Carbonylated Proteins as Key Regulators in the Progression of Metabolic Syndrome
Abstract
1. Introduction
2. Materials and Methods
2.1. Experiments of Animals
2.2. Measurements of Blood Pressure and Biochemical Tests
2.3. Preparation of Protein Samples
2.4. 2D-DIGE Analysis
2.5. 2D-Oxyblot
2.6. Protein Identification
2.7. UniProt Analysis
2.8. Statistical Analysis
3. Results
3.1. Changes in Body and Epididymal Adipose Tissue Weights, Blood Pressure, and Biochemical Data
3.2. Comparison and Identification of Protein Expression
3.3. Functional Categories of Identified Proteins
3.4. Detection and Identification of Carbonyl Modified Proteins
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA 2001, 285, 2486–2497. [Google Scholar] [CrossRef] [PubMed]
- Grundy, S.M. Metabolic syndrome update. Trends Cardiovasc. Med. 2015, 26, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Saklayen, M.G. The Global Epidemic of the Metabolic Syndrome. Curr. Hypertens. Rep. 2018, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.Y.; Goodman, D.L.; Willems, E.L.; Freedland, A.R.; Norden-Krichmar, T.M.; Santorico, S.A.; Edwards, K.L.; Boerwinkle, E.; Buse, J.; DeFronzo, R.; et al. Genome-wide association analysis of metabolic syndrome quantitative traits in the GENNID multiethnic family study. Diabetol. Metab. Syndr. 2021, 13, 59. [Google Scholar] [CrossRef]
- Dang, A.K.; Le, H.T.; Nguyen, G.T.; Mamun, A.A.; Do, K.N.; Nguyen, L.H.T.; Thai, P.K.; Phung, D. Prevalence of metabolic syndrome and its related factors among Vietnamese people: A systematic review and meta-analysis. Diabetes Metab. Syndr. Clin. Res. Rev. 2022, 16, 102477. [Google Scholar] [CrossRef]
- Hsieh, S.D.; Muto, T.; Tsuji, H.; Arase, Y.; Murase, T. Clustering of other metabolic risk factors in subjects with metabolic syndrome. Metabolism 2010, 59, 697–702. [Google Scholar] [CrossRef]
- Ambroselli, D.; Masciulli, F.; Romano, E.; Catanzaro, G.; Besharat, Z.M.; Massari, M.C.; Ferretti, E.; Migliaccio, S.; Izzo, L.; Ritieni, A.; et al. New Advances in Metabolic Syndrome, from Prevention to Treatment: The Role of Diet and Food. Nutrients 2023, 15, 640. [Google Scholar] [CrossRef]
- Karra, P.; Winn, M.; Pauleck, S.; Bulsiewicz-Jacobsen, A.; Peterson, L.; Coletta, A.; Doherty, J.; Ulrich, C.M.; Summers, S.A.; Gunter, M.; et al. Metabolic dysfunction and obesity-related cancer: Beyond obesity and metabolic syndrome. Obesity 2022, 30, 1323–1334. [Google Scholar] [CrossRef]
- Jialal, I.; Devaraj, S.; Adams-Huet, B.; Chen, X.; Kaur, H. Increased Cellular and Circulating Biomarkers of Oxidative Stress in Nascent Metabolic Syndrome. J. Clin. Endocrinol. Metab. 2012, 97, E1844–E1850. [Google Scholar] [CrossRef]
- Spahis, S.; Borys, J.-M.; Levy, E. Metabolic Syndrome as a Multifaceted Risk Factor for Oxidative Stress. Antioxidants Redox Signal. 2017, 26, 445–461. [Google Scholar] [CrossRef]
- Mizuno, Y.; Yamamotoya, T.; Nakatsu, Y.; Ueda, K.; Matsunaga, Y.; Inoue, M.-K.; Sakoda, H.; Fujishiro, M.; Ono, H.; Kikuchi, T.; et al. Xanthine Oxidase Inhibitor Febuxostat Exerts an Anti-Inflammatory Action and Protects against Diabetic Nephropathy Development in KK-Ay Obese Diabetic Mice. Int. J. Mol. Sci. 2019, 20, 4680. [Google Scholar] [CrossRef]
- Bullón-Vela, V.; Abete, I.; Tur, J.A.; Konieczna, J.; Romaguera, D.; Pintó, X.; Corbella, E.; Martínez-González, M.A.; Sayón-Orea, C.; Toledo, E.; et al. Relationship of visceral adipose tissue with surrogate insulin resistance and liver markers in individuals with metabolic syndrome chronic complications. Ther. Adv. Endocrinol. Metab. 2020, 11, 2042018820958298. [Google Scholar] [CrossRef] [PubMed]
- Parasiliti-Caprino, M.; Bollati, M.; Merlo, F.D.; Ghigo, E.; Maccario, M.; Bo, S. Adipose Tissue Dysfunction in Obesity: Role of Mineralocorticoid Receptor. Nutrients 2022, 14, 4735. [Google Scholar] [CrossRef] [PubMed]
- Saiki, A.; Oyama, T.; Endo, K.; Ebisuno, M.; Ohira, M.; Koide, N.; Murano, T.; Miyashita, Y.; Shirai, K. Preheparin serum lipoprotein lipase mass might be a biomarker of metabolic syndrome. Diabetes Res. Clin. Pract. 2007, 76, 93–101. [Google Scholar] [CrossRef]
- Chaves, L.O.; Carraro, J.C.C.; Vidigal, F.D.C.; Bressan, J. Higher Waist Circumference Is Related to Lower Plasma Polyunsaturated Fatty Acids in Healthy Participants: Metabolic Implications. J. Am. Coll. Nutr. 2018, 38, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Striffler, J.S.; Bhathena, S.J.; Michaelis, O.E.; Campbell, J.D.; Hansen, C.T.; Scalbert, E.; Thibault, N.; Velasquez, M.T. Long-term effects of perindopril on metabolic parameters and the heart in the spontaneously hypertensive/NIH-corpulent rat with non—Insulin-dependent diabetes mellitus and hypertension. Metabolism 1998, 47, 1199–1204. [Google Scholar] [CrossRef] [PubMed]
- Koletsky, S. Obese spontaneously hypertensive rats—A model for study of atherosclerosis. Exp. Mol. Pathol. 1973, 19, 53–60. [Google Scholar] [CrossRef]
- Russell, J.C.; Shillabeer, G.; Bar-Tana, J.; Lau, D.C.; Richardson, M.; Wenzel, L.M.; Graham, S.E.; Dolphin, P.J. Development of insulin resistance in the JCR:LA-cp rat: Role of triacylglycerols and effects of MEDICA 16. Diabetes 1998, 47, 770–778. [Google Scholar] [CrossRef]
- Kagota, S.; Tada, Y.; Nejime, N.; Nakamura, K.; Kunitomo, M.; Shinozuka, K. Chronic Production of Peroxynitrite in the Vascular Wall Impairs Vasorelaxation Function in SHR/NDmcr-cp Rats, an Animal Model of Metabolic Syndrome. J. Pharmacol. Sci. 2009, 109, 556–564. [Google Scholar] [CrossRef]
- Andreadou, I.; Schulz, R.; Badimon, L.; Adameová, A.; Kleinbongard, P.; Lecour, S.; Nikolaou, P.; Falcão-Pires, I.; Vilahur, G.; Woudberg, N.; et al. Hyperlipidaemia and cardioprotection: Animal models for translational studies. Br. J. Pharmacol. 2020, 177, 5287–5311. [Google Scholar] [CrossRef]
- Chang, J.; Oikawa, S.; Iwahashi, H.; Kitagawa, E.; Takeuchi, I.; Yuda, M.; Aoki, C.; Yamada, Y.; Ichihara, G.; Kato, M.; et al. Expression of proteins associated with adipocyte lipolysis was significantly changed in the adipose tissues of the obese spontaneously hypertensive/NDmcr-cp rat. Diabetol. Metab. Syndr. 2014, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Oikawa, S.; Ichihara, G.; Nanpei, Y.; Hotta, Y.; Yamada, Y.; Tada-Oikawa, S.; Iwahashi, H.; Kitagawa, E.; Takeuchi, I.; et al. Altered gene and protein expression in liver of the obese spontaneously hypertensive/NDmcr-cp rat. Nutr. Metab. 2012, 9, 87. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, S.; Kobayashi, H.; Kitamura, Y.; Zhu, H.; Obata, K.; Minabe, Y.; Dazortsava, M.; Ohashi, K.; Tada-Oikawa, S.; Takahashi, H.; et al. Proteomic analysis of carbonylated proteins in the monkey substantia nigra after ischemia-reperfusion. Free Radic. Res. 2014, 48, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Ducret, A.; Khoueiry, P.; Lignon, S.; Longhi, S.; Talla, E.; Dukan, S. Rules Governing Selective Protein Carbonylation. PLoS ONE 2009, 4, e7269. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Kitamori, K.; Ichihara, G.; Suzuki, Y.; Ochiai, M.; Yamada, Y.; Tada-Oikawa, S.; Tsuchikura, S.; Yamori, Y.; Ichihara, S. Serial changes in adipocytokines and cardiac function in a rat model of the metabolic syndrome. Clin. Exp. Pharmacol. Physiol. 2013, 40, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Rahaman, S.; Mise, N.; Ikegami, A.; Zong, C.; Ichihara, G.; Ichihara, S. The mechanism of low-level arsenic exposure-induced hypertension: Inhibition of the activity of the angiotensin-converting enzyme 2. Chemosphere 2023, 318, 137911. [Google Scholar] [CrossRef]
- Kondo, T.; Hirohashi, S. Application of highly sensitive fluorescent dyes (CyDye DIGE Fluor saturation dyes) to laser microdissection and two-dimensional difference gel electrophoresis (2D-DIGE) for cancer proteomics. Nat. Protoc. 2006, 1, 2940–2956. [Google Scholar] [CrossRef]
- Kitamura, Y.; Kojima, M.; Kurosawa, T.; Sasaki, R.; Ichihara, S.; Hiraku, Y.; Tomimoto, H.; Murata, M.; Oikawa, S. Proteomic Profiling of Exosomal Proteins for Blood-based Biomarkers in Parkinson’s Disease. Neuroscience 2018, 392, 121–128. [Google Scholar] [CrossRef]
- Ichihara, S.; Suzuki, Y.; Chang, J.; Kuzuya, K.; Inoue, C.; Kitamura, Y.; Oikawa, S. Involvement of oxidative modification of proteins related to ATP synthesis in the left ventricles of hamsters with cardiomyopathy. Sci. Rep. 2017, 7, 9243. [Google Scholar] [CrossRef]
- Suzuki, M.; Takeshita, K.; Kitamura, Y.; Kuribayashi, M.; Huang, Z.; Ichihara, G.; Oikawa, S.; Ichihara, S. In Vitro Exposure to Glucose Alters the Expression of Phosphorylated Proteins in Platelets. Biomedicines 2023, 11, 543. [Google Scholar] [CrossRef]
- Almansa-Ordonez, A.; Bellido, R.; Vassena, R.; Barragan, M.; Zambelli, F. Oxidative Stress in Reproduction: A Mitochondrial Perspective. Biology 2020, 9, 269. [Google Scholar] [CrossRef]
- Onukwufor, J.O.; Berry, B.J.; Wojtovich, A.P. Physiologic Implications of Reactive Oxygen Species Production by Mitochondrial Complex I Reverse Electron Transport. Antioxidants 2019, 8, 285. [Google Scholar] [CrossRef]
- Ruan, X.; Li, Z.; Zhang, Y.; Yang, L.; Pan, Y.; Wang, Z.; Feng, G.-S.; Chen, Y. Apolipoprotein A-I possesses an anti-obesity effect associated with increase of energy expenditure and up-regulation of UCP1 in brown fat. J. Cell. Mol. Med. 2011, 15, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, S.; Fujita, T.; Shimabukuro, M.; Iwaki, M.; Yamada, Y.; Nakajima, Y.; Nakayama, O.; Makishima, M.; Matsuda, M.; Shimomura, I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Investig. 2004, 114, 1752–1761. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, S.; Reed, T.T.; Venditti, P.; Victor, V.M. Role of ROS and RNS Sources in Physiological and Pathological Conditions. Oxid. Med. Cell. Longev. 2016, 2016, 1245049. [Google Scholar] [CrossRef] [PubMed]
- Masschelin, P.M.; Cox, A.R.; Chernis, N.; Hartig, S.M. The Impact of Oxidative Stress on Adipose Tissue Energy Balance. Front. Physiol. 2020, 10, 1638. [Google Scholar] [CrossRef]
- Quijano, C.; Trujillo, M.; Castro, L.; Trostchansky, A. Interplay between oxidant species and energy metabolism. Redox Biol. 2015, 8, 28–42. [Google Scholar] [CrossRef]
- Bedard, K.; Krause, K.-H. The NOX Family of ROS-Generating NADPH Oxidases: Physiology and Pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; Chandel, N.S. Warburg Effect and Redox Balance. Science 2011, 334, 1219–1220. [Google Scholar] [CrossRef]
- Tangvarasittichai, S. Oxidative stress, insulin resistance, dyslipidemia and type 2 diabetes mellitus. World J. Diabetes 2015, 6, 456–480. [Google Scholar] [CrossRef]
- Hauck, A.K.; Huang, Y.; Hertzel, A.V.; Bernlohr, D.A. Adipose oxidative stress and protein carbonylation. J. Biol. Chem. 2019, 294, 1083–1088. [Google Scholar] [CrossRef]
- Cangemi, R.; Angelico, F.; Loffredo, L.; Del Ben, M.; Pignatelli, P.; Martini, A.; Violi, F. Oxidative stress-mediated arterial dysfunction in patients with metabolic syndrome: Effect of ascorbic acid. Free Radic. Biol. Med. 2007, 43, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Avery, S.V. Molecular targets of oxidative stress. Biochem. J. 2011, 434, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Long, E.K.; Olson, D.M.; Bernlohr, D.A. High-fat diet induces changes in adipose tissue trans-4-oxo-2-nonenal and trans-4-hydroxy-2-nonenal levels in a depot-specific manner. Free Radic. Biol. Med. 2013, 63, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Hauck, A.K.; Bernlohr, D.A. Oxidative stress and lipotoxicity. J. Lipid Res. 2016, 57, 1976–1986. [Google Scholar] [CrossRef]
- Frohnert, B.; Sinaiko, A.R.; Serrot, F.J.; Foncea, R.E.; Moran, A.; Ikramuddin, S.; Choudry, U.; Bernlohr, D.A. Increased Adipose Protein Carbonylation in Human Obesity. Obesity 2011, 19, 1735–1741. [Google Scholar] [CrossRef] [PubMed]
- England, K.; Odriscoll, C.; Cotter, T.G. Carbonylation of glycolytic proteins is a key response to drug-induced oxidative stress and apoptosis. Cell Death Differ. 2004, 11, 252–260. [Google Scholar] [CrossRef]
- Curtis, J.M.; Hahn, W.; Stone, M.D.; Inda, J.J.; Droullard, D.J.; Kuzmicic, J.P.; Donoghue, M.A.; Long, E.K.; Armien, A.G.; Lavandero, S.; et al. Protein Carbonylation and Adipocyte Mitochondrial Function. J. Biol. Chem. 2012, 287, 32967–32980. [Google Scholar] [CrossRef]
- Mráček, T.; Drahota, Z.; Houštěk, J. The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochim. Biophys. Acta (BBA)—Bioenerg. 2013, 1827, 401–410. [Google Scholar] [CrossRef]
- Sun, H.Q.; Yamamoto, M.; Mejillano, M.; Yin, H.L. Gelsolin, a Multifunctional Actin Regulatory Protein. J. Biol. Chem. 1999, 274, 33179–33182. [Google Scholar] [CrossRef]
- Gupta, A.K.; Chopra, B.S.; Vaid, B.; Sagar, A.; Raut, S.; Badmalia, M.D.; Ashish; Khatri, N. Protective effects of gelsolin in acute pulmonary thromboembolism and thrombosis in the carotid artery of mice. PLoS ONE 2019, 14, e0215717. [Google Scholar] [CrossRef] [PubMed]
- Suhler, E.; Lin, W.; Yin, H.L.; Lee, W.M. Decreased plasma gelsolin concentrations in acute liver failure, myocardial infarction, septic shock, and myonecrosis. Crit. Care Med. 1997, 25, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.-C.; Luo, B.-Y.; Li, X.-F.; Yang, D.-G.; Zheng, X.-N.; Zhang, K. Plasma gelsolin levels and 1-year mortality after first-ever ischemic stroke. J. Crit. Care 2011, 26, 608–612. [Google Scholar] [CrossRef] [PubMed]
- Gertz, K.; Uhlemann, R.; Foryst-Ludwig, A.; Barrientos, R.M.; Kappert, K.; Thöne-Reineke, C.; Djoufack, P.; Kirschbaum, C.; Fink, K.B.; Heinz, A.; et al. The cytoskeleton in ‘couch potato-ism’: Insights from a murine model of impaired actin dynamics. Exp. Neurol. 2018, 306, 34–44. [Google Scholar] [CrossRef]
- Bai, Y.; Sun, Q. Macrophage recruitment in obese adipose tissue. Obes. Rev. 2015, 16, 127–136. [Google Scholar] [CrossRef]
- Kalupahana, N.S.; Massiera, F.; Quignard-Boulange, A.; Ailhaud, G.; Voy, B.H.; Wasserman, D.H.; Moustaid-Moussa, N. Overproduction of Angiotensinogen from Adipose Tissue Induces Adipose Inflammation, Glucose Intolerance, and Insulin Resistance. Obesity 2012, 20, 48–56. [Google Scholar] [CrossRef]
- Wang, L.; Chen, L.; Liu, Z.; Liu, Y.; Luo, M.; Chen, N.; Deng, X.; Luo, Y.; He, J.; Zhang, L.; et al. PAI-1 Exacerbates White Adipose Tissue Dysfunction and Metabolic Dysregulation in High Fat Diet-Induced Obesity. Front. Pharmacol. 2018, 9, 1087. [Google Scholar] [CrossRef]
- Behl, T.; Gupta, A.; Chigurupati, S.; Singh, S.; Sehgal, A.; Badavath, V.N.; Alhowail, A.; Mani, V.; Bhatia, S.; Al-Harrasi, A.; et al. Natural and Synthetic Agents Targeting Reactive Carbonyl Species against Metabolic Syndrome. Molecules 2022, 27, 1583. [Google Scholar] [CrossRef]

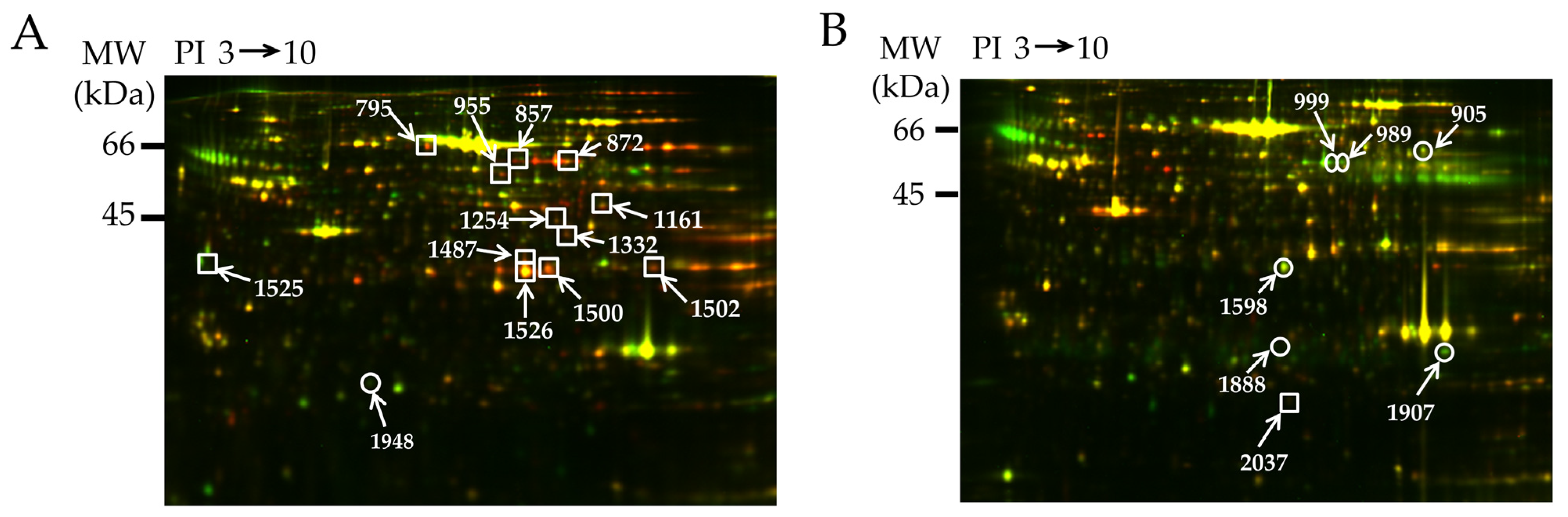
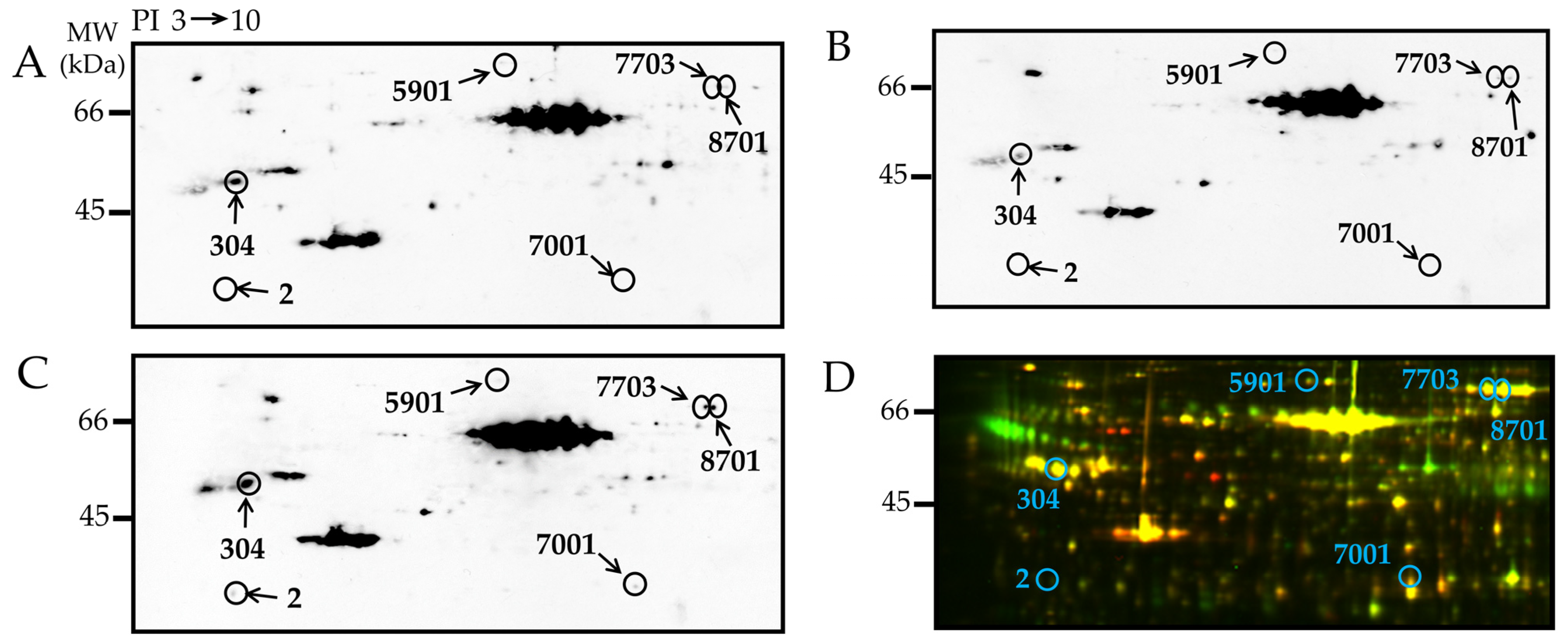
| Spot | Uniprot ID | Protein Name | % Cov. | Peptides (95%) | Fold Change Up (+) or Down (−) | |
|---|---|---|---|---|---|---|
| CP/WKY | CP/Lean | |||||
| 795 | P08461 | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase (DALT) | 19.5 | 7 | 1.39 | 1.54 |
| 857 | P47942 | Dihydropyrimidinase-related protein 2 (DRP-2) | 19.2 | 7 | 1.85 | 1.83 |
| 872 | P13697 | NADP-dependent malic enzyme (ME-1) | 11.9 | 5 | 1.62 | 1.50 |
| 955 | P05370 | Glucose-6-phosphate 1 dehydrogenase (G6PDX) | 21.2 | 8 | 1.77 | 1.88 |
| 1161 | P85968 | 6-phosphogluconate dehydrogenase decarboxylating (PGD) | 19.5 | 6 | 1.58 | 1.67 |
| 1254 | P85834 | Elongation factor Tu (TUFM) | 21.5 | 7 | 1.62 | 1.42 |
| 1332 | P15650 | Long chain specific acetyl CoA dehydrogenase (ACADL) | 16.1 | 5 | 1.71 | 1.54 |
| 1487 | O35077 | Glycerol-3-phosphate dehydrogenase [NAD+] (GPD1) | 35.0 | 9 | 1.42 | 1.61 |
| 1500 | Q91W30 | Aldose reductase (AKR1B8) | 39.9 | 9 | 1.78 | 1.88 |
| 1502 | P04797 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | 23.4 | 5 | 1.62 | 1.61 |
| 1525 | O35077 | Glycerol-3-phosphate dehydrogenase [NAD+] | 42.1 | 9 | 1.52 | 1.73 |
| 1526 | O35077 | Glycerol-3-phosphate dehydrogenase [NAD+] | 14.9 | 5 | 1.61 | 1.72 |
| 1948 | P04639 | Apolipoprotein A-1 (APOA1) | 22.0 | 4 | −1.60 | −1.81 |
| Spot | Uniprot ID | Protein Name | % Cov. | Peptides (95%) | Fold Change Up (+) or Down (−) | |
|---|---|---|---|---|---|---|
| CP/WKY | CP/Lean | |||||
| 905 | P04762 | Catalase (CAT) | 16.9 | 7 | −1.46 | −1.39 |
| 989 | Q68FS4 | Cytosol aminopeptidase (LAP3) | 7.9 | 3 | −2.39 | −2.58 |
| 999 | O08651 | D-3-phosphoglycerate dehydrogenase (PHGDH) | 15.0 | 4 | −1.69 | −2.24 |
| 1598 | O88989 | Malate dehydrogenase (MDH1) | 26.4 | 7 | −1.66 | −1.93 |
| 1888 | Q9Z0V6 | Thioredoxin-dependent peroxide reductase (PRDX3) | 23.7 | 4 | −1.47 | −1.27 |
| 1907 | P08010 | Glutathione S-transferase Mu-2 (GSTM2) | 36.7 | 7 | −1.61 | −2.10 |
| 2037 | P02793 | Ferritin light chain 1 (FTL1) | 31.7 | 4 | 1.78 | 1.59 |
| Protein Name | Molecular Function | Biological Process | Cellular Component |
|---|---|---|---|
| Up-regulation | |||
| Oxidoreductases | |||
| ME-1 | Malate dehydrogenase (decarboxylating) (NAD+) activity | NADH metabolic process, NADP metabolic process, Pyruvate metabolic process | Mitochondria |
| G6PDX | Glucose-6-phosphate dehydrogenase activity | Glucose metabolic process, NADP biosynthetic process | Nucleus, Cytosol |
| GPD1 | Glycerol-3-phosphate dehydrogenase [NAD(P)+] activity | NADH metabolic process, Glycerol-3-phosphate metabolic process, NADH oxidation, Gluconeogenesis | Cytosol |
| PGD | Phosphogluconate dehydrogenase (decarboxylating) activity | Carbohydrate metabolic process, NADP metabolic process | Cytosol |
| GAPDH | Microtubule binding, Glyceraldehyde-3-phosphate dehydrogenase (NAD+) activity | Carbohydrate metabolic process, Microtubule cytoskeleton organization | Nucleus, Cytosol, Cytoskeleton |
| ACADL | Acyl-CoA dehydrogenase activity, Fatty-acyl-CoA binding | Fatty acid catabolic process, Regulation of cholesterol metabolic process | Mitochondria |
| AKR1B8 | Alditol: NADP+ 1-oxidoreductase activity | - | Mitochondria |
| Transferase | |||
| DALT | Dihydrolipoyllysine-residue acetyltransferase activity | Tricarboxylic acid cycle, Acetyl-CoA biosynthetic process from pyruvate | Mitochondria |
| Cytoskeleton | |||
| DRP-2 | Microtubule binding, Hydrolase activity | Cytoskeleton organization | Cytosol |
| Elongation factor | |||
| TUFM | Translation elongation factor activity | Mitochondrial translational elongation | Mitochondria |
| Down-regulation | |||
| Lipid transport | |||
| APOA1 | Lipid transporter activity | Cholesterol metabolic process, Lipoprotein metabolic process, Reverse cholesterol transport | High-density lipoprotein particle |
| Protein Name | Molecular Function | Biological Process | Cellular Component |
|---|---|---|---|
| Down-regulation | |||
| Antioxidants | |||
| CAT | Antioxidant activity, Catalase activity | Hydrogen peroxide catabolic process, Response to hydrogen peroxide, Response to oxidative stress | Peroxisome |
| PRDX3 | Thioredoxin peroxidase activity | Cellular response to oxidative stress, Hydrogen peroxide catabolic process, Cell redox homeostasis | Mitochondria, Cytosol |
| GSTM2 | Glutathione peroxidase activity, Glutathione transferase activity, | Glutathione metabolic process | Cytosol, Cytoplasm |
| Oxidoreductases | |||
| PHGDH | Phosphoglycerate dehydrogenase activity | Glutamine metabolic process, Threonine metabolic process | - |
| MDH1 | Hydroxyphenylpyruvate reductase activity, Malate dehydrogenase activity | Tricarboxylic acid cycle, NADH metabolic process | Cytoplasm |
| Hydrolase | |||
| LAP3 | Carboxypeptidase activity, Metalloaminopeptidase activity | Proteolysis | Cytoplasm |
| Up-regulation | |||
| Iron storage | |||
| FTL1 | Ferric iron binding | Iron ion transport, Cellular iron ion homeostasis | Cytoplasm |
| Spot | Uniprot ID | Protein Name | % Cov. | Peptides (95%) | Fold Change Up (+) or Down (−) | p Value | |
|---|---|---|---|---|---|---|---|
| CP/WKY | CP/Lean | ||||||
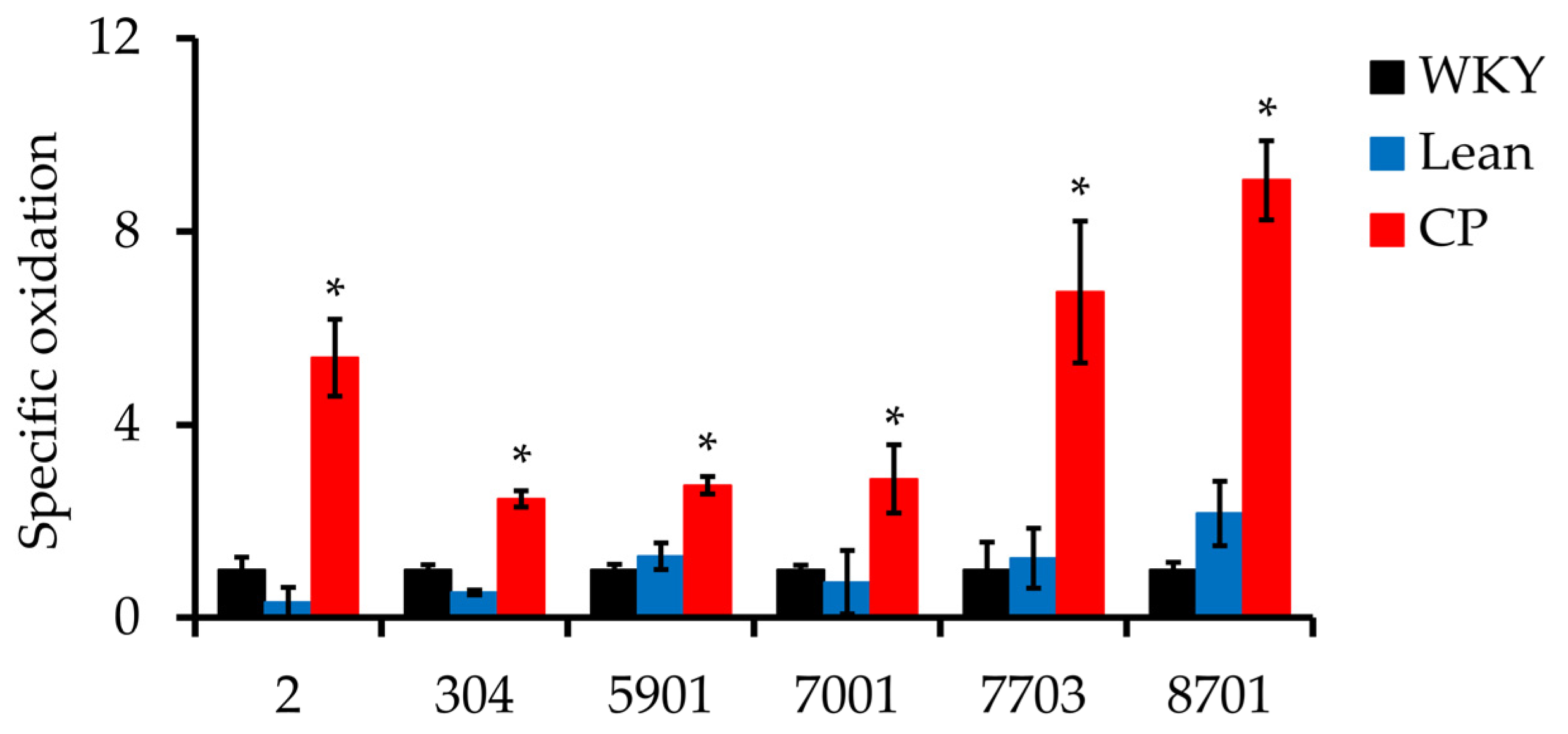
| 2 | P14668 | Annexin A5 (ANXA5) | 31.4 | 7 | 5.39 | 16.6 | 0.006 |
| 304 | Q3KRE8 | Tubulin beta-2B chain (TUBB2B) | 24.9 | 7 | 2.46 | 4.70 | 0.003 |
| 5901 | Q68FP1 | Gelsolin (GSN) | 11.3 | 6 | 2.74 | 2.14 | 0.017 |
| 7001 | O35077 | Glycerol-3-phosphate dehydrogenase [NAD+] (GPD1) | 34.1 | 7 | 2.87 | 3.89 | 0.029 |
| 7703 | P12346 | Serotransferrin (TF) | 18.6 | 5 | 6.74 | 5.45 | 0.042 |
| 8701 | P12346 | Serotransferrin | 19.8 | 7 | 9.06 | 4.19 | 0.005 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kitamura, Y.; Oikawa, S.; Chang, J.; Mori, Y.; Ichihara, G.; Ichihara, S. Carbonylated Proteins as Key Regulators in the Progression of Metabolic Syndrome. Antioxidants 2023, 12, 844. https://doi.org/10.3390/antiox12040844
Kitamura Y, Oikawa S, Chang J, Mori Y, Ichihara G, Ichihara S. Carbonylated Proteins as Key Regulators in the Progression of Metabolic Syndrome. Antioxidants. 2023; 12(4):844. https://doi.org/10.3390/antiox12040844
Chicago/Turabian StyleKitamura, Yuki, Shinji Oikawa, Jie Chang, Yurie Mori, Gaku Ichihara, and Sahoko Ichihara. 2023. "Carbonylated Proteins as Key Regulators in the Progression of Metabolic Syndrome" Antioxidants 12, no. 4: 844. https://doi.org/10.3390/antiox12040844
APA StyleKitamura, Y., Oikawa, S., Chang, J., Mori, Y., Ichihara, G., & Ichihara, S. (2023). Carbonylated Proteins as Key Regulators in the Progression of Metabolic Syndrome. Antioxidants, 12(4), 844. https://doi.org/10.3390/antiox12040844






