NO and Heme Proteins: Cross-Talk between Heme and Cysteine Residues
Abstract
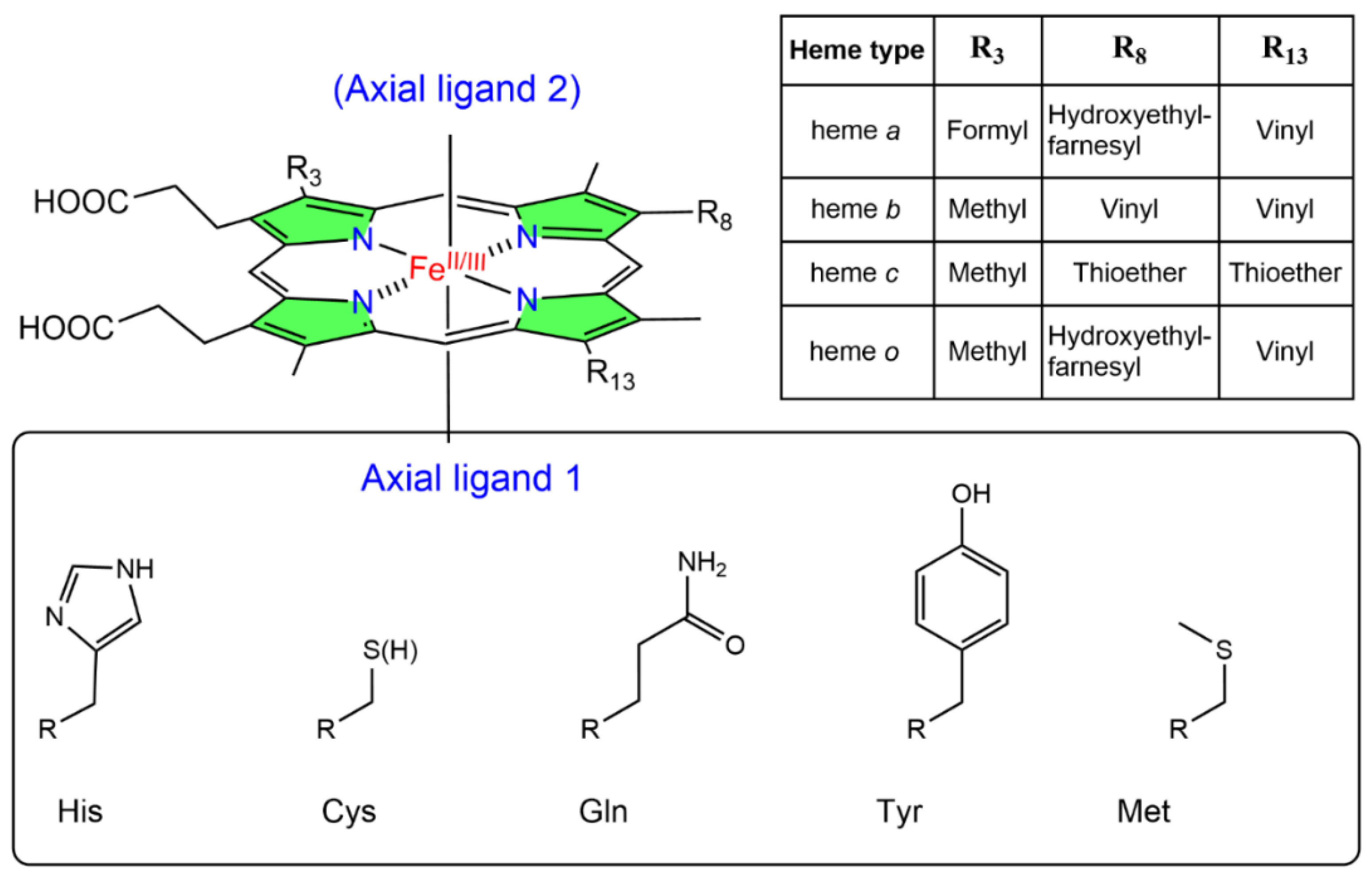
1. NO and Heme Proteins
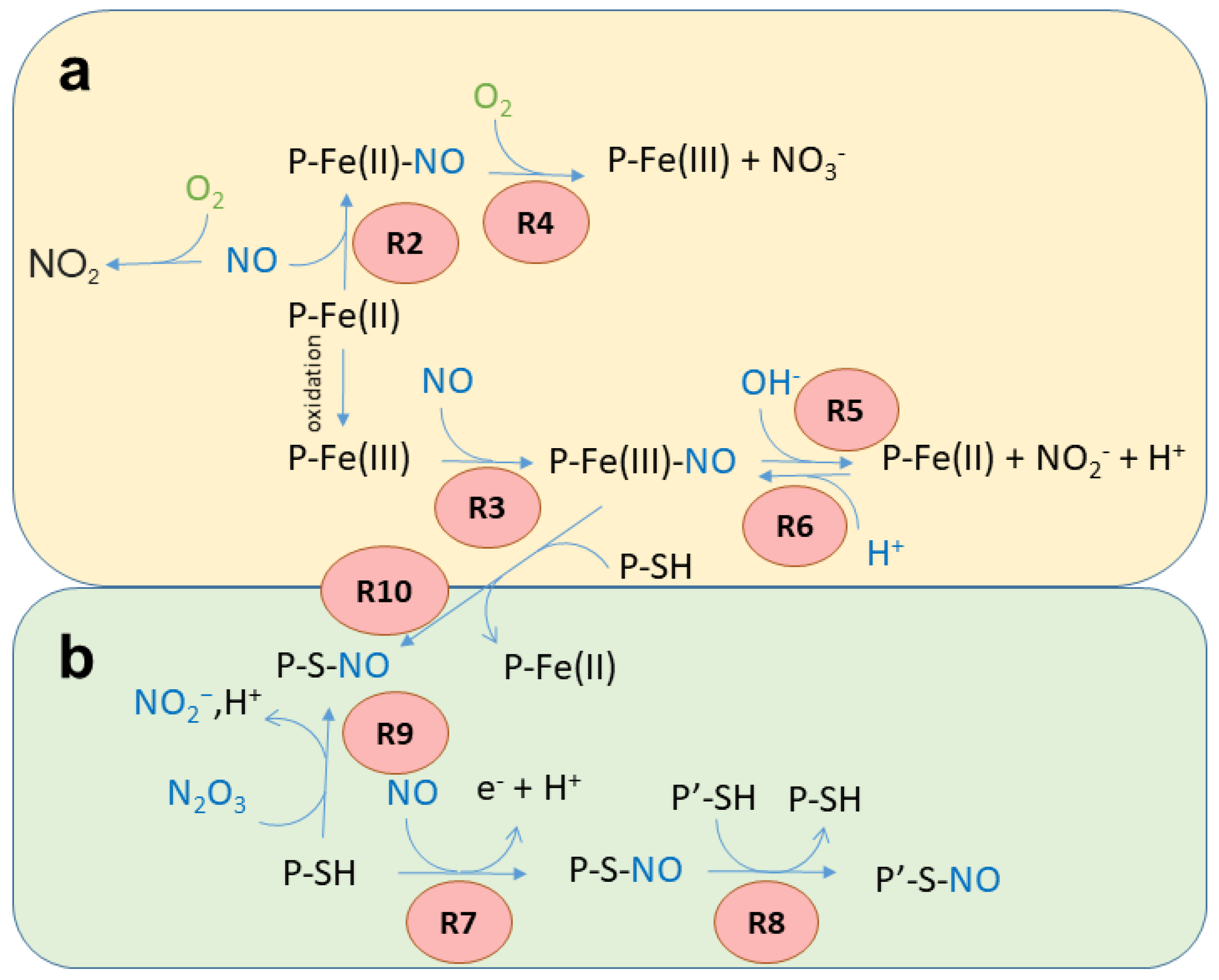
2. General Reactivity of Heme Proteins with NO
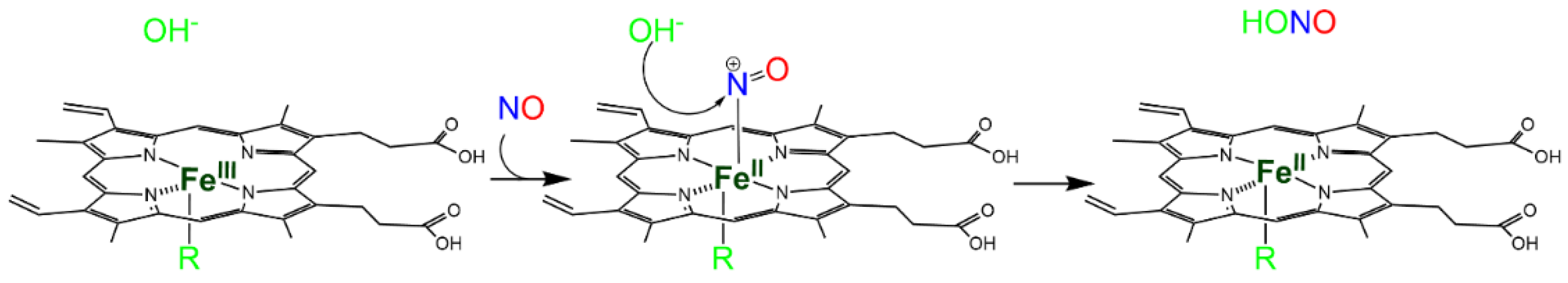
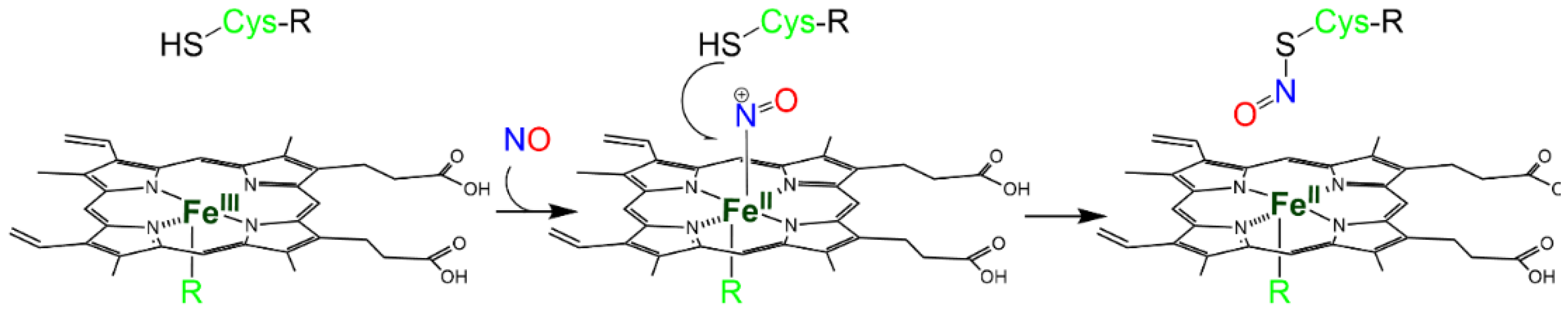
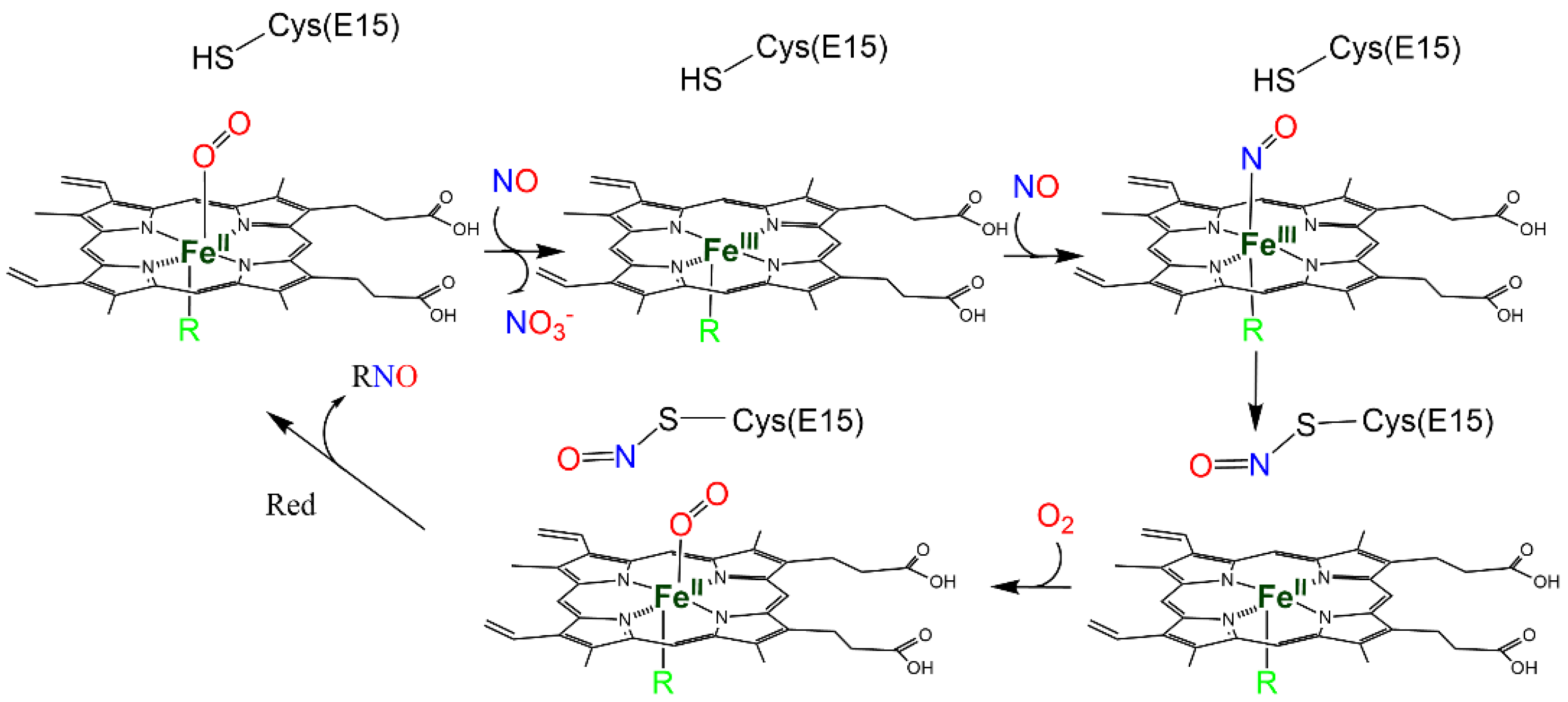
2.1. Reactivity of NO with the Heme Group
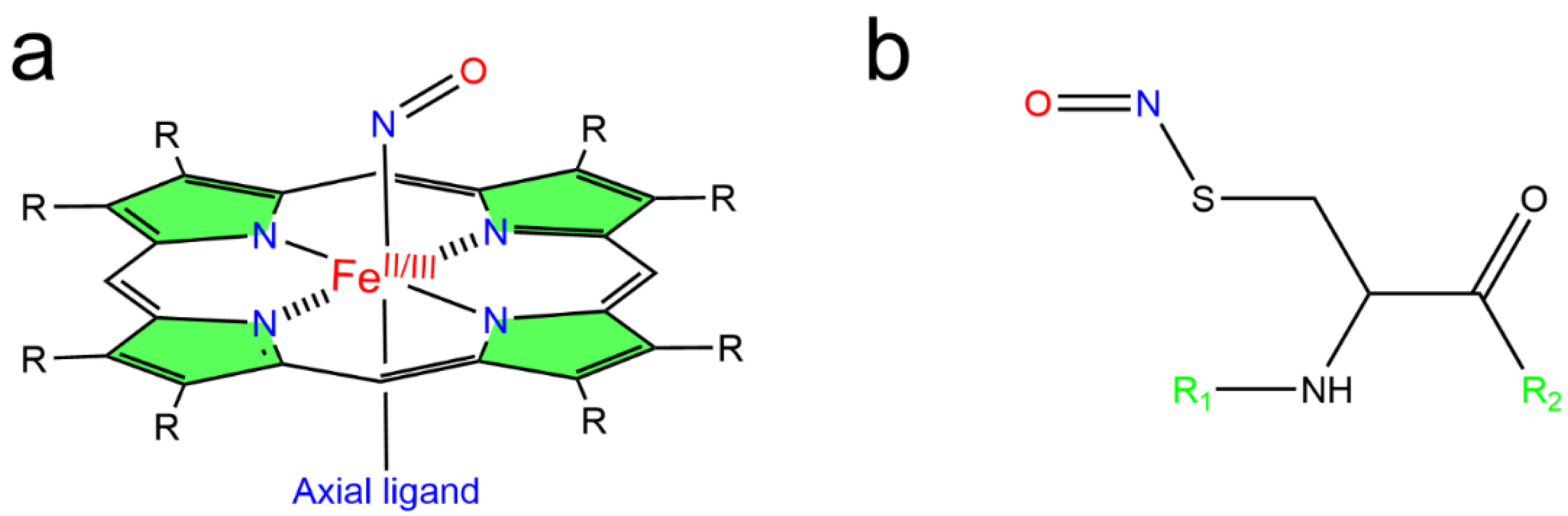
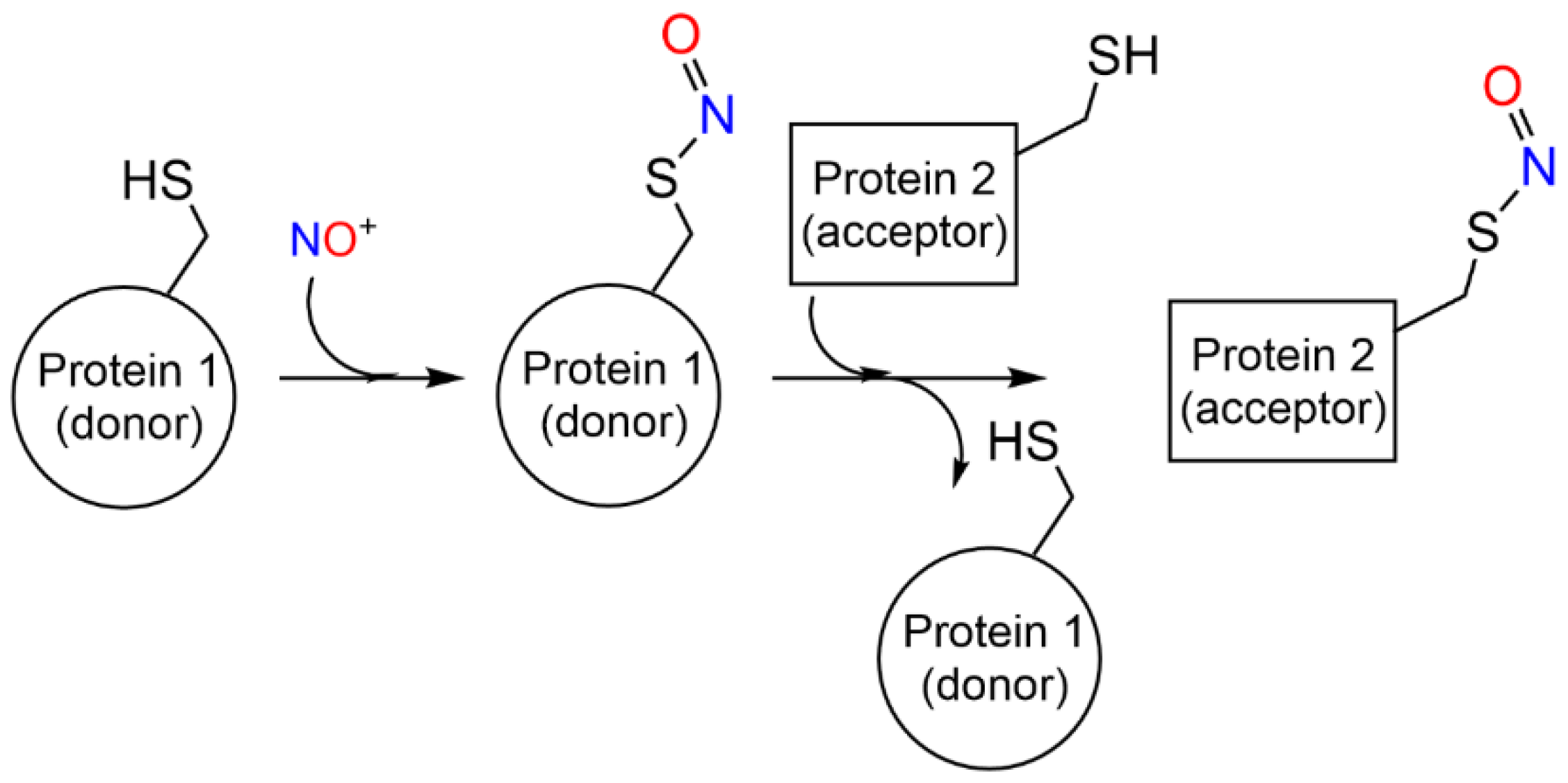
2.2. S-Nitrosylation
3. Heme Reactivity and S-Nitrosylation of Heme Proteins
3.1. Hemoglobin
3.1.1. Heme Reactivity with NO
3.1.2. S-Nitrosylation
3.2. Ascaris Hemoglobin
3.2.1. Heme Reactivity with NO
3.2.2. S-Nitrosylation
3.3. Myoglobin
3.3.1. Heme Reactivity with NO
3.3.2. S-Nitrosylation
3.4. Neuroglobin
3.4.1. Heme Reactivity with NO
3.4.2. S-Nitrosylation
3.5. Cytoglobin
3.5.1. Heme Reactivity with NO
3.5.2. S-Nitrosylation
3.6. NO Synthase
3.6.1. Heme Reactivity with NO
3.6.2. S-Nitrosylation
3.7. Guanylyl Cyclase
3.7.1. Heme Reactivity with NO
3.7.2. S-Nitrosylation
3.8. Cyclooxygenases
3.8.1. Heme Reactivity with NO
3.8.2. S-Nitrosylation
3.9. Nitrophorins (Cimex lectularius)
3.9.1. Heme Reactivity with NO
3.9.2. S-Nitrosylation
3.10. Plant Ascorbate Peroxidase
3.10.1. Heme Reactivity with NO
3.10.2. S-Nitrosylation
3.11. Catalase
3.11.1. Heme Reactivity with NO
3.11.2. S-Nitrosylation
3.12. Cytochrome c
3.12.1. Heme Reactivity with NO
3.12.2. S-Nitrosylation
3.13. Cytochrome c Oxidase
3.13.1. Heme Reactivity with NO
3.13.2. S-Nitrosylation
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Granick, S.; Beale, S.I. Hemes, chlorophylls, and related compounds: Biosynthesis and metabolic regulation. Adv. Enzymol. Relat. Areas Mol. Biol. 1978, 46, 33–203. [Google Scholar] [CrossRef]
- Paoli, M.; Marles-Wright, J.; Smith, A. Structure-function relationships in heme-proteins. DNA Cell Biol. 2002, 21, 271–280. [Google Scholar] [CrossRef]
- Burmester, T.; Hankeln, T. Function and evolution of vertebrate globins. Acta Physiol. 2014, 211, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Weber, R.E.; Vinogradov, S.N. Nonvertebrate hemoglobins: Functions and molecular adaptations. Physiol. Rev. 2001, 81, 569–628. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.J.; Kahraman, A.; Thornton, J.M. Heme proteins--diversity in structural characteristics, function, and folding. Proteins 2010, 78, 2349–2368. [Google Scholar] [CrossRef]
- De Simone, G.; Ascenzi, P.; di Masi, A.; Polticelli, F. Nitrophorins and nitrobindins: Structure and function. Biomol. Concepts 2017, 8, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Reeder, B.J.; Ukeri, J. Strong modulation of nitrite reductase activity of cytoglobin by disulfide bond oxidation: Implications for nitric oxide homeostasis. Nitric Oxide Biol. Chem. 2018, 72, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Tejero, J.; Gladwin, M.T. The globin superfamily: Functions in nitric oxide formation and decay. Biol. Chem. 2014, 395, 631–639. [Google Scholar] [CrossRef]
- Rydberg, P.; Sigfridsson, E.; Ryde, U. On the role of the axial ligand in heme proteins: A theoretical study. J. Biol. Inorg. Chem. JBIC A Publ. Soc. Biol. Inorg. Chem. 2004, 9, 203–223. [Google Scholar] [CrossRef]
- Burmester, T.; Hankeln, T. What is the function of neuroglobin? J. Exp. Biol. 2009, 212, 1423–1428. [Google Scholar] [CrossRef]
- Ford, P.C.; Lorkovic, I.M. Mechanistic aspects of the reactions of nitric oxide with transition-metal complexes. Chem. Rev. 2002, 102, 993–1018. [Google Scholar] [CrossRef]
- Palmer, R.M.; Ferrige, A.G.; Moncada, S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature 1987, 327, 524–526. [Google Scholar] [CrossRef]
- Olson, K.R.; Donald, J.A.; Dombkowski, R.A.; Perry, S.F. Evolutionary and comparative aspects of nitric oxide, carbon monoxide and hydrogen sulfide. Respir. Physiol. Neurobiol. 2012, 184, 117–129. [Google Scholar] [CrossRef]
- Alderton, W.K.; Cooper, C.E.; Knowles, R.G. Nitric oxide synthases: Structure, function and inhibition. Biochem. J. 2001, 357, 593–615. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Keceli, G.; Cao, R.; Su, J.; Mi, Z. Molybdenum-containing nitrite reductases: Spectroscopic characterization and redox mechanism. Redox Rep. Commun. Free Radic. Res. 2017, 22, 17–25. [Google Scholar] [CrossRef]
- Zheng, Y.; Deng, W.; Liu, D.; Li, Y.; Peng, K.; Lorimer, G.H.; Wang, J. Redox and spectroscopic properties of mammalian nitrite reductase-like hemoproteins. J. Inorg. Biochem. 2022, 237, 111982. [Google Scholar] [CrossRef]
- Lancaster, J.R., Jr. A tutorial on the diffusibility and reactivity of free nitric oxide. Nitric Oxide Biol. Chem. 1997, 1, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, J.R., Jr.; Gaston, B. NO and nitrosothiols: Spatial confinement and free diffusion. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004, 287, L465–L466. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thomas, D.D.; Liu, X.; Kantrow, S.P.; Lancaster, J.R., Jr. The biological lifetime of nitric oxide: Implications for the perivascular dynamics of NO and O2. Proc. Natl. Acad. Sci. USA 2001, 98, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Wink, D.A.; Grisham, M.B.; Mitchell, J.B.; Ford, P.C. Direct and indirect effects of nitric oxide in chemical reactions relevant to biology. Methods Enzymol. 1996, 268, 12–31. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, J.O.; Weitzberg, E. Nitric oxide signaling in health and disease. Cell 2022, 185, 2853–2878. [Google Scholar] [CrossRef] [PubMed]
- Hess, D.T.; Matsumoto, A.; Kim, S.O.; Marshall, H.E.; Stamler, J.S. Protein S-nitrosylation: Purview and parameters. Nat. Rev. Mol. Cell Biol. 2005, 6, 150–166. [Google Scholar] [CrossRef] [PubMed]
- Tamba, M.; Simone, G.; Quintiliani, M. Interactions of thiyl free radicals with oxygen: A pulse radiolysis study. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1986, 50, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Lehnert, N.; Kim, E.; Dong, H.T.; Harland, J.B.; Hunt, A.P.; Manickas, E.C.; Oakley, K.M.; Pham, J.; Reed, G.C.; Alfaro, V.S. The Biologically Relevant Coordination Chemistry of Iron and Nitric Oxide: Electronic Structure and Reactivity. Chem. Rev. 2021, 121, 14682–14905. [Google Scholar] [CrossRef]
- Foley, E.L.; Hvitved, A.N.; Eich, R.F.; Olson, J.S. Mechanisms of nitric oxide reactions with globins using mammalian myoglobin as a model system. J. Inorg. Biochem. 2022, 233, 111839. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.S.; Foley, E.W.; Rogge, C.; Tsai, A.L.; Doyle, M.P.; Lemon, D.D. No scavenging and the hypertensive effect of hemoglobin-based blood substitutes. Free Radic. Biol. Med. 2004, 36, 685–697. [Google Scholar] [CrossRef]
- Wanat, A.; Schneppensieper, T.; Stochel, G.; van Eldik, R.; Bill, E.; Wieghardt, K. Kinetics, mechanism, and spectroscopy of the reversible binding of nitric oxide to aquated iron(II). An undergraduate text book reaction revisited. Inorg. Chem. 2002, 41, 4–10. [Google Scholar] [CrossRef]
- Kharitonov, V.G.; Sharma, V.S.; Magde, D.; Koesling, D. Kinetics of nitric oxide dissociation from five- and six-coordinate nitrosyl hemes and heme proteins, including soluble guanylate cyclase. Biochemistry 1997, 36, 6814–6818. [Google Scholar] [CrossRef] [PubMed]
- Tsai, A.-l. How does NO activate hemeproteins? FEBS Lett. 1994, 341, 141–145. [Google Scholar] [CrossRef]
- Eich, R.F.; Li, T.; Lemon, D.D.; Doherty, D.H.; Curry, S.R.; Aitken, J.F.; Mathews, A.J.; Johnson, K.A.; Smith, R.D.; Phillips, G.N., Jr.; et al. Mechanism of NO-induced oxidation of myoglobin and hemoglobin. Biochemistry 1996, 35, 6976–6983. [Google Scholar] [CrossRef]
- Walker, F.A. Nitric oxide interaction with insect nitrophorins and thoughts on the electron configuration of the FeNO6 complex. J. Inorg. Biochem. 2005, 99, 216–236. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.W.; Winterrowd, C.A.; Friedman, A.R.; Thompson, D.P.; Klein, R.D.; Davis, J.P.; Maule, A.G.; Blair, K.L.; Geary, T.G. Nitric oxide mediates the inhibitory effects of SDPNFLRFamide, a nematode FMRFamide-related neuropeptide, in Ascaris suum. J. Neurophysiol. 1995, 74, 1880–1888. [Google Scholar] [CrossRef] [PubMed]
- Smagghe, B.J.; Trent, J.T., 3rd; Hargrove, M.S. NO dioxygenase activity in hemoglobins is ubiquitous in vitro, but limited by reduction in vivo. PLoS ONE 2008, 3, e2039. [Google Scholar] [CrossRef]
- Hoshino, M.; Maeda, M.; Konishi, R.; Seki, H.; Ford, P.C. Studies on the Reaction Mechanism for Reductive Nitrosylation of Ferrihemoproteins in Buffer Solutions. J. Am. Chem. Soc. 1996, 118, 5702–5707. [Google Scholar] [CrossRef]
- Cosby, K.; Partovi, K.S.; Crawford, J.H.; Patel, R.P.; Reiter, C.D.; Martyr, S.; Yang, B.K.; Waclawiw, M.A.; Zalos, G.; Xu, X.; et al. Nitrite reduction to nitric oxide by deoxyhemoglobin vasodilates the human circulation. Nat. Med. 2003, 9, 1498–1505. [Google Scholar] [CrossRef]
- Gladwin, M.T.; Kim-Shapiro, D.B. The functional nitrite reductase activity of the heme-globins. Blood 2008, 112, 2636–2647. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tong, J.; Zweier, J.R.; Follmer, D.; Hemann, C.; Ismail, R.S.; Zweier, J.L. Differences in oxygen-dependent nitric oxide metabolism by cytoglobin and myoglobin account for their differing functional roles. FEBS J. 2013, 280, 3621–3631. [Google Scholar] [CrossRef]
- Bryan, N.S.; Fernandez, B.; Bauer, S.M.; Garcia-Saura, M.F.; Milsom, A.B.; Rassaf, T.; Maloney, R.E.; Bharti, A.; Rodriguez, J.; Feelisch, M. Nitrite is a signaling molecule and regulator of gene expression in mammalian tissues. Nat. Chem. Biol. 2005, 1, 290–297. [Google Scholar] [CrossRef]
- Tsuchiya, K.; Kanematsu, Y.; Yoshizumi, M.; Ohnishi, H.; Kirima, K.; Izawa, Y.; Shikishima, M.; Ishida, T.; Kondo, S.; Kagami, S.; et al. Nitrite is an alternative source of NO in vivo. Am. J. Physiol. Heart Circ. Physiol. 2005, 288, H2163–H2170. [Google Scholar] [CrossRef]
- Coelho, C.; Romao, M.J. Structural and mechanistic insights on nitrate reductases. Protein Sci. A Publ. Protein Soc. 2015, 24, 1901–1911. [Google Scholar] [CrossRef]
- DeMartino, A.W.; Kim-Shapiro, D.B.; Patel, R.P.; Gladwin, M.T. Nitrite and nitrate chemical biology and signalling. Br. J. Pharmacol. 2019, 176, 228–245. [Google Scholar] [CrossRef]
- Lundberg, J.O.; Weitzberg, E.; Gladwin, M.T. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 2008, 7, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Broniowska, K.A.; Diers, A.R.; Hogg, N. S-nitrosoglutathione. Biochim. Et Biophys. Acta 2013, 1830, 3173–3181. [Google Scholar] [CrossRef] [PubMed]
- Marino, S.M.; Gladyshev, V.N. Structural analysis of cysteine S-nitrosylation: A modified acid-based motif and the emerging role of trans-nitrosylation. J. Mol. Biol. 2010, 395, 844–859. [Google Scholar] [CrossRef]
- Wolhuter, K.; Eaton, P. How widespread is stable protein S-nitrosylation as an end-effector of protein regulation? Free Radic. Biol. Med. 2017, 109, 156–166. [Google Scholar] [CrossRef]
- Derakhshan, B.; Wille, P.C.; Gross, S.S. Unbiased identification of cysteine S-nitrosylation sites on proteins. Nat. Protoc. 2007, 2, 1685–1691. [Google Scholar] [CrossRef]
- Stomberski, C.T.; Hess, D.T.; Stamler, J.S. Protein S-Nitrosylation: Determinants of Specificity and Enzymatic Regulation of S-Nitrosothiol-Based Signaling. Antioxid. Redox Signal. 2019, 30, 1331–1351. [Google Scholar] [CrossRef] [PubMed]
- Marchesani, F.; Gianquinto, E.; Autiero, I.; Michielon, A.; Campanini, B.; Faggiano, S.; Bettati, S.; Mozzarelli, A.; Spyrakis, F.; Bruno, S. The allosteric interplay between S-nitrosylation and glycine binding controls the activity of human serine racemase. FEBS J. 2021, 288, 3034–3054. [Google Scholar] [CrossRef]
- Stamler, J.S.; Lamas, S.; Fang, F.C. Nitrosylation. the prototypic redox-based signaling mechanism. Cell 2001, 106, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Arif, A.; Terenzi, F.; Willard, B.; Plow, E.F.; Hazen, S.L.; Fox, P.L. Target-selective protein S-nitrosylation by sequence motif recognition. Cell 2014, 159, 623–634. [Google Scholar] [CrossRef]
- Nakamura, T.; Lipton, S.A. Emerging role of protein-protein transnitrosylation in cell signaling pathways. Antioxid. Redox Signal. 2013, 18, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Stomberski, C.T.; Zhou, H.L.; Wang, L.; van den Akker, F.; Stamler, J.S. Molecular recognition of S-nitrosothiol substrate by its cognate protein denitrosylase. J. Biol. Chem. 2019, 294, 1568–1578. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Stamler, J.S. Enzymatic mechanisms regulating protein S-nitrosylation: Implications in health and disease. J. Mol. Med. 2012, 90, 233–244. [Google Scholar] [CrossRef]
- Liu, X.; Miller, M.J.; Joshi, M.S.; Thomas, D.D.; Lancaster, J.R., Jr. Accelerated reaction of nitric oxide with O2 within the hydrophobic interior of biological membranes. Proc. Natl. Acad. Sci. USA 1998, 95, 2175–2179. [Google Scholar] [CrossRef]
- Cooper, C.E. Nitric oxide and iron proteins. Biochim. Et Biophys. Acta 1999, 1411, 290–309. [Google Scholar] [CrossRef]
- Azizi, F.; Kielbasa, J.E.; Adeyiga, A.M.; Maree, R.D.; Frazier, M.; Yakubu, M.; Shields, H.; King, S.B.; Kim-Shapiro, D.B. Rates of nitric oxide dissociation from hemoglobin. Free Radic. Biol. Med. 2005, 39, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Gibson, Q.H.; Roughton, F.J. The kinetics and equilibria of the reactions of nitric oxide with sheep haemoglobin. J. Physiol. 1957, 136, 507–524. [Google Scholar] [CrossRef]
- Luchsinger, B.P.; Rich, E.N.; Gow, A.J.; Williams, E.M.; Stamler, J.S.; Singel, D.J. Routes to S-nitroso-hemoglobin formation with heme redox and preferential reactivity in the beta subunits. Proc. Natl. Acad. Sci. USA 2003, 100, 461–466. [Google Scholar] [CrossRef]
- Pezacki, J.P.; Ship, N.J.; Kluger, R. Release of Nitric Oxide from S-Nitrosohemoglobin. Electron Transfer as a Response to Deoxygenation. J. Am. Chem. Soc. 2001, 123, 4615–4616. [Google Scholar] [CrossRef]
- Xu, X.; Cho, M.; Spencer, N.Y.; Patel, N.; Huang, Z.; Shields, H.; King, S.B.; Gladwin, M.T.; Hogg, N.; Kim-Shapiro, D.B. Measurements of nitric oxide on the heme iron and beta-93 thiol of human hemoglobin during cycles of oxygenation and deoxygenation. Proc. Natl. Acad. Sci. USA 2003, 100, 11303–11308. [Google Scholar] [CrossRef]
- Kim, H.W.; Greenburg, A.G. Artificial Oxygen Carriers as Red Blood Cell Substitutes: A Selected Review and Current Status. Artif. Organs 2004, 28, 813–828. [Google Scholar] [CrossRef]
- Haldane, J. The Red Colour of Salted Meat. J. Hyg. 1901, 1, 115–122. [Google Scholar] [CrossRef]
- Gladwin, M.T.; Shelhamer, J.H.; Schechter, A.N.; Pease-Fye, M.E.; Waclawiw, M.A.; Panza, J.A.; Ognibene, F.P.; Cannon, R.O., 3rd. Role of circulating nitrite and S-nitrosohemoglobin in the regulation of regional blood flow in humans. Proc. Natl. Acad. Sci. USA 2000, 97, 11482–11487. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kloek, A.P.; Goldberg, D.E.; Mathews, F.S. The structure of Ascaris hemoglobin domain I at 2.2 A resolution: Molecular features of oxygen avidity. Proc. Natl. Acad. Sci. USA 1995, 92, 4224–4228. [Google Scholar] [CrossRef]
- Minning, D.M.; Gow, A.J.; Bonaventura, J.; Braun, R.; Dewhirst, M.; Goldberg, D.E.; Stamler, J.S. Ascaris haemoglobin is a nitric oxide-activated ‘deoxygenase’. Nature 1999, 401, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Gardner, P.R. Nitric oxide dioxygenase function and mechanism of flavohemoglobin, hemoglobin, myoglobin and their associated reductases. J. Inorg. Biochem. 2005, 99, 247–266. [Google Scholar] [CrossRef]
- Rayner, B.S.; Wu, B.J.; Raftery, M.; Stocker, R.; Witting, P.K. Human S-nitroso oxymyoglobin is a store of vasoactive nitric oxide. J. Biol. Chem. 2005, 280, 9985–9993. [Google Scholar] [CrossRef]
- Shiva, S.; Huang, Z.; Grubina, R.; Sun, J.; Ringwood, L.A.; MacArthur, P.H.; Xu, X.; Murphy, E.; Darley-Usmar, V.M.; Gladwin, M.T. Deoxymyoglobin is a nitrite reductase that generates nitric oxide and regulates mitochondrial respiration. Circ. Res. 2007, 100, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Schreiter, E.R.; Rodriguez, M.M.; Weichsel, A.; Montfort, W.R.; Bonaventura, J. S-nitrosylation-induced conformational change in blackfin tuna myoglobin. J. Biol. Chem. 2007, 282, 19773–19780. [Google Scholar] [CrossRef]
- Turan, H.T.; Meuwly, M. Spectroscopy, Dynamics, and Hydration of S-Nitrosylated Myoglobin. J. Phys. Chem. B 2021, 125, 4262–4273. [Google Scholar] [CrossRef]
- Helbo, S.; Gow, A.J.; Jamil, A.; Howes, B.D.; Smulevich, G.; Fago, A. Oxygen-linked S-nitrosation in fish myoglobins: A cysteine-specific tertiary allosteric effect. PLoS ONE 2014, 9, e97012. [Google Scholar] [CrossRef] [PubMed]
- Helbo, S.; Fago, A. Allosteric modulation by S-nitrosation in the low-O(2) affinity myoglobin from rainbow trout. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, R101–R108. [Google Scholar] [CrossRef] [PubMed]
- Herold, S.; Fago, A.; Weber, R.E.; Dewilde, S.; Moens, L. Reactivity studies of the Fe(III) and Fe(II)NO forms of human neuroglobin reveal a potential role against oxidative stress. J. Biol. Chem. 2004, 279, 22841–22847. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.G.; Dewilde, S.; Fago, A. Reactions of ferrous neuroglobin and cytoglobin with nitrite under anaerobic conditions. J. Inorg. Biochem. 2008, 102, 1777–1782. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hemann, C.; Abdelghany, T.M.; El-Mahdy, M.A.; Zweier, J.L. Characterization of the Mechanism and Magnitude of Cytoglobin-mediated Nitrite Reduction and Nitric Oxide Generation under Anaerobic Conditions. J. Biol. Chem. 2012, 287, 36623–36633. [Google Scholar] [CrossRef] [PubMed]
- Corti, P.; Ieraci, M.; Tejero, J. Characterization of zebrafish neuroglobin and cytoglobins 1 and 2: Zebrafish cytoglobins provide insights into the transition from six-coordinate to five-coordinate globins. Nitric Oxide Biol. Chem. 2016, 53, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Tejero, J.; Santolini, J.; Stuehr, D.J. Fast ferrous heme–NO oxidation in nitric oxide synthases. FEBS J. 2009, 276, 4505–4514. [Google Scholar] [CrossRef] [PubMed]
- Tejero, J.; Hunt, A.P.; Santolini, J.; Lehnert, N.; Stuehr, D.J. Mechanism and regulation of ferrous heme-nitric oxide (NO) oxidation in NO synthases. J. Biol. Chem. 2019, 294, 7904–7916. [Google Scholar] [CrossRef]
- Zhang, Y.; Hogg, N. Formation and stability of S-nitrosothiols in RAW 264.7 cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004, 287, L467–L474. [Google Scholar] [CrossRef]
- Tummala, M.; Ryzhov, V.; Ravi, K.; Black, S.M. Identification of the cysteine nitrosylation sites in human endothelial nitric oxide synthase. DNA Cell Biol. 2008, 27, 25–33. [Google Scholar] [CrossRef]
- Taldone, F.S.; Tummala, M.; Goldstein, E.J.; Ryzhov, V.; Ravi, K.; Black, S.M. Studying the S-nitrosylation of model peptides and eNOS protein by mass spectrometry. Nitric Oxide Biol. Chem. 2005, 13, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Dierks, E.A.; Hu, S.; Vogel, K.M.; Yu, A.E.; Spiro, T.G.; Burstyn, J.N. Demonstration of the Role of Scission of the Proximal Histidine−Iron Bond in the Activation of Soluble Guanylyl Cyclase through Metalloporphyrin Substitution Studies. J. Am. Chem. Soc. 1997, 119, 7316–7323. [Google Scholar] [CrossRef]
- Liu, R.; Kang, Y.; Chen, L. Activation mechanism of human soluble guanylate cyclase by stimulators and activators. Nat. Commun. 2021, 12, 5492. [Google Scholar] [CrossRef]
- Kang, Y.; Liu, R.; Wu, J.X.; Chen, L. Structural insights into the mechanism of human soluble guanylate cyclase. Nature 2019, 574, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Fernhoff, N.B.; Derbyshire, E.R.; Underbakke, E.S.; Marletta, M.A. Heme-assisted S-nitrosation desensitizes ferric soluble guanylate cyclase to nitric oxide. J. Biol. Chem. 2012, 287, 43053–43062. [Google Scholar] [CrossRef] [PubMed]
- Sayed, N.; Baskaran, P.; Ma, X.; van den Akker, F.; Beuve, A. Desensitization of soluble guanylyl cyclase, the NO receptor, by S-nitrosylation. Proc. Natl. Acad. Sci. USA 2007, 104, 12312–12317. [Google Scholar] [CrossRef]
- Tsai, A.L.; Wei, C.; Kulmacz, R.J. Interaction between nitric oxide and prostaglandin H synthase. Arch. Biochem. Biophys. 1994, 313, 367–372. [Google Scholar] [CrossRef]
- Kim, S.F.; Huri, D.A.; Snyder, S.H. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science 2005, 310, 1966–1970. [Google Scholar] [CrossRef]
- Christmann, R.; Auerbach, H.; Berry, R.E.; Walker, F.A.; Schünemann, V. Nitric oxide heme interactions in nitrophorin from Cimex lectularius. Hyperfine Interact. 2016, 237, 19. [Google Scholar] [CrossRef]
- Weichsel, A.; Maes, E.M.; Andersen, J.F.; Valenzuela, J.G.; Shokhireva, T.; Walker, F.A.; Montfort, W.R. Heme-assisted S-nitrosation of a proximal thiolate in a nitric oxide transport protein. Proc. Natl. Acad. Sci. USA 2005, 102, 594–599. [Google Scholar] [CrossRef]
- Clark, D.; Durner, J.; Navarre, D.A.; Klessig, D.F. Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol. Plant-Microbe Interact. MPMI 2000, 13, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Mu, J.; Chen, L.; Feng, J.; Hu, J.; Li, L.; Zhou, J.M.; Zuo, J. S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 2015, 167, 1604–1615. [Google Scholar] [CrossRef]
- de Pinto, M.C.; Locato, V.; Sgobba, A.; Romero-Puertas Mdel, C.; Gadaleta, C.; Delledonne, M.; De Gara, L. S-nitrosylation of ascorbate peroxidase is part of programmed cell death signaling in tobacco Bright Yellow-2 cells. Plant Physiol. 2013, 163, 1766–1775. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhu, T.; Chen, X.; Wang, Z.; Yang, Z.; Ren, A.; Shi, L.; Yu, H.; Zhao, M. GSNOR regulates ganoderic acid content in Ganoderma lucidum under heat stress through S-nitrosylation of catalase. Commun. Biol. 2022, 5, 32. [Google Scholar] [CrossRef]
- Schonhoff, C.M.; Gaston, B.; Mannick, J.B. Nitrosylation of cytochrome c during apoptosis. J. Biol. Chem. 2003, 278, 18265–18270. [Google Scholar] [CrossRef]
- Ascenzi, P.; Coletta, M.; Santucci, R.; Polizio, F.; Desideri, A. Nitric oxide binding to ferrous native horse heart cytochrome c and to its carboxymethylated derivative: A spectroscopic and thermodynamic Study. J. Inorg. Biochem. 1994, 53, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Keszler, A.; Azarova, N.A.; Nwanze, N.; Perlegas, A.; Shiva, S.; Broniowska, K.A.; Hogg, N.; Kim-Shapiro, D.B. A novel role for cytochrome c: Efficient catalysis of S-nitrosothiol formation. Free Radic. Biol. Med. 2010, 48, 255–263. [Google Scholar] [CrossRef]
- Broniowska, K.A.; Keszler, A.; Basu, S.; Kim-Shapiro, D.B.; Hogg, N. Cytochrome c-mediated formation of S-nitrosothiol in cells. Biochem. J. 2012, 442, 191–197. [Google Scholar] [CrossRef]
- Pearce, L.L.; Kanai, A.J.; Birder, L.A.; Pitt, B.R.; Peterson, J. The catabolic fate of nitric oxide: The nitric oxide oxidase and peroxynitrite reductase activities of cytochrome oxidase. J. Biol. Chem. 2002, 277, 13556–13562. [Google Scholar] [CrossRef]
- Collman, J.P.; Dey, A.; Decreau, R.A.; Yang, Y.; Hosseini, A.; Solomon, E.I.; Eberspacher, T.A. Interaction of nitric oxide with a functional model of cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 2008, 105, 9892–9896. [Google Scholar] [CrossRef]
- Chang, A.H.; Sancheti, H.; Garcia, J.; Kaplowitz, N.; Cadenas, E.; Han, D. Respiratory substrates regulate S-nitrosylation of mitochondrial proteins through a thiol-dependent pathway. Chem. Res. Toxicol. 2014, 27, 794–804. [Google Scholar] [CrossRef]
- Zhang, J.; Jin, B.; Li, L.; Block, E.R.; Patel, J.M. Nitric oxide-induced persistent inhibition and nitrosylation of active site cysteine residues of mitochondrial cytochrome-c oxidase in lung endothelial cells. Am. J. Physiol. Cell Physiol. 2005, 288, C840–C849. [Google Scholar] [CrossRef]
- Perutz, M.F. Relation between structure and sequence of haemoglobin. Nature 1962, 194, 914–917. [Google Scholar] [CrossRef]
- Perutz, M.F.; Wilkinson, A.J.; Paoli, M.; Dodson, G.G. The stereochemical mechanism of the cooperative effects in hemoglobin revisited. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 1–34. [Google Scholar] [CrossRef]
- Mozzarelli, A.; Ronda, L.; Faggiano, S.; Bettati, S.; Bruno, S. Haemoglobin-based oxygen carriers: Research and reality towards an alternative to blood transfusions. Blood Transfus. Trasfus. Del Sangue 2010, 8 (Suppl. S3), s59–s68. [Google Scholar] [CrossRef]
- Gladwin, M.T.; Ognibene, F.P.; Pannell, L.K.; Nichols, J.S.; Pease-Fye, M.E.; Shelhamer, J.H.; Schechter, A.N. Relative role of heme nitrosylation and beta-cysteine 93 nitrosation in the transport and metabolism of nitric oxide by hemoglobin in the human circulation. Proc. Natl. Acad. Sci. USA 2000, 97, 9943–9948. [Google Scholar] [CrossRef]
- Jia, L.; Bonaventura, C.; Bonaventura, J.; Stamler, J.S. S-nitrosohaemoglobin: A dynamic activity of blood involved in vascular control. Nature 1996, 380, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Premont, R.T.; Stamler, J.S. Essential Role of Hemoglobin betaCys93 in Cardiovascular Physiology. Physiology 2020, 35, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Roche, C.J.; Cassera, M.B.; Dantsker, D.; Hirsch, R.E.; Friedman, J.M. Generating S-nitrosothiols from hemoglobin: Mechanisms, conformational dependence, and physiological relevance. J. Biol. Chem. 2013, 288, 22408–22425. [Google Scholar] [CrossRef]
- Bonaventura, C.; Henkens, R.; Alayash, A.I.; Banerjee, S.; Crumbliss, A.L. Molecular controls of the oxygenation and redox reactions of hemoglobin. Antioxid. Redox Signal. 2013, 18, 2298–2313. [Google Scholar] [CrossRef]
- Alayash, A.I. betaCysteine 93 in human hemoglobin: A gateway to oxidative stability in health and disease. Lab. Investig. A J. Tech. Methods Pathol. 2021, 101, 4–11. [Google Scholar] [CrossRef]
- Vitturi, D.A.; Sun, C.W.; Harper, V.M.; Thrash-Williams, B.; Cantu-Medellin, N.; Chacko, B.K.; Peng, N.; Dai, Y.; Wyss, J.M.; Townes, T.; et al. Antioxidant functions for the hemoglobin beta93 cysteine residue in erythrocytes and in the vascular compartment in vivo. Free Radic. Biol. Med. 2013, 55, 119–129. [Google Scholar] [CrossRef]
- Kuhn, V.; Diederich, L.; Keller, T.C.S.t.; Kramer, C.M.; Luckstadt, W.; Panknin, C.; Suvorava, T.; Isakson, B.E.; Kelm, M.; Cortese-Krott, M.M. Red Blood Cell Function and Dysfunction: Redox Regulation, Nitric Oxide Metabolism, Anemia. Antioxid. Redox Signal. 2017, 26, 718–742. [Google Scholar] [CrossRef] [PubMed]
- Stamler, J.S.; Jia, L.; Eu, J.P.; McMahon, T.J.; Demchenko, I.T.; Bonaventura, J.; Gernert, K.; Piantadosi, C.A. Blood flow regulation by S-nitrosohemoglobin in the physiological oxygen gradient. Science 1997, 276, 2034–2037. [Google Scholar] [CrossRef]
- Pawloski, J.R.; Hess, D.T.; Stamler, J.S. Impaired vasodilation by red blood cells in sickle cell disease. Proc. Natl. Acad. Sci. USA 2005, 102, 2531–2536. [Google Scholar] [CrossRef] [PubMed]
- Irwin, D.; Buehler, P.W.; Alayash, A.I.; Jia, Y.; Bonventura, J.; Foreman, B.; White, M.; Jacobs, R.; Piteo, B.; TissotvanPatot, M.C.; et al. Mixed S-nitrosylated polymerized bovine hemoglobin species moderate hemodynamic effects in acutely hypoxic rats. Am. J. Respir. Cell Mol. Biol. 2010, 42, 200–209. [Google Scholar] [CrossRef]
- Sun, C.W.; Yang, J.; Kleschyov, A.L.; Zhuge, Z.; Carlstrom, M.; Pernow, J.; Wajih, N.; Isbell, T.S.; Oh, J.Y.; Cabrales, P.; et al. Hemoglobin beta93 Cysteine Is Not Required for Export of Nitric Oxide Bioactivity From the Red Blood Cell. Circulation 2019, 139, 2654–2663. [Google Scholar] [CrossRef]
- Isbell, T.S.; Sun, C.W.; Wu, L.C.; Teng, X.; Vitturi, D.A.; Branch, B.G.; Kevil, C.G.; Peng, N.; Wyss, J.M.; Ambalavanan, N.; et al. SNO-hemoglobin is not essential for red blood cell-dependent hypoxic vasodilation. Nat. Med. 2008, 14, 773–777. [Google Scholar] [CrossRef]
- Srihirun, S.; Sriwantana, T.; Unchern, S.; Kittikool, D.; Noulsri, E.; Pattanapanyasat, K.; Fucharoen, S.; Piknova, B.; Schechter, A.N.; Sibmooh, N. Platelet inhibition by nitrite is dependent on erythrocytes and deoxygenation. PLoS ONE 2012, 7, e30380. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Popel, A.S. Nitric oxide production pathways in erythrocytes and plasma. Biorheology 2009, 46, 107–119. [Google Scholar] [CrossRef]
- Dold, C.; Holland, C.V. Ascaris and ascariasis. Microbes Infect. 2011, 13, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Darawshe, S.; Tsafadyah, Y.; Daniel, E. Quaternary structure of erythrocruorin from the nematode Ascaris suum. Evidence for unsaturated haem-binding sites. Biochem. J. 1987, 242, 689–694. [Google Scholar] [CrossRef]
- Flogel, U.; Fago, A.; Rassaf, T. Keeping the heart in balance: The functional interactions of myoglobin with nitrogen oxides. J. Exp. Biol. 2010, 213, 2726–2733. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, M.D.; Macqueen, D.J. Evolution and Expression of Tissue Globins in Ray-Finned Fishes. Genome Biol. Evol. 2017, 9, 32–47. [Google Scholar] [CrossRef]
- Moylan, T.J.; Sidell, B.D. Concentrations of myoglobin and myoglobin mRNA in heart ventricles from Antarctic fishes. J. Exp. Biol. 2000, 203, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Cossins, A.R.; Williams, D.R.; Foulkes, N.S.; Berenbrink, M.; Kipar, A. Diverse cell-specific expression of myoglobin isoforms in brain, kidney, gill and liver of the hypoxia-tolerant carp and zebrafish. J. Exp. Biol. 2009, 212, 627–638. [Google Scholar] [CrossRef]
- Gorr, T.A.; Wichmann, D.; Pilarsky, C.; Theurillat, J.P.; Fabrizius, A.; Laufs, T.; Bauer, T.; Koslowski, M.; Horn, S.; Burmester, T.; et al. Old proteins—New locations: Myoglobin, haemoglobin, neuroglobin and cytoglobin in solid tumours and cancer cells. Acta Physiol. 2011, 202, 563–581. [Google Scholar] [CrossRef] [PubMed]
- Helbo, S.; Weber, R.E.; Fago, A. Expression patterns and adaptive functional diversity of vertebrate myoglobins. Biochim. Et Biophys. Acta 2013, 1834, 1832–1839. [Google Scholar] [CrossRef]
- Garry, D.J.; Ordway, G.A.; Lorenz, J.N.; Radford, N.B.; Chin, E.R.; Grange, R.W.; Bassel-Duby, R.; Williams, R.S. Mice without myoglobin. Nature 1998, 395, 905–908. [Google Scholar] [CrossRef]
- Hendgen-Cotta, U.B.; Kelm, M.; Rassaf, T. Myoglobin’s novel role in nitrite-induced hypoxic vasodilation. Trends Cardiovasc. Med. 2014, 24, 69–74. [Google Scholar] [CrossRef]
- Kleschyov, A.L. The NO-heme signaling hypothesis. Free Radic. Biol. Med. 2017, 112, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Hendgen-Cotta, U.B.; Merx, M.W.; Shiva, S.; Schmitz, J.; Becher, S.; Klare, J.P.; Steinhoff, H.J.; Goedecke, A.; Schrader, J.; Gladwin, M.T.; et al. Nitrite reductase activity of myoglobin regulates respiration and cellular viability in myocardial ischemia-reperfusion injury. Proc. Natl. Acad. Sci. USA 2008, 105, 10256–10261. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, S.R.; Hendrickson, W.A.; Lambright, D.G.; Boxer, S.G. X-ray crystal structure of a recombinant human myoglobin mutant at 2.8 A resolution. J. Mol. Biol. 1990, 213, 215–218. [Google Scholar] [CrossRef]
- Rayner, B.S.; Wu, B.J.; Raftery, M.; Stocker, R.; Witting, P.K. Regulation of vascular tone by S-nitroso-myoglobin. Redox Rep. Commun. Free Radic. Res. 2004, 9, 382–386. [Google Scholar] [CrossRef]
- Witting, P.K.; Douglas, D.J.; Mauk, A.G. Reaction of human myoglobin and nitric oxide. Heme iron or protein sulfhydryl (s) nitrosation dependence on the absence or presence of oxygen. J. Biol. Chem. 2001, 276, 3991–3998. [Google Scholar] [CrossRef] [PubMed]
- Godecke, A.; Flogel, U.; Zanger, K.; Ding, Z.; Hirchenhain, J.; Decking, U.K.; Schrader, J. Disruption of myoglobin in mice induces multiple compensatory mechanisms. Proc. Natl. Acad. Sci. USA 1999, 96, 10495–10500. [Google Scholar] [CrossRef]
- Marcinek, D.J.; Bonaventura, J.; Wittenberg, J.B.; Block, B.A. Oxygen affinity and amino acid sequence of myoglobins from endothermic and ectothermic fish. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001, 280, R1123–R1133. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, G.I.; Evans, S.V.; Przybylska, M.; Rose, D.R. 1.70 A resolution structure of myoglobin from yellowfin tuna. An example of a myoglobin lacking the D helix. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994, 50, 283–289. [Google Scholar] [CrossRef]
- Exertier, C.; Montemiglio, L.C.; Freda, I.; Gugole, E.; Parisi, G.; Savino, C.; Vallone, B. Neuroglobin, clues to function and mechanism. Mol. Asp. Med. 2022, 84, 101055. [Google Scholar] [CrossRef]
- Abbruzzetti, S.; Faggiano, S.; Bruno, S.; Spyrakis, F.; Mozzarelli, A.; Dewilde, S.; Moens, L.; Viappiani, C. Ligand migration through the internal hydrophobic cavities in human neuroglobin. Proc. Natl. Acad. Sci. USA 2009, 106, 18984–18989. [Google Scholar] [CrossRef]
- Boron, I.; Capece, L.; Pennacchietti, F.; Wetzler, D.E.; Bruno, S.; Abbruzzetti, S.; Chisari, L.; Luque, F.J.; Viappiani, C.; Marti, M.A.; et al. Engineered chimeras reveal the structural basis of hexacoordination in globins: A case study of neuroglobin and myoglobin. Biochim. Et Biophys. Acta 2015, 1850, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Burmester, T.; Ebner, B.; Weich, B.; Hankeln, T. Cytoglobin: A Novel Globin Type Ubiquitously Expressed inVertebrate Tissues. Mol. Biol. Evol. 2002, 19, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Gerlach, F.; Avivi, A.; Laufs, T.; Wystub, S.; Simpson, J.C.; Nevo, E.; Saaler-Reinhardt, S.; Reuss, S.; Hankeln, T.; et al. Cytoglobin is a respiratory protein in connective tissue and neurons, which is up-regulated by hypoxia. J. Biol. Chem. 2004, 279, 8063–8069. [Google Scholar] [CrossRef]
- Gabba, M.; Abbruzzetti, S.; Spyrakis, F.; Forti, F.; Bruno, S.; Mozzarelli, A.; Luque, F.J.; Viappiani, C.; Cozzini, P.; Nardini, M.; et al. CO rebinding kinetics and molecular dynamics simulations highlight dynamic regulation of internal cavities in human cytoglobin. PLoS ONE 2013, 8, e49770. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Follmer, D.; Zweier, J.R.; Huang, X.; Hemann, C.; Liu, K.; Druhan, L.J.; Zweier, J.L. Characterization of the function of cytoglobin as an oxygen-dependent regulator of nitric oxide concentration. Biochemistry 2012, 51, 5072–5082. [Google Scholar] [CrossRef]
- McRonald, F.E.; Risk, J.M.; Hodges, N.J. Protection from intracellular oxidative stress by cytoglobin in normal and cancerous oesophageal cells. PLoS ONE 2012, 7, e30587. [Google Scholar] [CrossRef]
- Reeder, B.J.; Svistunenko, D.A.; Wilson, M.T. Lipid binding to cytoglobin leads to a change in haem co-ordination: A role for cytoglobin in lipid signalling of oxidative stress. Biochem. J. 2011, 434, 483–492. [Google Scholar] [CrossRef]
- de Sanctis, D.; Dewilde, S.; Pesce, A.; Moens, L.; Ascenzi, P.; Hankeln, T.; Burmester, T.; Bolognesi, M. Mapping protein matrix cavities in human cytoglobin through Xe atom binding. Biochem. Biophys. Res. Commun. 2004, 316, 1217–1221. [Google Scholar] [CrossRef]
- Forstermann, U.; Sessa, W.C. Nitric oxide synthases: Regulation and function. Eur. Heart J. 2012, 33, 829–837. [Google Scholar] [CrossRef]
- Gonzalez-Domenech, C.M.; Munoz-Chapuli, R. Molecular evolution of nitric oxide synthases in metazoans. Comp. Biochem. Physiol. Part D Genom. Proteom. 2010, 5, 295–301. [Google Scholar] [CrossRef]
- Negrerie, M.; Berka, V.; Vos, M.H.; Liebl, U.; Lambry, J.C.; Tsai, A.L.; Martin, J.L. Geminate recombination of nitric oxide to endothelial nitric-oxide synthase and mechanistic implications. J. Biol. Chem. 1999, 274, 24694–24702. [Google Scholar] [CrossRef] [PubMed]
- Abu-Soud, H.M.; Wang, J.; Rousseau, D.L.; Fukuto, J.M.; Ignarro, L.J.; Stuehr, D.J. Neuronal nitric oxide synthase self-inactivates by forming a ferrous-nitrosyl complex during aerobic catalysis. J. Biol. Chem. 1995, 270, 22997–23006. [Google Scholar] [CrossRef] [PubMed]
- Santolini, J.; Meade, A.L.; Stuehr, D.J. Differences in Three Kinetic Parameters Underpin the Unique Catalytic Profiles of Nitric-oxide Synthases, I., II, and III. J. Biol. Chem. 2001, 276, 48887–48898. [Google Scholar] [CrossRef]
- Moller, J.K.; Skibsted, L.H. Mechanism of nitrosylmyoglobin autoxidation: Temperature and oxygen pressure effects on the two consecutive reactions. Chemistry 2004, 10, 2291–2300. [Google Scholar] [CrossRef] [PubMed]
- Herold, S.; Rock, G. Mechanistic studies of the oxygen-mediated oxidation of nitrosylhemoglobin. Biochemistry 2005, 44, 6223–6231. [Google Scholar] [CrossRef] [PubMed]
- Ravi, K.; Brennan, L.A.; Levic, S.; Ross, P.A.; Black, S.M. S-nitrosylation of endothelial nitric oxide synthase is associated with monomerization and decreased enzyme activity. Proc. Natl. Acad. Sci. USA 2004, 101, 2619–2624. [Google Scholar] [CrossRef]
- Erwin, P.A.; Lin, A.J.; Golan, D.E.; Michel, T. Receptor-regulated dynamic S-nitrosylation of endothelial nitric-oxide synthase in vascular endothelial cells. J. Biol. Chem. 2005, 280, 19888–19894. [Google Scholar] [CrossRef]
- Erwin, P.A.; Mitchell, D.A.; Sartoretto, J.; Marletta, M.A.; Michel, T. Subcellular targeting and differential S-nitrosylation of endothelial nitric-oxide synthase. J. Biol. Chem. 2006, 281, 151–157. [Google Scholar] [CrossRef]
- Beuve, A. Thiol-Based Redox Modulation of Soluble Guanylyl Cyclase, the Nitric Oxide Receptor. Antioxid. Redox Signal. 2017, 26, 137–149. [Google Scholar] [CrossRef]
- Arnold, W.P.; Mittal, C.K.; Katsuki, S.; Murad, F. Nitric oxide activates guanylate cyclase and increases guanosine 3′:5′-cyclic monophosphate levels in various tissue preparations. Proc. Natl. Acad. Sci. USA 1977, 74, 3203–3207. [Google Scholar] [CrossRef]
- Derbyshire, E.R.; Marletta, M.A. Structure and regulation of soluble guanylate cyclase. Annu. Rev. Biochem. 2012, 81, 533–559. [Google Scholar] [CrossRef] [PubMed]
- Montfort, W.R.; Wales, J.A.; Weichsel, A. Structure and Activation of Soluble Guanylyl Cyclase, the Nitric Oxide Sensor. Antioxid. Redox Signal. 2017, 26, 107–121. [Google Scholar] [CrossRef]
- Zhao, Y.; Brandish, P.E.; Di Valentin, M.; Schelvis, J.P.; Babcock, G.T.; Marletta, M.A. Inhibition of soluble guanylate cyclase by ODQ. Biochemistry 2000, 39, 10848–10854. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Alapa, M.; Shu, P.; Nagarajan, N.; Wu, C.; Sadoshima, J.; Kholodovych, V.; Li, H.; Beuve, A. Guanylyl cyclase sensitivity to nitric oxide is protected by a thiol oxidation-driven interaction with thioredoxin-1. J. Biol. Chem. 2017, 292, 14362–14370. [Google Scholar] [CrossRef]
- Rouzer, C.A.; Marnett, L.J. Cyclooxygenases: Structural and functional insights. J. Lipid Res. 2009, 50, S29–S34. [Google Scholar] [CrossRef] [PubMed]
- Salvemini, D.; Masferrer, J.L. Interactions of nitric oxide with cyclooxygenase: In vitro, ex vivo, and in vivo studies. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1996; Volume 269, pp. 12–25. [Google Scholar]
- Qiao, J.; Ma, L.; Roth, J.; Li, Y.; Liu, Y. Kinetic basis for the activation of human cyclooxygenase-2 rather than cyclooxygenase-1 by nitric oxide. Org. Biomol. Chem. 2018, 16, 765–770. [Google Scholar] [CrossRef]
- Hajjar, D.P.; Lander, H.M.; Pearce, S.F.A.; Upmacis, R.; Pomerantz, K.B. Nitric oxide enhances prostaglandin-H synthase-1 activity by a heme-independent mechanism: Evidence implicating nitrosothiols. J. Am. Chem. Soc. 1995, 117, 3340–3346. [Google Scholar] [CrossRef]
- Alexanian, A.; Sorokin, A. Cyclooxygenase 2: Protein-protein interactions and posttranslational modifications. Physiol. Genom. 2017, 49, 667–681. [Google Scholar] [CrossRef]
- Tian, J.; Kim, S.F.; Hester, L.; Snyder, S.H. S-nitrosylation/activation of COX-2 mediates NMDA neurotoxicity. Proc. Natl. Acad. Sci. USA 2008, 105, 10537–10540. [Google Scholar] [CrossRef]
- Atar, S.; Ye, Y.; Lin, Y.; Freeberg, S.Y.; Nishi, S.P.; Rosanio, S.; Huang, M.H.; Uretsky, B.F.; Perez-Polo, J.R.; Birnbaum, Y. Atorvastatin-induced cardioprotection is mediated by increasing inducible nitric oxide synthase and consequent S-nitrosylation of cyclooxygenase-2. Am. J. Physiol. Heart Circ. Physiol. 2006, 290, H1960–H1968. [Google Scholar] [CrossRef]
- Dalli, J.; Chiang, N.; Serhan, C.N. Elucidation of novel 13-series resolvins that increase with atorvastatin and clear infections. Nat. Med. 2015, 21, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, A. Nitric Oxide Synthase and Cyclooxygenase Pathways: A Complex Interplay in Cellular Signaling. Curr. Med. Chem. 2016, 23, 2559–2578. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Shimizu, T.; Higashi, Y.; Nakamura, K.; Taniuchi, K.; Dimitriadis, F.; Shimizu, S.; Yokotani, K.; Saito, M. Central bombesin possibly induces S-nitrosylation of cyclooxygenase-1 in pre-sympathetic neurons of rat hypothalamic paraventricular nucleus. Life Sci. 2014, 100, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Shimizu, T.; Lu, L.; Yokotani, K. Possible involvement of S-nitrosylation of brain cyclooxygenase-1 in bombesin-induced central activation of adrenomedullary outflow in rats. Eur. J. Pharmacol. 2012, 679, 40–50. [Google Scholar] [CrossRef]
- Xiao, C.Y.; Hara, A.; Yuhki, K.; Fujino, T.; Ma, H.; Okada, Y.; Takahata, O.; Yamada, T.; Murata, T.; Narumiya, S.; et al. Roles of prostaglandin I(2) and thromboxane A(2) in cardiac ischemia-reperfusion injury: A study using mice lacking their respective receptors. Circulation 2001, 104, 2210–2215. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, J.M.; Hazzard, J.M.; Nussenzveig, R.H.; Champagne, D.E.; Walker, F.A. Reversible binding of nitric oxide by a salivary heme protein from a bloodsucking insect. Science 1993, 260, 539–541. [Google Scholar] [CrossRef]
- Weichsel, A.; Andersen, J.F.; Champagne, D.E.; Walker, F.A.; Montfort, W.R. Crystal structures of a nitric oxide transport protein from a blood-sucking insect. Nat. Struct. Biol. 1998, 5, 304–309. [Google Scholar] [CrossRef]
- Mollace, V.; Salvemini, D.; Anggard, E.; Vane, J. Nitric oxide from vascular smooth muscle cells: Regulation of platelet reactivity and smooth muscle cell guanylate cyclase. Br. J. Pharmacol. 1991, 104, 633–638. [Google Scholar] [CrossRef]
- Valenzuela, J.G.; Walker, F.A.; Ribeiro, J.M. A salivary nitrophorin (nitric-oxide-carrying hemoprotein) in the bedbug Cimex lectularius. J. Exp. Biol. 1995, 198, 1519–1526. [Google Scholar] [CrossRef]
- Galinato, M.G.I.; Spolitak, T.; Ballou, D.P.; Lehnert, N. Elucidating the Role of the Proximal Cysteine Hydrogen-Bonding Network in Ferric Cytochrome P450cam and Corresponding Mutants Using Magnetic Circular Dichroism Spectroscopy. Biochemistry 2011, 50, 1053–1069. [Google Scholar] [CrossRef]
- Kelly, G.J.; Latzko, E. Soluble ascorbate peroxidase. Sci. Nat. 1979, 66, 617–618. [Google Scholar] [CrossRef]
- Anjum, N.A.; Sharma, P.; Gill, S.S.; Hasanuzzaman, M.; Khan, E.A.; Kachhap, K.; Mohamed, A.A.; Thangavel, P.; Devi, G.D.; Vasudhevan, P.; et al. Catalase and ascorbate peroxidase-representative H2O2-detoxifying heme enzymes in plants. Environ. Sci. Pollut. Res. Int. 2016, 23, 19002–19029. [Google Scholar] [CrossRef]
- Yu, M.; Lamattina, L.; Spoel, S.H.; Loake, G.J. Nitric oxide function in plant biology: A redox cue in deconvolution. New Phytol. 2014, 202, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Frohlich, A.; Durner, J. The hunt for plant nitric oxide synthase (NOS): Is one really needed? Plant Sci. Int. J. Exp. Plant Biol. 2011, 181, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Zaffagnini, M.; De Mia, M.; Morisse, S.; Di Giacinto, N.; Marchand, C.H.; Maes, A.; Lemaire, S.D.; Trost, P. Protein S-nitrosylation in photosynthetic organisms: A comprehensive overview with future perspectives. Biochim. Biophys. Acta 2016, 1864, 952–966. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Gordo, S.; Rodriguez-Ruiz, M.; Lopez-Jaramillo, J.; Munoz-Vargas, M.A.; Palma, J.M.; Corpas, F.J. Nitric Oxide (NO) Differentially Modulates the Ascorbate Peroxidase (APX) Isozymes of Sweet Pepper (Capsicum annuum L.) Fruits. Antioxidants 2022, 11, 765. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Van Breusegem, F.; Noctor, G. Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010, 61, 4197–4220. [Google Scholar] [CrossRef]
- Padovani, D.; Hessani, A.; Castillo, F.T.; Liot, G.; Andriamihaja, M.; Lan, A.; Pilati, C.; Blachier, F.; Sen, S.; Galardon, E.; et al. Sulfheme formation during homocysteine S-oxygenation by catalase in cancers and neurodegenerative diseases. Nat. Commun. 2016, 7, 13386. [Google Scholar] [CrossRef]
- Regelsberger, G.; Jakopitsch, C.; Plasser, L.; Schwaiger, H.; Furtmüller, P.G.; Peschek, G.A.; Zámocký, M.; Obinger, C. Occurrence and biochemistry of hydroperoxidases in oxygenic phototrophic prokaryotes (cyanobacteria). Plant Physiol. Biochem. 2002, 40, 479–490. [Google Scholar] [CrossRef]
- Frugoli, J.A.; Zhong, H.H.; Nuccio, M.L.; McCourt, P.; McPeek, M.A.; Thomas, T.L.; McClung, C.R. Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996, 112, 327–336. [Google Scholar] [CrossRef]
- Brown, G.C. Reversible binding and inhibition of catalase by nitric oxide. Eur. J. Biochem. 1995, 232, 188–191. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, M.; Ozawa, K.; Seki, H.; Ford, P.C. Photochemistry of nitric oxide adducts of water-soluble iron(III) porphyrin and ferrihemoproteins studied by nanosecond laser photolysis. J. Am. Chem. Soc. 1993, 115, 9568–9575. [Google Scholar] [CrossRef]
- Purwar, N.; McGarry, J.M.; Kostera, J.; Pacheco, A.A.; Schmidt, M. Interaction of nitric oxide with catalase: Structural and kinetic analysis. Biochemistry 2011, 50, 4491–4503. [Google Scholar] [CrossRef]
- Ortega-Galisteo, A.P.; Rodríguez-Serrano, M.; Pazmiño, D.M.; Gupta, D.K.; Sandalio, L.M.; Romero-Puertas, M.C. S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: Changes under abiotic stress. J. Exp. Bot. 2012, 63, 2089–2103. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; Sanchez-Calvo, B.; Chaki, M.; Valderrama, R.; Mata-Perez, C.; Padilla, M.N.; Corpas, F.J.; Barroso, J.B. Antioxidant Systems are Regulated by Nitric Oxide-Mediated Post-translational Modifications (NO-PTMs). Front. Plant Sci. 2016, 7, 152. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chu, C. S-Nitrosylation Control of ROS and RNS Homeostasis in Plants: The Switching Function of Catalase. Mol. Plant 2020, 13, 946–948. [Google Scholar] [CrossRef]
- Hannibal, L.; Tomasina, F.; Capdevila, D.A.; Demicheli, V.; Tortora, V.; Alvarez-Paggi, D.; Jemmerson, R.; Murgida, D.H.; Radi, R. Alternative Conformations of Cytochrome c: Structure, Function, and Detection. Biochemistry 2016, 55, 407–428. [Google Scholar] [CrossRef]
- Guerra-Castellano, A.; Marquez, I.; Perez-Mejias, G.; Diaz-Quintana, A.; De la Rosa, M.A.; Diaz-Moreno, I. Post-Translational Modifications of Cytochrome c in Cell Life and Disease. Int. J. Mol. Sci. 2020, 21, 8483. [Google Scholar] [CrossRef]
- Ow, Y.P.; Green, D.R.; Hao, Z.; Mak, T.W. Cytochrome c: Functions beyond respiration. Nat. Rev. Mol. Cell Biol. 2008, 9, 532–542. [Google Scholar] [CrossRef]
- Schweitzer-Stenner, R.; Hagarman, A.; Verbaro, D.; Soffer, J.B. Chapter Six—Conformational Stability of Cytochrome c Probed by Optical Spectroscopy. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2009; Volume 466, pp. 109–153. [Google Scholar]
- Schweitzer-Stenner, R. Cytochrome c: A Multifunctional Protein Combining Conformational Rigidity with Flexibility. New J. Sci. 2014, 2014, 484538. [Google Scholar] [CrossRef]
- Kluck, R.M.; Bossy-Wetzel, E.; Green, D.R.; Newmeyer, D.D. The release of cytochrome c from mitochondria: A primary site for Bcl-2 regulation of apoptosis. Science 1997, 275, 1132–1136. [Google Scholar] [CrossRef]
- Liu, X.; Kim, C.N.; Yang, J.; Jemmerson, R.; Wang, X. Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c. Cell 1996, 86, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Jemmerson, R.; Liu, J.; Hausauer, D.; Lam, K.P.; Mondino, A.; Nelson, R.D. A conformational change in cytochrome c of apoptotic and necrotic cells is detected by monoclonal antibody binding and mimicked by association of the native antigen with synthetic phospholipid vesicles. Biochemistry 1999, 38, 3599–3609. [Google Scholar] [CrossRef] [PubMed]
- Arnelle, D.R.; Stamler, J.S. NO+, NO, and NO- donation by S-nitrosothiols: Implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch. Biochem. Biophys. 1995, 318, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.J.; Hogg, N.; Joseph, J.; Kalyanaraman, B. Mechanism of nitric oxide release from S-nitrosothiols. J. Biol. Chem. 1996, 271, 18596–18603. [Google Scholar] [CrossRef] [PubMed]
- Capaldi, R.A. Structure and function of cytochrome c oxidase. Annu. Rev. Biochem. 1990, 59, 569–596. [Google Scholar] [CrossRef]
- Cleeter, M.W.; Cooper, J.M.; Darley-Usmar, V.M.; Moncada, S.; Schapira, A.H. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide. Implications for neurodegenerative diseases. FEBS Lett. 1994, 345, 50–54. [Google Scholar] [CrossRef]
- Stubauer, G.; Giuffre, A.; Brunori, M.; Sarti, P. Cytochrome c oxidase does not catalyze the anaerobic reduction of NO. Biochem. Biophys. Res. Commun. 1998, 245, 459–465. [Google Scholar] [CrossRef]
- Mason, M.G.; Nicholls, P.; Wilson, M.T.; Cooper, C.E. Nitric oxide inhibition of respiration involves both competitive (heme) and noncompetitive (copper) binding to cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 2006, 103, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Clementi, E.; Brown, G.C.; Feelisch, M.; Moncada, S. Persistent inhibition of cell respiration by nitric oxide: Crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc. Natl. Acad. Sci. USA 1998, 95, 7631–7636. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.T.; Moncada, S. Nitric oxide, cytochrome C oxidase, and the cellular response to hypoxia. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 643–647. [Google Scholar] [CrossRef] [PubMed]
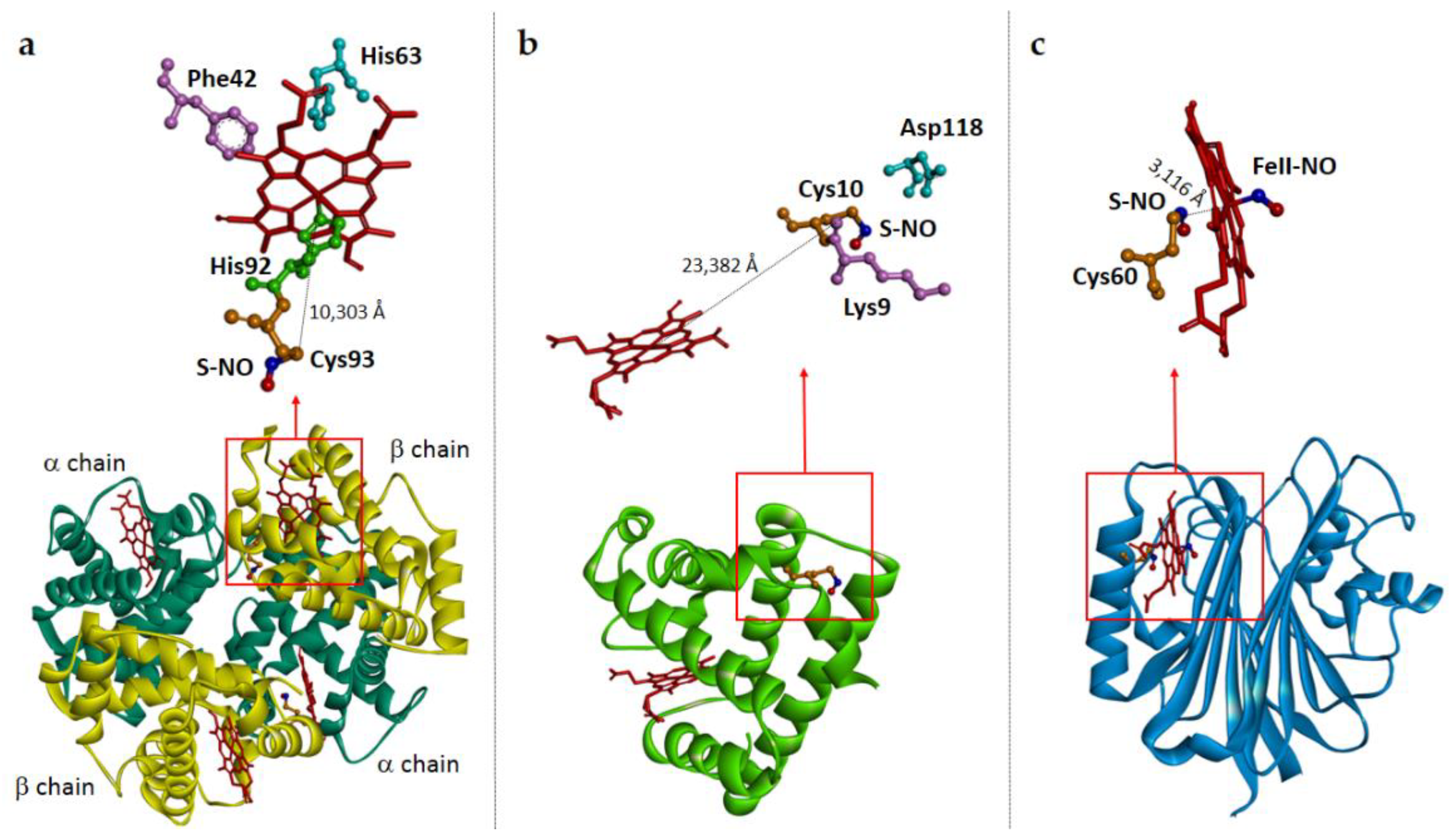

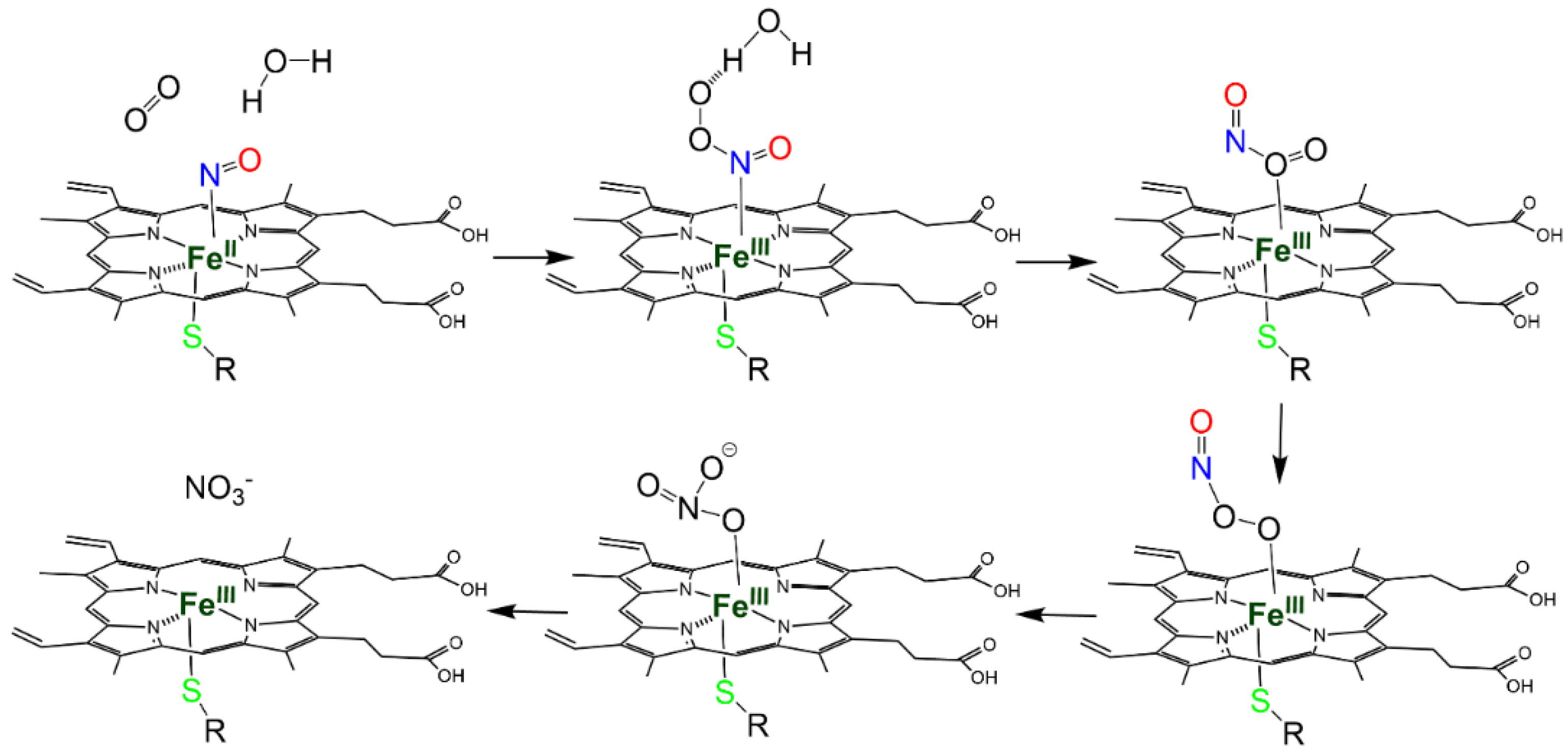
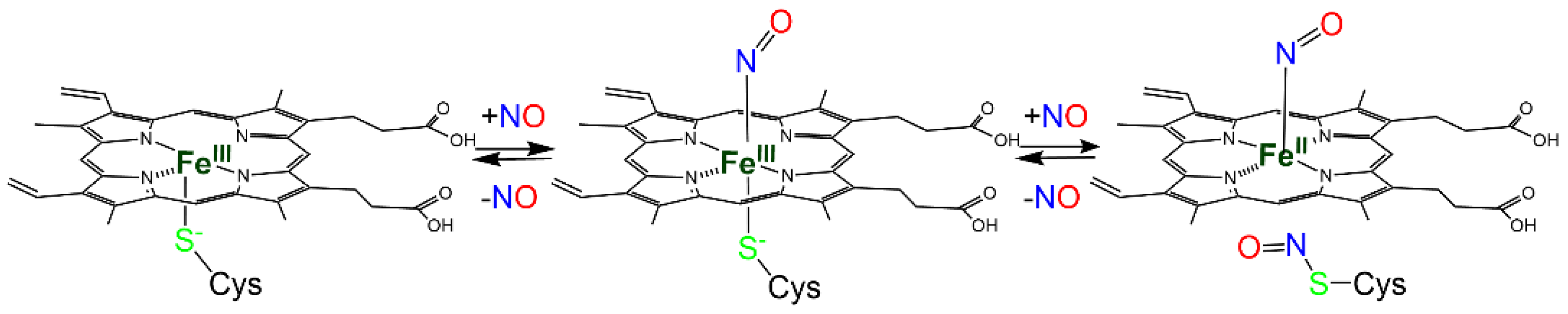
| Protein | Heme-NO Binding [Ref] | S-Nitrosylation: Cys Involved; Biological Effect [Ref]; PDB Entry | Heme-Dependent SNO [Ref] |
|---|---|---|---|
| Mammalian Hemoglobin | Reaction 2 [55,56,57] | Cysβ93; NO release [58]; PDB 1BUW (Figure 4a) | Yes [59]; Disputed [60] |
| Reaction 4 [8,16,36,37,61] | |||
| Reaction 5 [62] | |||
| Reaction 6 [63] | |||
| Ascaris Hemoglobin | Reaction 3 [64] | Cys72 in vitro evidence; NO-related functions in dioxygenation and oxygenation [64] | Yes [65] |
| Myoglobin | Reaction 4 [66,67] Reaction 6 [68] | Human Hb: Cys110, in vitro evidence [67]; Thunnus atlanticus (blackfin tuna) Mb: Cys10, in vitro evidence; Changes in the structure and dynamics [69,70]; PDB 2NRM (Figure 4b); Salmonid Mbs: Cys107, in vitro evidence; Increase in O2 affinity [71,72] | Yes (human) [67] |
| Neuroglobin | Reaction 2 [73] Reaction 4 [73] Reaction 5 [73] Reaction 6 [74] | Murine Ngb: Cys55 [74] | Yes [74] |
| Cytoglobin | Reaction 2 [75] Reaction 4 [33] Reaction 6 [75,76] | Cys38 and Cys83 [74] | Yes [74] |
| NO synthase | Reaction 2 Reaction 3 Reaction 4 [77,78] | Cys101, in vitro evidence; Regulation of NO signaling pathways in the vascular endothelium [79] | ND 1 |
| Cys93-Cys98, in vitro evidence; Formation of eNOS homodimer [80,81] | |||
| Guanilyl cyclase | Reaction 2 [82,83,84] | βCys122 and αCys243; Desensitization of NO [85,86] | Yes [85] |
| Cyclooxygenase | Reaction 2 [87] | COX2: Cys526; Activation of the enzyme [88] | ND 1 |
| Nitrophorin (Cimex lectularius) | Reaction 3 [31,89] | Cys60; NO release [90]; PDB 1Y21 (Figure 4c) | Yes [90] |
| Ascorbate peroxidase (Plant) | Reaction 3 [91] | Arabdopsis thaliana APX1: Cys32 in vivo and Cys49 in vitro evidence; Increase in the enzymatic activity; regulation of immune response [92,93] | ND 1 |
| Catalase | Reaction 3 [91] | Ganoderma lucidum: Cys401, Cys642, Cys653 in vivo and in vitro evidence; Alteration in catalase (CAT) activity [94] | ND 1 |
| Cytochrome c | Reaction 2 [95] Reaction 3 [96] | Trans-nitrosation of other thiols; Mediator of S-nitrosation in biological systems [97,98] | ND 1 |
| Cytochrome c oxidase | Reaction 2 [99,100] | Key Cys residues in subunit II of CcOX in vitro evidence Cys196, Cys200; Inhibition of cell respiration [101,102] | ND 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verde, C.; Giordano, D.; Bruno, S. NO and Heme Proteins: Cross-Talk between Heme and Cysteine Residues. Antioxidants 2023, 12, 321. https://doi.org/10.3390/antiox12020321
Verde C, Giordano D, Bruno S. NO and Heme Proteins: Cross-Talk between Heme and Cysteine Residues. Antioxidants. 2023; 12(2):321. https://doi.org/10.3390/antiox12020321
Chicago/Turabian StyleVerde, Cinzia, Daniela Giordano, and Stefano Bruno. 2023. "NO and Heme Proteins: Cross-Talk between Heme and Cysteine Residues" Antioxidants 12, no. 2: 321. https://doi.org/10.3390/antiox12020321
APA StyleVerde, C., Giordano, D., & Bruno, S. (2023). NO and Heme Proteins: Cross-Talk between Heme and Cysteine Residues. Antioxidants, 12(2), 321. https://doi.org/10.3390/antiox12020321








