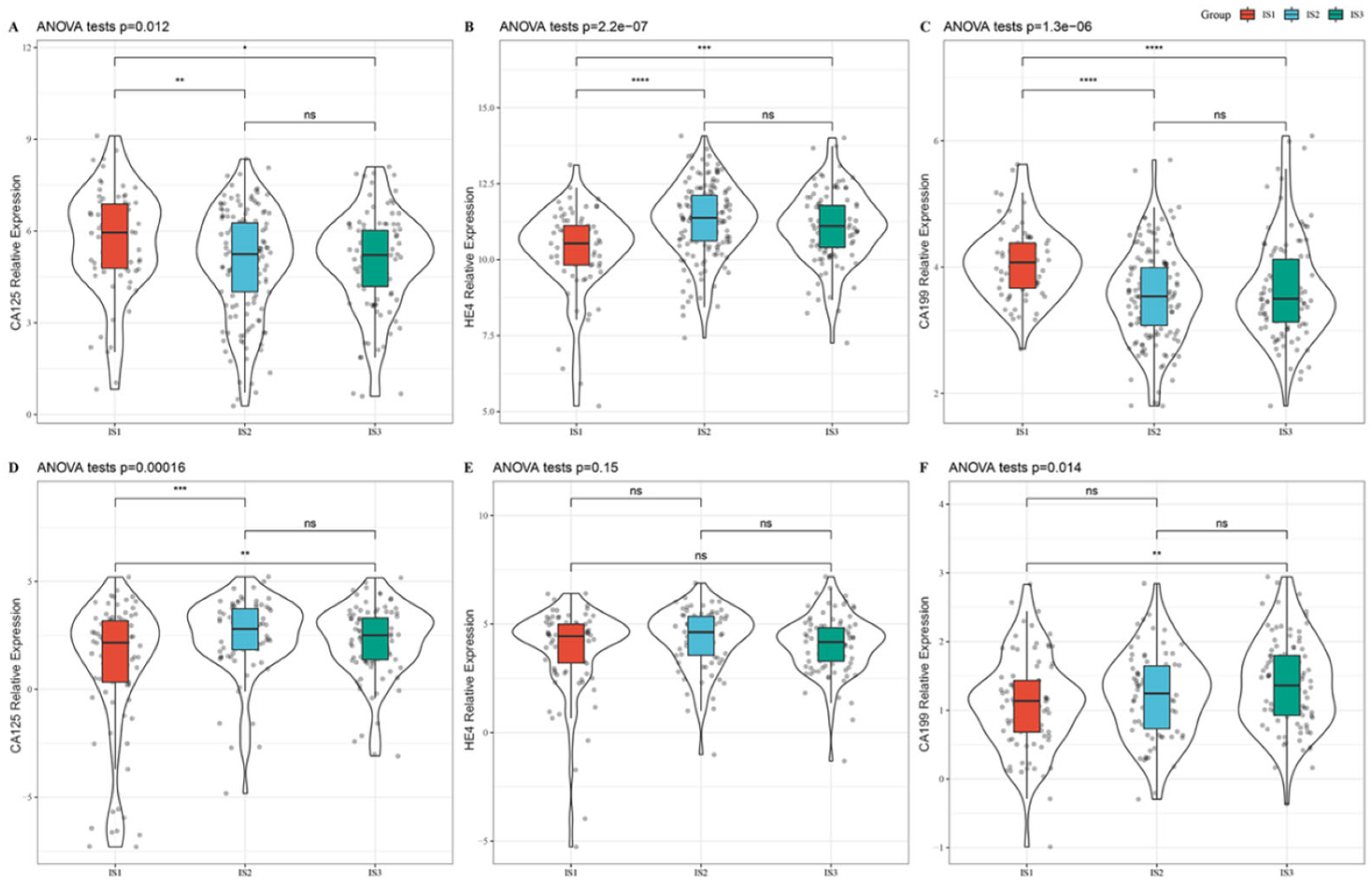
3.4. Association between Immune Subtypes and Tumor Biomarkers
3.5. Characteristics of TIICs of the Immune Subtypes
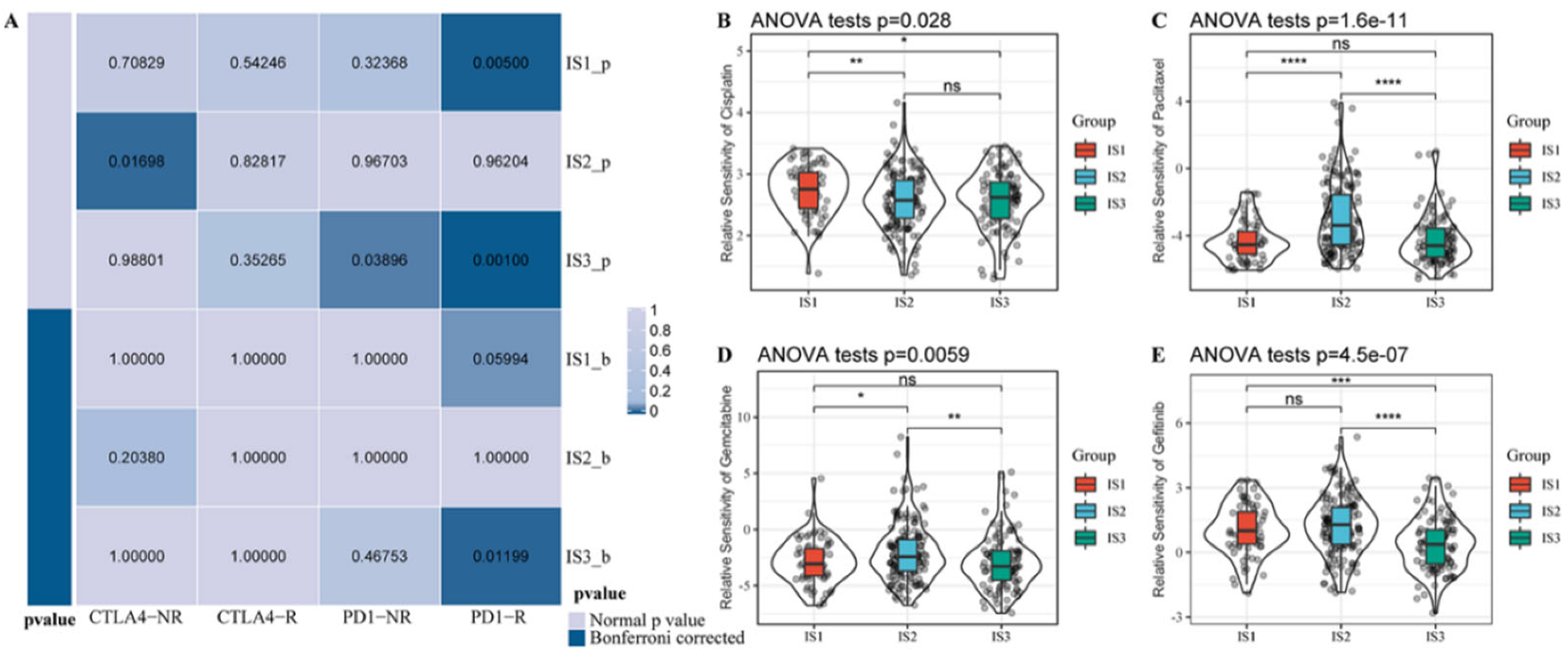
3.6. Association between Immune Subtypes and Immunotherapy/Chemotherapy Response
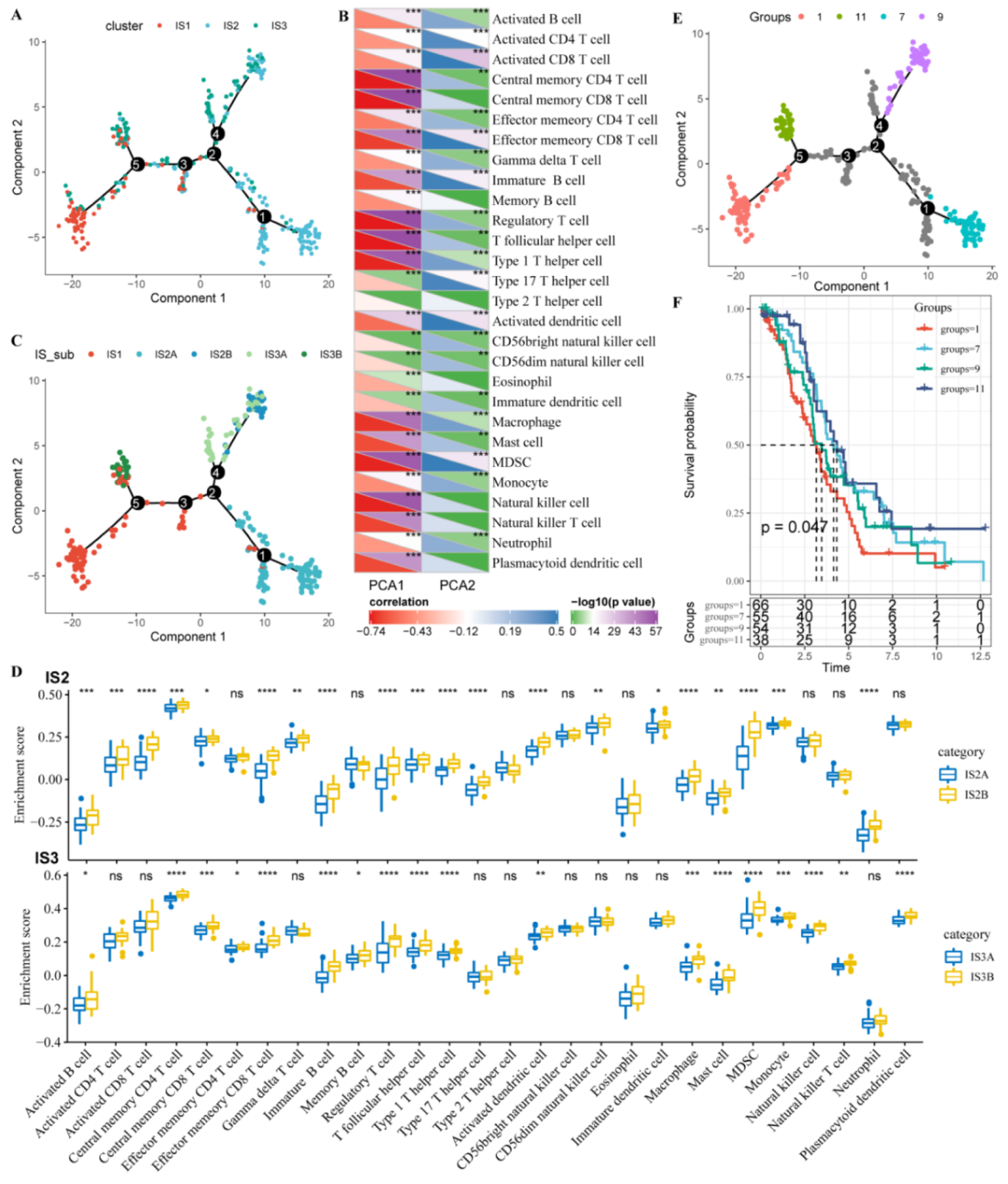
3.7. Immune Landscape of OC in the TCGA Cohort
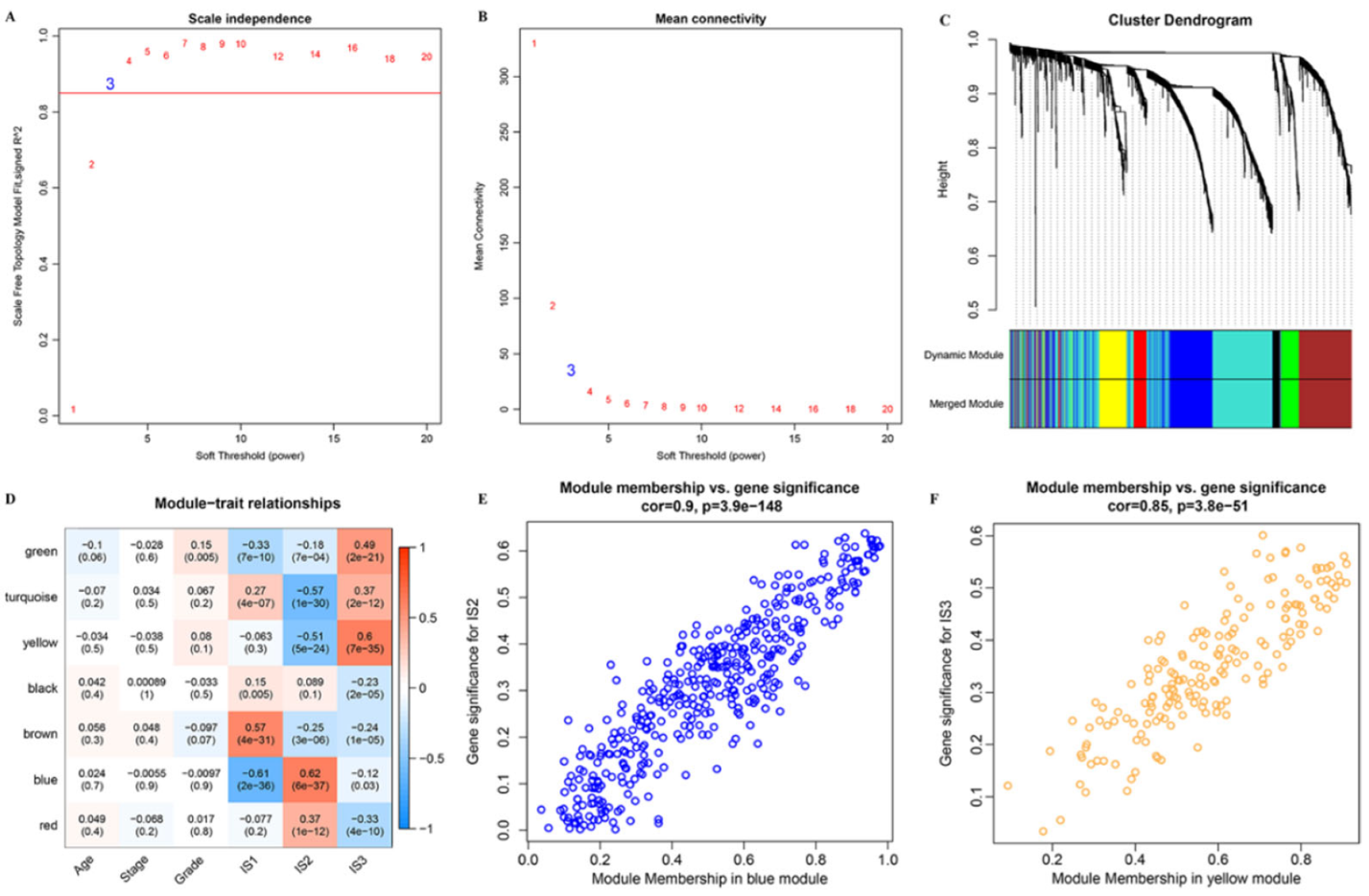
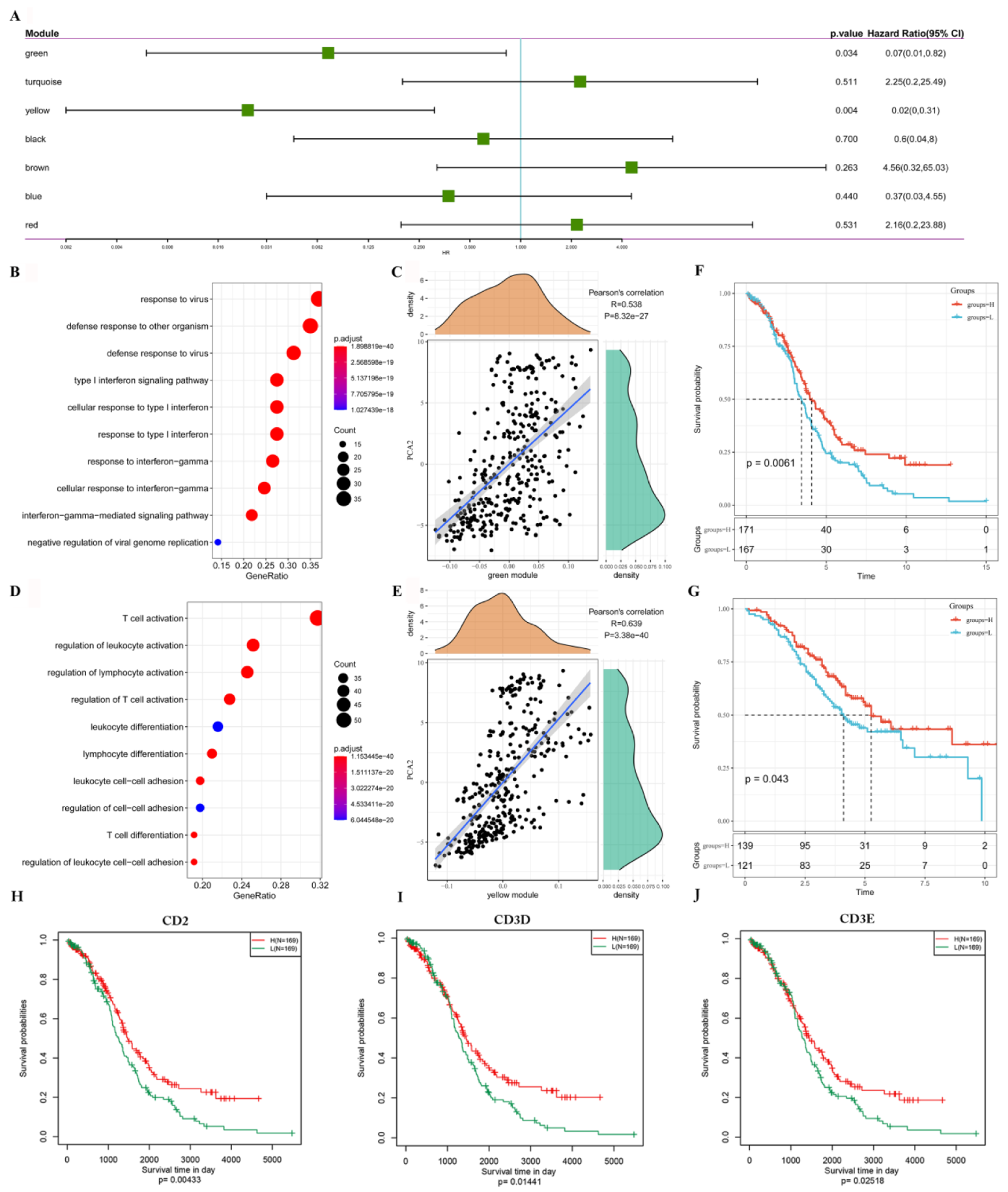
3.8. Identification of Immune Gene Co-Expression Modules and Immune Hub Genes
3.9. Verification of the Prognostic Characteristics of CD2, CD3D and CD3E in the ICGC Cohort and Independent Cohort
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Van Den Bulk, J.; Verdegaal, E.M.; De Miranda, N.F. Cancer Immunotherapy: Broadening The Scope Of Targetable Tumours. Open Biol. 2018, 8, 180037. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, J.; Page, D.b.; Li, B.t.; Connell, L.C.; Schindler, K.; Lacouture, M.E.; Postow, M.A.; Wolchok, J.D. Toxicities Of The Anti-Pd-1 And Anti-Pd-L1 Immune Checkpoint Antibodies. Ann. Oncol. 2016, 27, 1362. [Google Scholar] [CrossRef] [PubMed]
- Odunsi, K. Immunotherapy In Ovarian Cancer. Ann. Oncol. 2017, 28, Viii1–Viii7. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, Activity, And Immune Correlates Of Anti-Pd-1 Antibody In Cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, A.; Kulus, M.; Wieczorkiewicz, M.; Pienkowski, W.; Stefanska, K.; Skupin-Mrugalska, P.; Bryl, R.; Mozdziak, P.; Kempisty, B. Ovarian Cancer And Cancer Stem Cells-Cellular And Molecular Characteristics, Signaling Pathways, And Usefulness As A Diagnostic Tool In Medicine And Oncology. Cancers 2021, 13, 4178. [Google Scholar] [CrossRef]
- Pfisterer, J. Recurrent Ovarian Cancer. Onkologie 2004, 27, 7–8. [Google Scholar] [CrossRef]
- Herzog, T.J.; Pothuri, B. Ovarian Cancer: A Focus On Management Of Recurrent Disease. Nat. Clin. Pract. Oncol. 2006, 3, 604–611. [Google Scholar] [CrossRef]
- Morand, S.; Devanaboyina, M.; Staats, H.; Stanbery, L.; Nemunaitis, J. Ovarian Cancer Immunotherapy And Personalized Medicine. Int. J. Mol. Sci. 2021, 22, 6532. [Google Scholar] [CrossRef]
- Rodriguez, G.; Galpin, K.; McCloskey, C.; Vanderhyden, B. The Tumor Microenvironment of Epithelial Ovarian Cancer and Its Influence on Response to Immunotherapy. Cancers 2018, 10, 242. [Google Scholar] [CrossRef]
- Ning, F.; Cole, C.B.; Annunziata, C.M. Driving Immune Responses in the Ovarian Tumor Microenvironment. Front. Oncol. 2020, 10, 604084. [Google Scholar] [CrossRef]
- Breuer, K.; Foroushani, A.K.; Laird, M.R.; Chen, C.; Sribnaia, A.; Lo, R.; Winsor, G.L.; Hancock, R.E.; Brinkman, F.S.; Lynn, D.J. Innatedb: Systems Biology Of Innate Immunity and Beyond—Recent Updates And Continuing Curation. Nucleic Acids Res. 2013, 41, D1228–D1233. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, G.; Tang, T.; Liang, T. Identification Of Tumor Antigens And Immune Subtypes Of Pancreatic Adenocarcinoma For Mrna Vaccine Development. Mol. Cancer 2021, 20, 44. [Google Scholar] [CrossRef] [PubMed]
- Wilkerson, M.D.; Hayes, D.N. Consensusclusterplus: A Class Discovery Tool with Confidence Assessments and Item Tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. Gsva: Gene Set Variation Analysis For Microarray And Rna-Seq Data. Bmc Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-Cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships And Predictors Of Response To Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated Genomic Analyses of Ovarian Carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Hu, Y.; Taylor-Harding, B.; Raz, Y.; Haro, M.; Recouvreux, M.S.; Taylan, E.; Lester, J.; Millstein, J.; Walts, A.E.; Karlan, B.Y.; et al. Are Epithelial Ovarian Cancers of The Mesenchymal Subtype Actually Intraperitoneal Metastases to the Ovary? Front. Cell Dev. Biol. 2020, 8, 647. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e814. [Google Scholar] [CrossRef]
- Wang, L.; Mao, Q. Probabilistic Dimensionality Reduction Via Structure Learning. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 41, 205–219. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The Dynamics and Regulators of Cell Fate Decisions Are Revealed by Pseudotemporal Ordering of Single Cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Yao, J.; Nowak, N.J.; Goodison, S. Cancer Progression Modeling Using Static Sample Data. Genome Biol. 2014, 15, 440. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. Wgcna: An R Package For Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.-Q.; Zhang, M.-J.; Cheng, Z.-J.; Yu, J.; Yuan, Q.; Zhang, J.; Cai, Y.; Yang, L.Y.; Zhang, Y.; Hao, J.J.; et al. Fkbp10 Promotes Proliferation of Glioma Cells via Activating Akt-Creb-Pcna Axis. J. Biomed. Sci. 2021, 28, 13. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Ba, Y.; Ji, H.; Wang, F.; Du, J.; Hu, S. Recognition of Tumor-Associated Antigens and Immune Subtypes in Glioma for Mrna Vaccine Development. Front. Immunol. 2021, 12, 738435. [Google Scholar] [CrossRef]
- Li, Y.; Shi, X.; Mao, B.; Wang, L.; Wu, L.; Li, J.; Jiao, S. The Genomic Mutational Landscape and Its Correlation with Tmb, Pd-L1 Expression and Cd8(+) T Cell Infiltration in Chinese Lung Large Cell Neuroendocrine Carcinoma. Lung Cancer 2022, 166, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Jiang, P. A Different Facet of P53 Function: Regulation of Immunity and Inflammation During Tumor Development. Front. Cell Dev. Biol. 2021, 9, 762651. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Päivinen, P.J.; Tripathi, S.; Domenech-Moreno, E.; Wong, I.P.L.; Vaahtomeri, K.; Nagaraj, A.S.; Talwelkar, S.S.; Foretz, M.; Verschuren, E.W.; et al. Inactivation of Ampk Leads to Attenuation of Antigen Presentation and Immune Evasion in Lung Adenocarcinoma. Clin. Cancer Res. 2022, 28, 227–237. [Google Scholar] [CrossRef]
- Danilova, L.; Ho, W.J.; Zhu, Q.; Vithayathil, T.; De Jesus-Acosta, A.; Azad, N.S.; Laheru, D.A.; Fertig, E.J.; Anders, R.; Jaffee, E.M.; et al. Programmed Cell Death Ligand-1 (Pd-L1) And Cd8 Expression Profiling Identify an Immunologic Subtype of Pancreatic Ductal Adenocarcinomas with Favorable Survival. Cancer Immunol. Res. 2019, 7, 886–895. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Z.; Ma, J.; Hou, Y.; Zhao, J.; Dong, B.; Tu, S.; Wang, L.; Guo, Y. Elevated Serum Level of Ca125 Is a Biomarker That Can Be Used to Alter Prognosis Determined by Brca Mutation and Family History in Ovarian Cancer. Genet. Test Mol. Biomark. 2017, 21, 547–554. [Google Scholar] [CrossRef]
- Shilpi, A.; Kandpal, M.; Ji, Y.; Seagle, B.L.; Shahabi, S.; Davuluri, R.V. Platform-Independent Classification System to Predict Molecular Subtypes of High-Grade Serous Ovarian Carcinoma. JCO Clin. Cancer Inform. 2019, 3, 1–9. [Google Scholar] [CrossRef]
- Riaz, N.; Havel, J.J.; Makarov, V.; Desrichard, A.; Urba, W.J.; Sims, J.S.; Hodi, F.S.; Martin-Algarra, S.; Mandal, R.; Sharfman, W.H.; et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017, 171, 934–949.e916. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the Tumor Immune Microenvironment (Time) For Effective Therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, S.; Zhang, Q.; Pan, Y.; Lv, Y.; Chen, X.; Zuo, Y.; Hao, D. Clinical Significance of the Immune Microenvironment in Ovarian Cancer Patients. Mol. Omics 2018, 14, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Grabiner, B.C.; Nardi, V.; Birsoy, K.; Possemato, R.; Shen, K.; Sinha, S.; Jordan, A.; Beck, A.H.; Sabatini, D.M. A Diverse Array of Cancer-Associated Mtor Mutations Are Hyperactivating and Can Predict Rapamycin Sensitivity. Cancer Discov. 2014, 4, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Fumet, J.-D.; Limagne, E.; Thibaudin, M.; Ghiringhelli, F. Immunogenic Cell Death and Elimination of Immunosuppressive Cells: A Double-Edged Sword of Chemotherapy. Cancers 2020, 12, 2637. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Choi, Y.; Yang, S.; Lee, J.; Choi, J.; Moon, Y.; Kim, J.; Shim, N.; Cho, H.; Shim, M.K.; et al. Sustained and Long-Term Release of Doxorubicin From Plga Nanoparticles for Eliciting Anti-Tumor Immune Responses. Pharmaceutics 2022, 14, 474. [Google Scholar] [CrossRef]
- Chen, F.; Shen, J.; Wang, J.; Cai, P.; Huang, Y. Clinical Analysis of Four Serum Tumor Markers in 458 Patients with Ovarian Tumors: Diagnostic Value of the Combined Use of He4, Ca125, Ca19-9, and Cea in Ovarian Tumors. Cancer Manag. Res. 2018, 10, 1313–1318. [Google Scholar] [CrossRef]
- Palucka, K.; Banchereau, J. Dendritic-Cell-Based Therapeutic Cancer Vaccines. Immunity 2013, 39, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Kortekaas, K.E.; Santegoets, S.J.; Sturm, G.; Ehsan, I.; van Egmond, S.L.; Finotello, F.; Trajanoski, Z.; Welters, M.J.P.; van Poelgeest, M.I.E.; van der Burg, S.H. Cd39 Identifies the Cd4(+) Tumor-Specific T-Cell Population in Human Cancer. Cancer Immunol. Res. 2020, 8, 1311–1321. [Google Scholar] [CrossRef]
- Farhood, B.; Najafi, M.; Mortezaee, K. Cd8(+) Cytotoxic T Lymphocytes in Cancer Immunotherapy: A Review. J. Cell. Physiol. 2019, 234, 8509–8521. [Google Scholar] [CrossRef]
- Liu, S.-Y.; Zhu, R.-H.; Wang, Z.-T.; Tan, W.; Zhang, L.; Wang, Y.-Q.; Dai, F.-F.; Yuan, M.-Q.; Zheng, Y.-J.; Yang, D.-Y.; et al. Landscape Of Immune Microenvironment in Epithelial Ovarian Cancer and Establishing Risk Model by Machine Learning. J. Oncol. 2021, 2021, 5523749. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.-C.; Cheng, A.-L.; Brotto, M.; Chuang, C.-Y. Visual Gene-Network Analysis Reveals The Cancer Gene Co-Expression In Human Endometrial Cancer. Bmc Genom. 2014, 15, 300. [Google Scholar] [CrossRef]
- Matsui, T.; Connolly, J.E.; Michnevitz, M.; Chaussabel, D.; Yu, C.I.; Glaser, C.; Tindle, S.; Pypaert, M.; Freitas, H.; Piqueras, B.; et al. Cd2 Distinguishes Two Subsets of Human Plasmacytoid Dendritic Cells with Distinct Phenotype and Functions. J. Immunol. 2009, 182, 6815–6823. [Google Scholar] [CrossRef] [PubMed]
- Springer, T.A.; Dustin, M.; Kishimoto, T.K.; Marlin, S.D. The Lymphocyte Function-Associated Lfa-1, Cd2, and Lfa-3 Molecules: Cell Adhesion Receptors of the Immune System. Annu. Rev. Immunol. 1987, 5, 223–252. [Google Scholar] [CrossRef]
- Call, M.E.; Wucherpfennig, K.W. Molecular Mechanisms for the Assembly of the T Cell Receptor-Cd3 Complex. Mol. Immunol. 2004, 40, 1295–1305. [Google Scholar] [CrossRef]
- Yang, Y.; Zang, Y.; Zheng, C.; Li, Z.; Gu, X.; Zhou, M.; Wang, Z.; Xiang, J.; Chen, Z.; Zhou, Y. Cd3d Is Associated with Immune Checkpoints and Predicts Favorable Clinical Outcome in Colon Cancer. Immunotherapy 2020, 12, 25–35. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, H.; Gao, S.; Wang, J.; Li, C.; Shu, J.; Lin, J. Cd3e as a New Predictive Biomarker of Response to Omalizumab Treatment in Asthma Patients: Evidence from Bioinformatic Analysis. Int. Immunopharmacol. 2021, 93, 107423. [Google Scholar] [CrossRef]
- Huo, X.; Sun, H.; Liu, S.; Liang, B.; Bai, H.; Wang, S.; Li, S. Identification of a Prognostic Signature for Ovarian Cancer Based on the Microenvironment Genes. Front. Genet. 2021, 12, 680413. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Shi, M.; Yu, Y.; Sang, J.; Wang, H.; Shi, J.; Duan, P.; Ge, R. The Immune Subtypes and Landscape of Advanced-Stage Ovarian Cancer. Vaccines 2022, 10, 1451. https://doi.org/10.3390/vaccines10091451
Zhang M, Shi M, Yu Y, Sang J, Wang H, Shi J, Duan P, Ge R. The Immune Subtypes and Landscape of Advanced-Stage Ovarian Cancer. Vaccines. 2022; 10(9):1451. https://doi.org/10.3390/vaccines10091451
Chicago/Turabian StyleZhang, Minjie, Mengna Shi, Yang Yu, Jianmin Sang, Hong Wang, Jianhong Shi, Ping Duan, and Renshan Ge. 2022. "The Immune Subtypes and Landscape of Advanced-Stage Ovarian Cancer" Vaccines 10, no. 9: 1451. https://doi.org/10.3390/vaccines10091451
APA StyleZhang, M., Shi, M., Yu, Y., Sang, J., Wang, H., Shi, J., Duan, P., & Ge, R. (2022). The Immune Subtypes and Landscape of Advanced-Stage Ovarian Cancer. Vaccines, 10(9), 1451. https://doi.org/10.3390/vaccines10091451





