Pterostilbene Promotes Spinal Cord Injury Recovery by Inhibiting Ferroptosis via Keap1/Nrf2/SLC7A11/GPX4 Axis Activation
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal
2.2. SCI Model and Pte Treatment
2.3. Behavioral Assessment
2.4. Histological Analyses
2.5. Perls Staining
2.6. Measurement of Malondialdehyde (MDA) Levels and Glutathione (GSH) Activity
2.7. Screening Differentially Expressed Genes in SCI
2.8. Screening the Target Genes of Pte
2.9. Screening the Characteristic Genes of Oligodendrocytes
2.10. Construction of the Protein–Protein Interaction (PPI) Network
2.11. Screening the Characteristic Genes of PPI Network by Using cytoHubb
2.12. Cell Culture and Drug Treatment
2.13. Cell Viability Assay
2.14. Immunofluorescence
2.15. Western Blot
2.16. FerroOrange Detection
2.17. Lipid Peroxides Assay
2.18. Transmission Electron Microscopy
2.19. Molecular Docking (MD)
2.20. Molecular Dynamics Simulation (MDS)
2.21. Statistical Analysis
3. Results
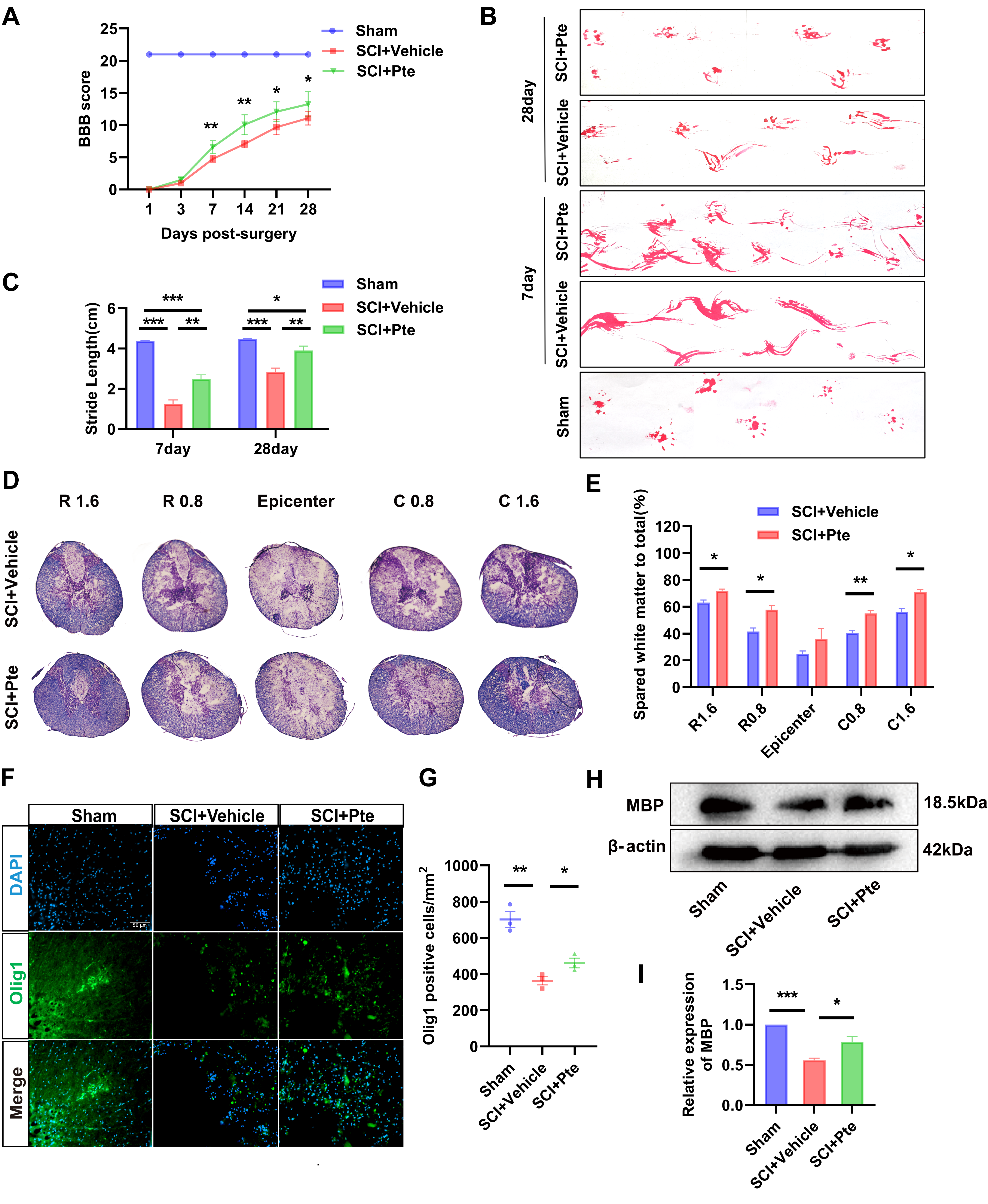
3.1. Pte Promotes the Recovery of Hindlimb Motor Capacity in Rats
3.2. Pte Attenuates Spinal Cord Tissue Damage and Oligodendrocyte Loss
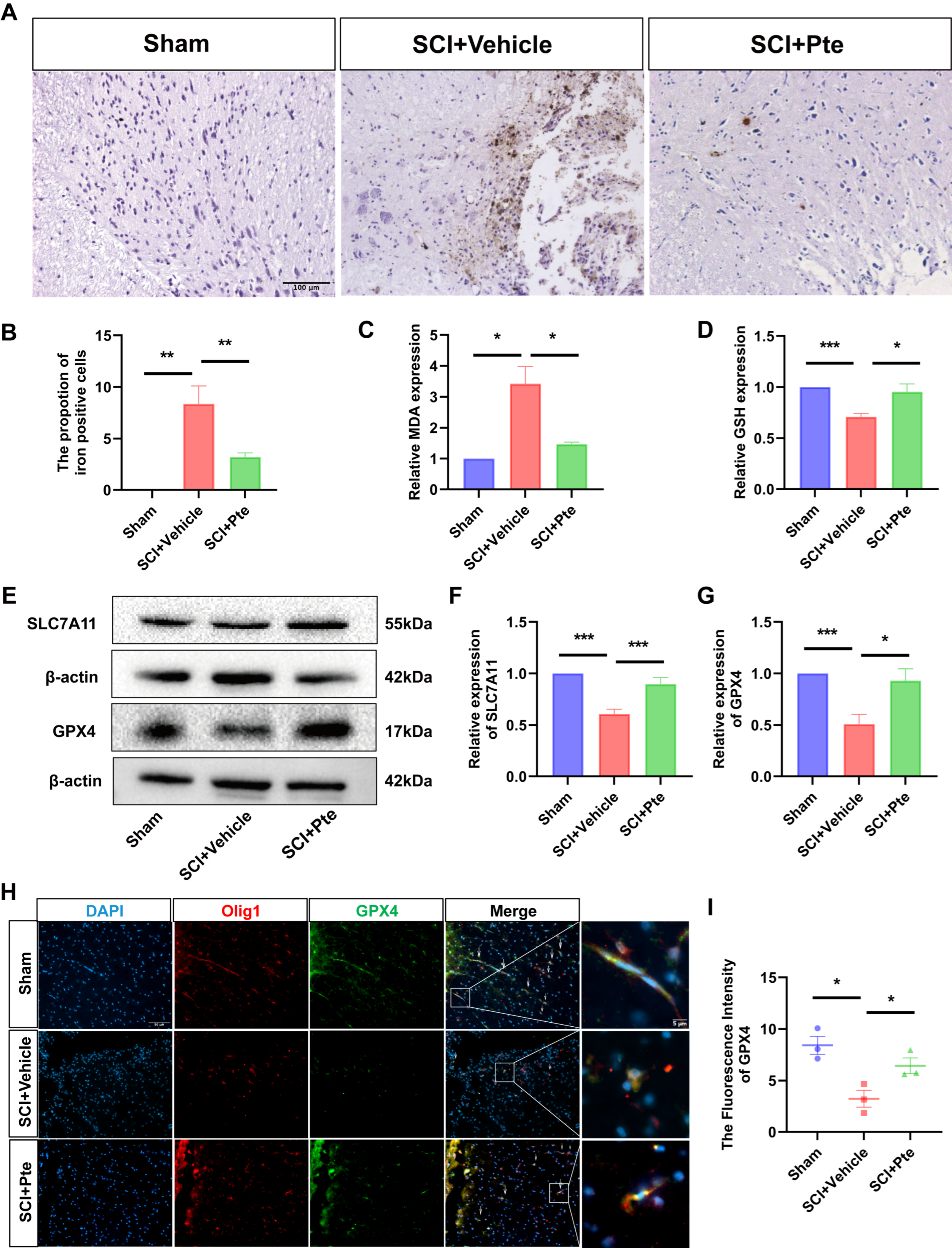
3.3. Pte Protects Spinal Cord Tissue by Suppressing Ferroptosis
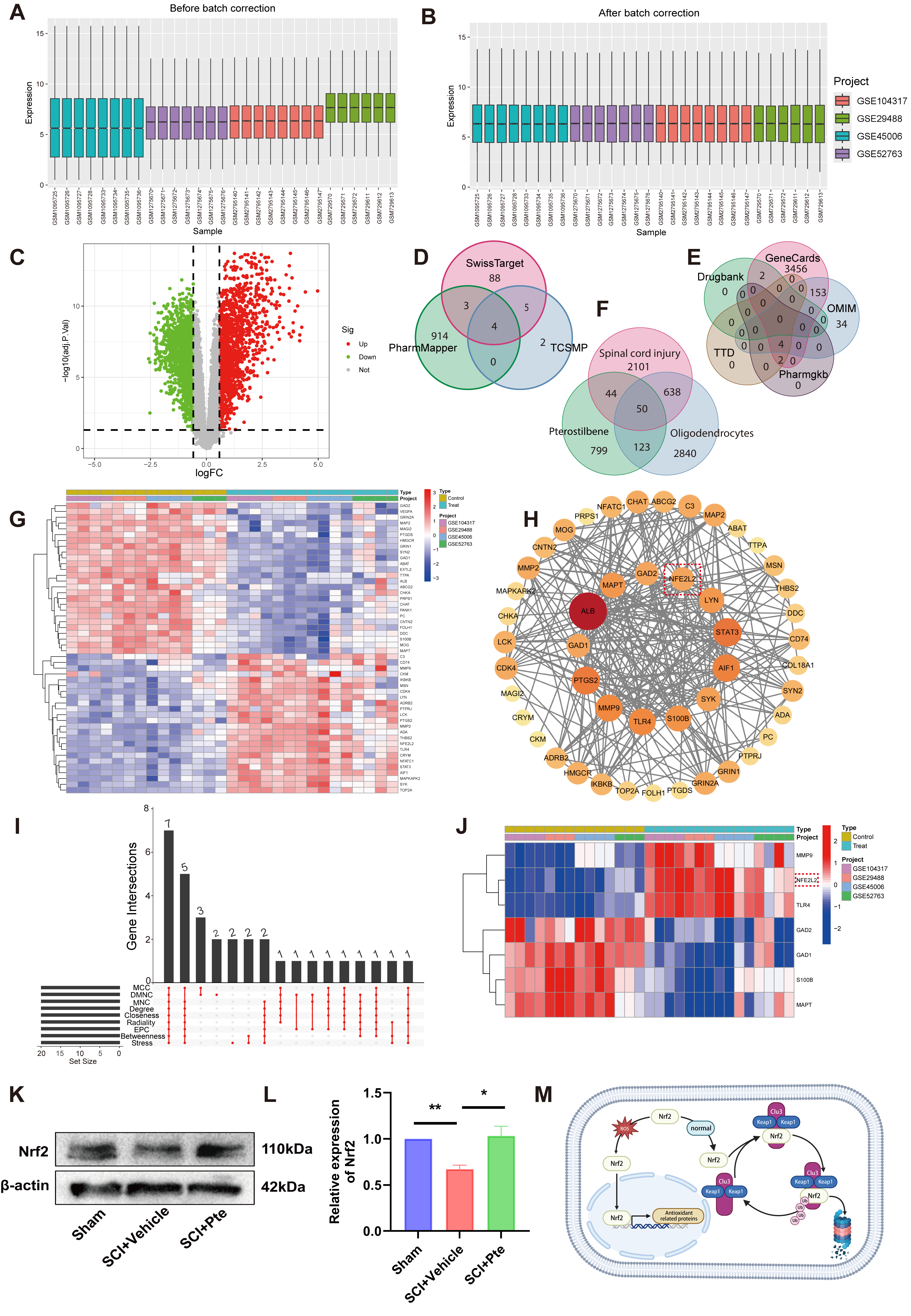
3.4. Nrf2 Is an Effective Target of Pte
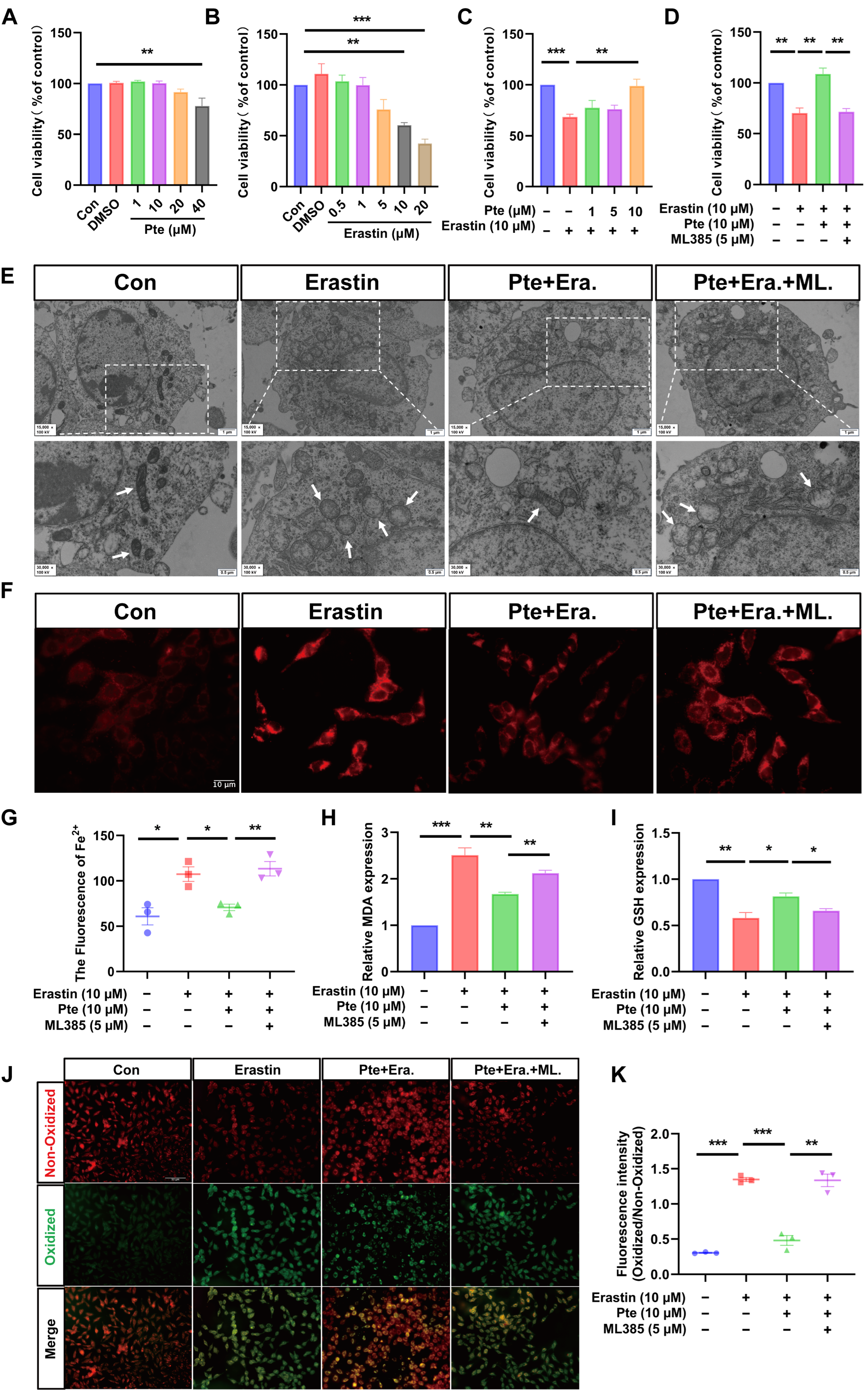
3.5. Pte Inhibits Erastin-Induced Oligodendroglial Cell Ferroptosis
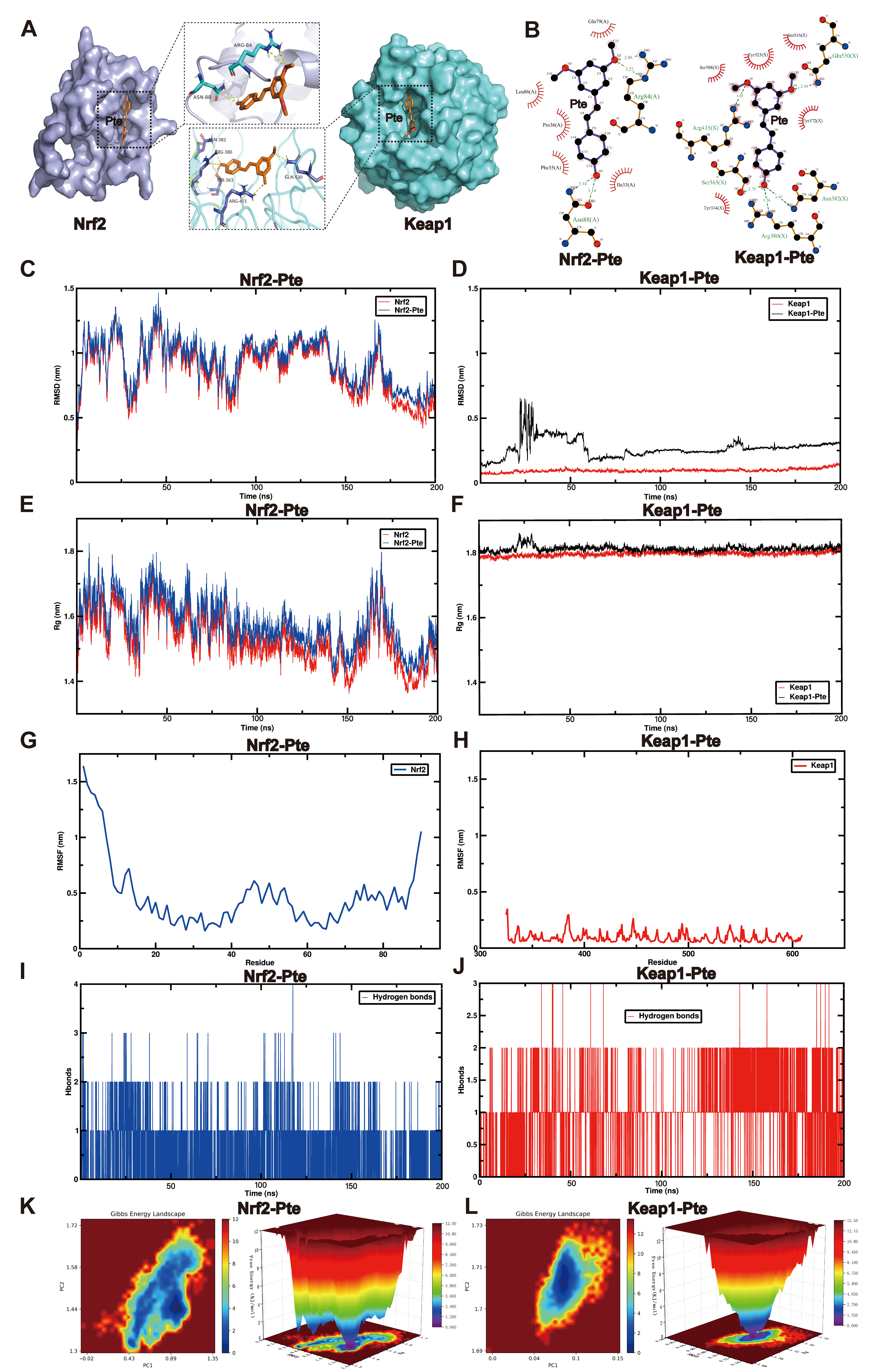
3.6. MD and MDS Show That Pte Had a Higher Affinity for Keap1
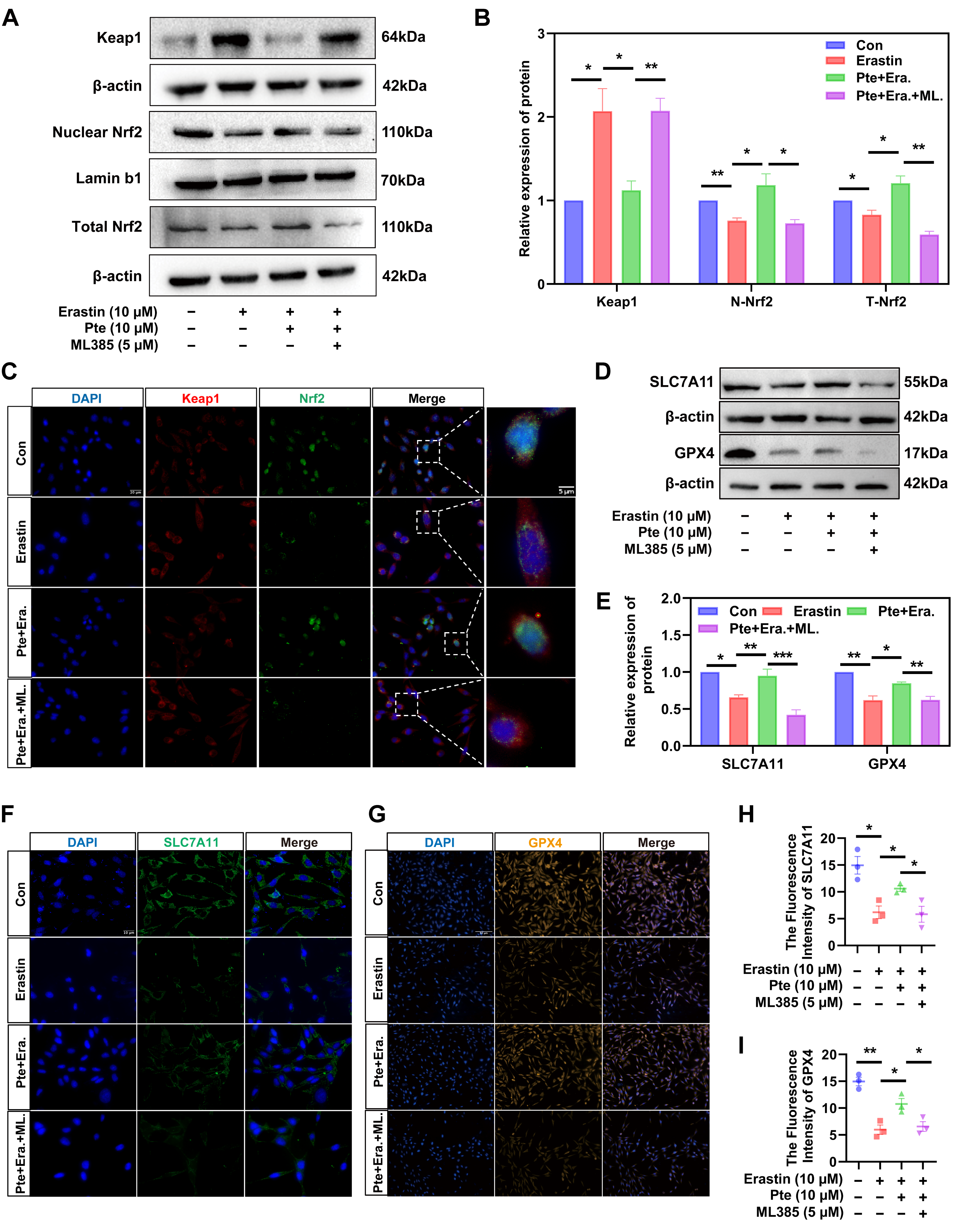
3.7. Pte Increases Nrf2 Nuclear Translocation by Promoting the Dissociation of Keap1 and Nrf2, Thereby Activating the Nrf2/SLC7A11/GPX4 Axis
4. Discussion
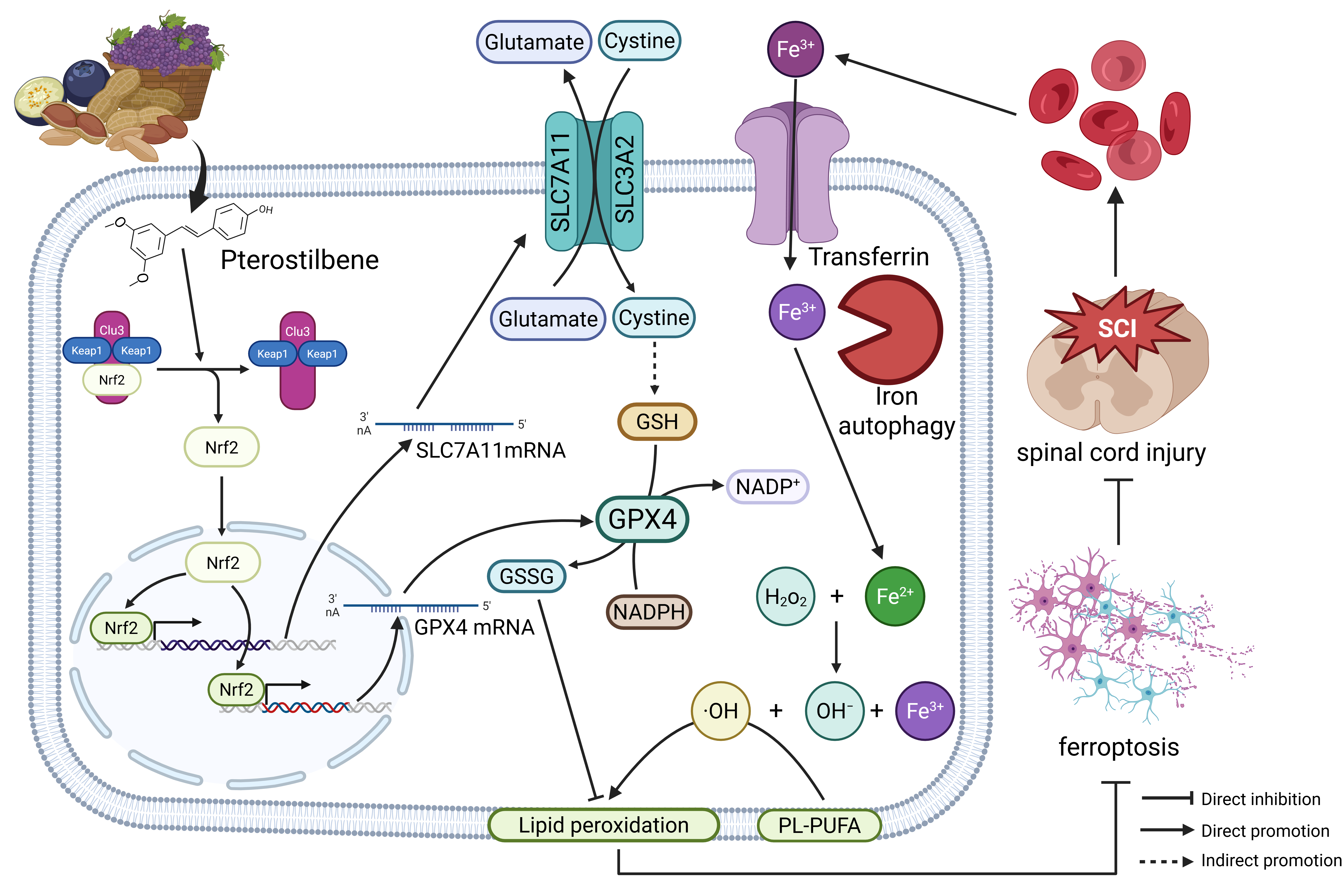
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SCI | Spinal cord injury |
| Pte | Pterostilbene |
| CNS | Central nervous system |
| GPX4 | Glutathione peroxidase |
| SLC7A11 | Golute Carrier Family 7 Member 11 |
| Nrf2 | Nuclear factor E2 |
| WM | White matter |
| EC | Iron-eriochrome cyanine R |
| BBB | Basso–Beattie–Bresnahan |
| GEO | Gene expression omnibus |
| TCMSP | Traditional Chinese Medicine Systems Pharmacology |
| PPI | Protein-protein interaction |
| MD | Molecular docking |
| MDS | Molecular dynamics simulation |
| RMSD | Root-mean-square deviation |
| Rg | Radius of gyration |
| SASA | Solve-accessible surface area |
| RMSF | Root-mean-square fluctuation |
| PCA | Principal component analysis |
| MDA | Malondialdehyde |
| GSH | Glutathione |
| FDR | False Discovery Rate |
| PDB | Protein data bank |
| TIP3P | Potential with 3 points |
| NVT | Normal volume and temperature |
| NPT | Normal pressure and temperature |
| FC | Log2 fold change |
| MBP | Myelin basic protein |
| Keap1 | Kelch Ech-associated protein 1 |
| GSSG | Synthesize oxidized glutathione |
| PLOOH | Phospholipid hydroperoxide |
| PLOH | Phosphatidylcholine |
| PUFAs | Polyunsaturated fatty acids |
| SLC3A2 | Solute carrier family 3 member 2 |
| AREs | Antioxidant response elements |
References
- McDonald, J.W.; Sadowsky, C. Spinal-cord injury. Lancet 2002, 359, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Safdarian, M.; Trinka, E.; Rahimi-Movaghar, V.; Thomschewski, A.; Aali, A.; Abady, G.G.; Abate, S.M.; Abd-Allah, F.; Abedi, A.; Adane, D.E.; et al. Global, regional, and national burden of spinal cord injury, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet Neurol. 2023, 22, 1026–1047. [Google Scholar] [CrossRef] [PubMed]
- Ramer, L.M.; Ramer, M.S.; Bradbury, E.J. Restoring function after spinal cord injury: Towards clinical translation of experimental strategies. Lancet Neurol. 2014, 13, 1241–1256. [Google Scholar] [CrossRef] [PubMed]
- Anjum, A.; Yazid, M.D.; Fauzi Daud, M.; Idris, J.; Ng, A.M.H.; Selvi Naicker, A.; Ismail, O.H.R.; Athi Kumar, R.K.; Lokanathan, Y. Spinal Cord Injury: Pathophysiology, Multimolecular Interactions, and Underlying Recovery Mechanisms. Int. J. Mol. Sci. 2020, 21, 7533. [Google Scholar] [CrossRef]
- Lipinski, M.M.; Wu, J.; Faden, A.I.; Sarkar, C. Function and Mechanisms of Autophagy in Brain and Spinal Cord Trauma. Antioxid. Redox Signal. 2015, 23, 565–577. [Google Scholar] [CrossRef]
- Almad, A.; Sahinkaya, F.R.; McTigue, D.M. Oligodendrocyte fate after spinal cord injury. Neurotherapeutics 2011, 8, 262–273. [Google Scholar] [CrossRef]
- Silva, N.A.; Sousa, N.; Reis, R.L.; Salgado, A.J. From basics to clinical: A comprehensive review on spinal cord injury. Prog. Neurobiol. 2014, 114, 25–57. [Google Scholar] [CrossRef]
- Miron, V.E.; Kuhlmann, T.; Antel, J.P. Cells of the oligodendroglial lineage, myelination, and remyelination. Biochim. Biophys. Acta 2011, 1812, 184–193. [Google Scholar] [CrossRef]
- Mekhail, M.; Almazan, G.; Tabrizian, M. Oligodendrocyte-protection and remyelination post-spinal cord injuries: A review. Prog. Neurobiol. 2012, 96, 322–339. [Google Scholar] [CrossRef]
- Kassmann, C.M.; Lappe-Siefke, C.; Baes, M.; Brügger, B.; Mildner, A.; Werner, H.B.; Natt, O.; Michaelis, T.; Prinz, M.; Frahm, J.; et al. Axonal loss and neuroinflammation caused by peroxisome-deficient oligodendrocytes. Nat. Genet. 2007, 39, 969–976. [Google Scholar] [CrossRef]
- Kassmann, C.M.; Nave, K.A. Oligodendroglial impact on axonal function and survival—A hypothesis. Curr. Opin. Neurol. 2008, 21, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cao, F.; Yin, H.L.; Huang, Z.J.; Lin, Z.T.; Mao, N.; Sun, B.; Wang, G. Ferroptosis: Past, present and future. Cell Death Dis. 2020, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.S.; Jia, Y.J. Ferroptosis: A critical player and potential therapeutic target in traumatic brain injury and spinal cord injury. Neural Regen. Res. 2023, 18, 506–512. [Google Scholar] [CrossRef]
- Shen, L.; Lin, D.; Li, X.; Wu, H.; Lenahan, C.; Pan, Y.; Xu, W.; Chen, Y.; Shao, A.; Zhang, J. Ferroptosis in Acute Central Nervous System Injuries: The Future Direction? Front. Cell Dev. Biol. 2020, 8, 594. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, S.; Li, J.; Li, Z.; Quan, J.; Liu, X.; Tang, Y.; Liu, B. The Latest View on the Mechanism of Ferroptosis and Its Research Progress in Spinal Cord Injury. Oxid. Med. Cell. Longev. 2020, 2020, 6375938. [Google Scholar] [CrossRef]
- Yao, X.; Zhang, Y.; Hao, J.; Duan, H.Q.; Zhao, C.X.; Sun, C.; Li, B.; Fan, B.Y.; Wang, X.; Li, W.X.; et al. Deferoxamine promotes recovery of traumatic spinal cord injury by inhibiting ferroptosis. Neural Regen. Res. 2019, 14, 532–541. [Google Scholar] [CrossRef]
- Hua, R.; Zhao, C.; Xu, Z.; Liu, D.; Shen, W.; Yuan, W.; Li, Y.; Ma, J.; Wang, Z.; Feng, S. ROS-responsive nanoparticle delivery of ferroptosis inhibitor prodrug to facilitate mesenchymal stem cell-mediated spinal cord injury repair. Bioact. Mater. 2024, 38, 438–454. [Google Scholar] [CrossRef]
- Shen, W.; Li, C.; Liu, Q.; Cai, J.; Wang, Z.; Pang, Y.; Ning, G.; Yao, X.; Kong, X.; Feng, S. Celastrol inhibits oligodendrocyte and neuron ferroptosis to promote spinal cord injury recovery. Phytomedicine 2024, 128, 155380. [Google Scholar] [CrossRef]
- Kang, Y.; Zhu, R.; Li, S.; Qin, K.P.; Tang, H.; Shan, W.S.; Yin, Z.S. Erythropoietin inhibits ferroptosis and ameliorates neurological function after spinal cord injury. Neural Regen. Res. 2023, 18, 881–888. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, W.; Zhang, Z.; Zhang, L.; Li, M. Metformin alleviates spinal cord injury by inhibiting nerve cell ferroptosis through upregulation of heme oxygenase-1 expression. Neural Regen. Res. 2024, 19, 2041–2049. [Google Scholar] [CrossRef]
- Ge, M.H.; Tian, H.; Mao, L.; Li, D.Y.; Lin, J.Q.; Hu, H.S.; Huang, S.C.; Zhang, C.J.; Mei, X.F. Zinc attenuates ferroptosis and promotes functional recovery in contusion spinal cord injury by activating Nrf2/GPX4 defense pathway. CNS Neurosci. Ther. 2021, 27, 1023–1040. [Google Scholar] [CrossRef] [PubMed]
- Fotiadis, D.; Kanai, Y.; Palacín, M. The SLC3 and SLC7 families of amino acid transporters. Mol. Asp. Med. 2013, 34, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R. Ferroptosis turns 10: Emerging mechanisms, physiological functions, and therapeutic applications. Cell 2022, 185, 2401–2421. [Google Scholar] [CrossRef] [PubMed]
- Dodson, M.; Castro-Portuguez, R.; Zhang, D.D. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 2019, 23, 101107. [Google Scholar] [CrossRef]
- Michaličková, D.; Hrnčíř, T.; Canová, N.K.; Slanař, O. Targeting Keap1/Nrf2/ARE signaling pathway in multiple sclerosis. Eur. J. Pharmacol. 2020, 873, 172973. [Google Scholar] [CrossRef]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The Keap1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef]
- Kim, H.; Seo, K.H.; Yokoyama, W. Chemistry of Pterostilbene and Its Metabolic Effects. J. Agric. Food Chem. 2020, 68, 12836–12841. [Google Scholar] [CrossRef]
- Qu, X.; Zhang, L.; Wang, L. Pterostilbene as a Therapeutic Alternative for Central Nervous System Disorders: A Review of the Current Status and Perspectives. J. Agric. Food Chem. 2023, 71, 14432–14457. [Google Scholar] [CrossRef]
- Dos Santos Lacerda, D.; Türck, P.; Gazzi de Lima-Seolin, B.; Colombo, R.; Duarte Ortiz, V.; Poletto Bonetto, J.H.; Campos-Carraro, C.; Bianchi, S.E.; Belló-Klein, A.; Linck Bassani, V.; et al. Pterostilbene reduces oxidative stress, prevents hypertrophy and preserves systolic function of right ventricle in cor pulmonale model. Br. J. Pharmacol. 2017, 174, 3302–3314. [Google Scholar] [CrossRef]
- Hseu, Y.C.; Gowrisankar, Y.V.; Wang, L.W.; Zhang, Y.Z.; Chen, X.Z.; Huang, P.J.; Yen, H.R.; Yang, H.L. The in vitro and in vivo depigmenting activity of pterostilbene through induction of autophagy in melanocytes and inhibition of UVA-irradiated α-MSH in keratinocytes via Nrf2-mediated antioxidant pathways. Redox Biol. 2021, 44, 102007. [Google Scholar] [CrossRef]
- Xue, E.X.; Lin, J.P.; Zhang, Y.; Sheng, S.R.; Liu, H.X.; Zhou, Y.L.; Xu, H. Pterostilbene inhibits inflammation and ROS production in chondrocytes by activating Nrf2 pathway. Oncotarget 2017, 8, 41988–42000. [Google Scholar] [CrossRef] [PubMed]
- Obrador, E.; Salvador, R.; Marchio, P.; López-Blanch, R.; Jihad-Jebbar, A.; Rivera, P.; Vallés, S.L.; Banacloche, S.; Alcácer, J.; Colomer, N.; et al. Nicotinamide Riboside and Pterostilbene Cooperatively Delay Motor Neuron Failure in ALS SOD1(G93A) Mice. Mol. Neurobiol. 2021, 58, 1345–1371. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, J.; Jia, L.; Zhou, Y.; Fu, Q.; Wang, Y.; Mu, D.; Wang, D.; Li, N.; Hou, Y. Pterostilbene nanoemulsion promotes Nrf2 signaling pathway to downregulate oxidative stress for treating Alzheimer’s disease. Int. J. Pharm. 2024, 655, 124002. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zeng, Z.; Zhang, J.; Li, X.; Yang, W.; Wei, Y.; Guo, X. Pterostilbene attenuates heart failure by inhibiting myocardial ferroptosis through SIRT1/GSK-3β/GPX4 signaling pathway. Heliyon 2024, 10, e24562. [Google Scholar] [CrossRef]
- Karsy, M.; Hawryluk, G. Modern Medical Management of Spinal Cord Injury. Curr. Neurol. Neurosci. Rep. 2019, 19, 65. [Google Scholar] [CrossRef]
- Bracken, M.B.; Shepard, M.J.; Holford, T.R.; Leo-Summers, L.; Aldrich, E.F.; Fazl, M.; Fehlings, M.G.; Herr, D.L.; Hitchon, P.W.; Marshall, L.F.; et al. Methylprednisolone or tirilazad mesylate administration after acute spinal cord injury: 1-year follow up. Results of the third National Acute Spinal Cord Injury randomized controlled trial. J. Neurosurg. 1998, 89, 699–706. [Google Scholar] [CrossRef]
- Wang, P.; Sang, S. Metabolism and pharmacokinetics of resveratrol and pterostilbene. Biofactors 2018, 44, 16–25. [Google Scholar] [CrossRef]
- Chen, L.; Wang, K.; Wang, L.; Wang, W.; Wang, L.; Wang, W.; Li, J.; Liu, X.; Wang, M.; Ruan, B. Design and synthesis of pterostilbene derivatives bearing triazole moiety that might treat DSS-induced colitis in mice through modulation of NF-κB/MAPK signaling pathways. Eur. J. Med. Chem. 2024, 263, 115949. [Google Scholar] [CrossRef]
- Tang, K.W.; Hsu, C.Y.; Aljuffali, I.A.; Alalaiwe, A.; Lai, W.N.; Gu, P.Y.; Tseng, C.H.; Fang, J.Y. Skin delivery of synthetic benzoyl pterostilbenes suppresses atopic dermatitis-like inflammation through the inhibition of keratinocyte and macrophage activation. Biomed. Pharmacother. 2024, 170, 116073. [Google Scholar] [CrossRef]
- Riche, D.M.; McEwen, C.L.; Riche, K.D.; Sherman, J.J.; Wofford, M.R.; Deschamp, D.; Griswold, M. Analysis of safety from a human clinical trial with pterostilbene. J. Toxicol. 2013, 2013, 463595. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, M.J.; Fernández, M.; Picó, Y.; Mañes, J.; Asensi, M.; Carda, C.; Asensio, G.; Estrela, J.M. Dietary administration of high doses of pterostilbene and quercetin to mice is not toxic. J. Agric. Food Chem. 2009, 57, 3180–3186. [Google Scholar] [CrossRef] [PubMed]
- Kosuru, R.; Singh, S. Pterostilbene ameliorates insulin sensitivity, glycemic control and oxidative stress in fructose-fed diabetic rats. Life Sci. 2017, 182, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, X.; Luo, J.; Wang, X.; Guo, H.; Feng, D.; Zhao, L.; Bai, H.; Song, M.; Liu, X.; et al. Pterostilbene Attenuates Astrocytic Inflammation and Neuronal Oxidative Injury After Ischemia-Reperfusion by Inhibiting NF-κB Phosphorylation. Front. Immunol. 2019, 10, 2408. [Google Scholar] [CrossRef]
- Zhang, Z.T.; Deng, S.M.; Chen, C.; He, Q.H.; Peng, X.W.; Liang, Q.F.; Zhuang, G.D.; Wang, S.M.; Tang, D. Pterostilbene could alleviate diabetic cognitive impairment by suppressing TLR4/NF-κB pathway through microbiota-gut-brain axis. Phytother. Res. 2023, 37, 3522–3542. [Google Scholar] [CrossRef]
- Shi, Z.; Yuan, S.; Shi, L.; Li, J.; Ning, G.; Kong, X.; Feng, S. Programmed cell death in spinal cord injury pathogenesis and therapy. Cell Prolif. 2021, 54, e12992. [Google Scholar] [CrossRef]
- Tator, C.H.; Koyanagi, I. Vascular mechanisms in the pathophysiology of human spinal cord injury. J. Neurosurg. 1997, 86, 483–492. [Google Scholar] [CrossRef]
- Huang, Y.; Bai, J. Ferroptosis in the neurovascular unit after spinal cord injury. Exp. Neurol. 2024, 381, 114943. [Google Scholar] [CrossRef]
- Ryan, F.; Blex, C.; Ngo, T.D.; Kopp, M.A.; Michalke, B.; Venkataramani, V.; Curran, L.; Schwab, J.M.; Ruprecht, K.; Otto, C.; et al. Ferroptosis inhibitor improves outcome after early and delayed treatment in mild spinal cord injury. Acta Neuropathol. 2024, 147, 106. [Google Scholar] [CrossRef]
- Yan, R.; Xie, E.; Li, Y.; Li, J.; Zhang, Y.; Chi, X.; Hu, X.; Xu, L.; Hou, T.; Stockwell, B.R.; et al. The structure of erastin-bound xCT-4F2hc complex reveals molecular mechanisms underlying erastin-induced ferroptosis. Cell Res. 2022, 32, 687–690. [Google Scholar] [CrossRef]
- Liu, Y.; Wan, Y.; Jiang, Y.; Zhang, L.; Cheng, W. GPX4: The hub of lipid oxidation, ferroptosis, disease and treatment. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188890. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Kobayashi, A.; Kang, M.I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef]
- McMahon, M.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003, 278, 21592–21600. [Google Scholar] [CrossRef]
- Zgorzynska, E.; Dziedzic, B.; Walczewska, A. An Overview of the Nrf2/ARE Pathway and Its Role in Neurodegenerative Diseases. Int. J. Mol. Sci. 2021, 22, 9592. [Google Scholar] [CrossRef]
- Baird, L.; Llères, D.; Swift, S.; Dinkova-Kostova, A.T. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proc. Natl. Acad. Sci. USA 2013, 110, 15259–15264. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, L.-X.; An, J.; Yan, L.; Liu, C.-C.; Zhao, J.-J.; Yang, H. Huangqin flavonoid extraction for spinal cord injury in a rat model. Neural Regen. Res. 2018, 13, 2200–2208. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Dong, Y.; Liu, Y.; Ji, Y.; Meng, W.; Cheng, X.; Zheng, X. Pterostilbene Promotes Spinal Cord Injury Recovery by Inhibiting Ferroptosis via Keap1/Nrf2/SLC7A11/GPX4 Axis Activation. Antioxidants 2026, 15, 188. https://doi.org/10.3390/antiox15020188
Dong Y, Liu Y, Ji Y, Meng W, Cheng X, Zheng X. Pterostilbene Promotes Spinal Cord Injury Recovery by Inhibiting Ferroptosis via Keap1/Nrf2/SLC7A11/GPX4 Axis Activation. Antioxidants. 2026; 15(2):188. https://doi.org/10.3390/antiox15020188
Chicago/Turabian StyleDong, Yadan, Yichen Liu, Yixuan Ji, Wen Meng, Xiaoxin Cheng, and Xu Zheng. 2026. "Pterostilbene Promotes Spinal Cord Injury Recovery by Inhibiting Ferroptosis via Keap1/Nrf2/SLC7A11/GPX4 Axis Activation" Antioxidants 15, no. 2: 188. https://doi.org/10.3390/antiox15020188
APA StyleDong, Y., Liu, Y., Ji, Y., Meng, W., Cheng, X., & Zheng, X. (2026). Pterostilbene Promotes Spinal Cord Injury Recovery by Inhibiting Ferroptosis via Keap1/Nrf2/SLC7A11/GPX4 Axis Activation. Antioxidants, 15(2), 188. https://doi.org/10.3390/antiox15020188






