The Peptide Methionine Sulfoxide Reductase (MsrAB) of Haemophilus influenzae Repairs Oxidatively Damaged Outer Membrane and Periplasmic Proteins Involved in Nutrient Acquisition and Virulence
Abstract
1. Introduction
2. Materials and Methods
2.1. Media and Bacterial Growth Conditions for E. coli and Haemophilus influenzae
2.2. Standard Molecular and Biochemical Methods
2.3. Construction of Protein Expression Plasmids and Optimisation of Protein Expression Protocols
2.4. Protein Expression in E. coli
2.5. Preparation of Cell-Free Extracts
2.6. Periplasm Preparation
2.7. Purification of Histidine-Tagged Proteins
2.8. Enzyme Assays
2.9. Thioredoxin Reductase (TrxR) Activity Assay
2.10. Thioredoxin Activity Assay
2.11. TrxR Thioredoxin Redox Relay Activity
2.12. MsrAB Assays—DTNB-Based Colourimetric Activity Assay
2.13. NADPH-Dependent MsrAB Activity Assay
2.14. MsrAB-Based Repair of Oxidised Proteins
2.15. Assessment of Trx Reduction State
2.16. H. influenzae Samples for Proteome Analyses
2.17. Outer Membrane Protein Isolation
2.18. MS/MS Sample Preparation
2.19. Preparation of Whole-Cell Samples for Shotgun Proteomics
2.20. Bioinformatic Analyses
2.21. Statistical Analysis
3. Results
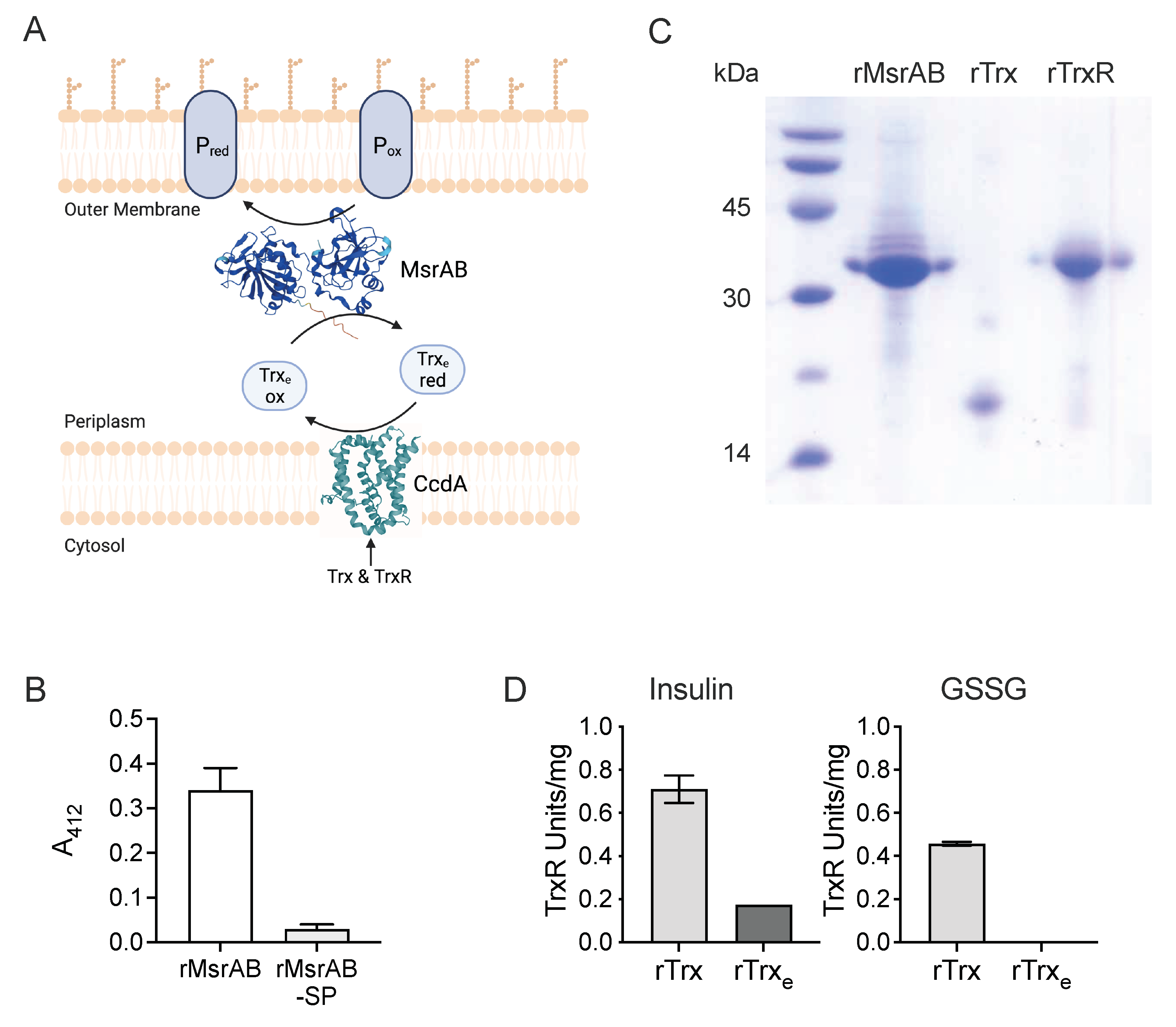
3.1. H. influenzae MsrAB Maturation Requires Export to the Periplasm
3.2. The H. influenzae Extracellular Thioredoxin Is Sensitive to Air-Oxidation and Has Low Reducing Activity
3.3. rTrx-rTrxR Form a Suitable “Redox Module” for Electron Donation in an rMsrAB Assay
| Plasmid | Protein | Protein Properties | E. coli Strain | Selective Markers (μg/mL) | IPTG (mM) | Post-Induction Temp. | Induction Time | Ref. |
|---|---|---|---|---|---|---|---|---|
| pProex-Htb MsrAB | rMsrAB | rMsrAB, expresses in E. coli periplasm, untagged | Rosetta | Amp 100, Cam 60 | 0.5 | 30 °C | 2 h | This study |
| pProex-Htb-MsrAB-sp | rMsrAB-sp | rMsrAB-sp, MsrAB protein without the N-terminal signal peptide (aa 22–353), N-terminal 6xHis tag | Rosetta | Amp 100, Cam 60 | 0.5 | 30 °C | 2 h | This study |
| pProexHtb Hi-eTrx | rTrxe | rTrxe, thioredoxin encoded in msrAB operon with N-terminal 6xHis tag | DH5α | Amp 100 | 0.1 | 37 °C | 16–18 h | This study |
| pProexHtb-Hi-Trx | rTrx | rTrx, cytoplasmic thioredoxin with N-terminal 6xHis tag | DH5α | Amp 100 | 0.1 | 37 °C | 16–18 h | This study |
| pProexHtb Hi-TrxR | rTrxR | rTrx-R, Thioredoxin reductase with N-terminal 6xHis tag | DH5α | Amp 100 | 0.5 | 37 °C | 16–18 h | This study |
| pET_22_P4 | eP4 | eP4, P4 adhesin (aa 22–274), C-terminal 6xHis tag | BL21(DE3) | Amp 100 | 0.1 | 37 °C | 2 h | This study; [43] |
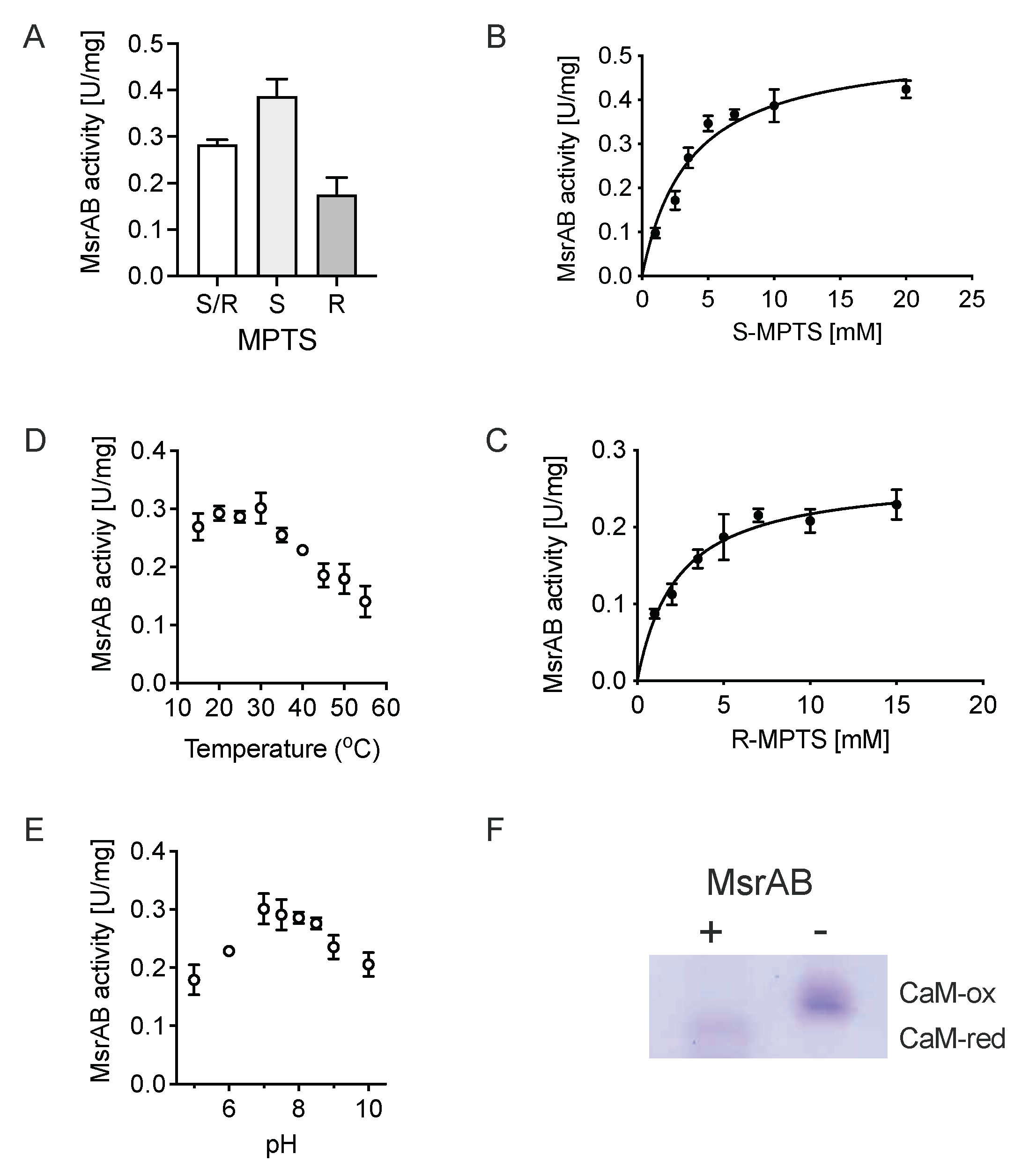
3.4. rMsrAB Converts Both S- and R-Diastereoisomers of Sulfoxide Substrates with High Efficiency
3.5. H. influenzae MsrAB Can Repair MetSO Damage in Proteins
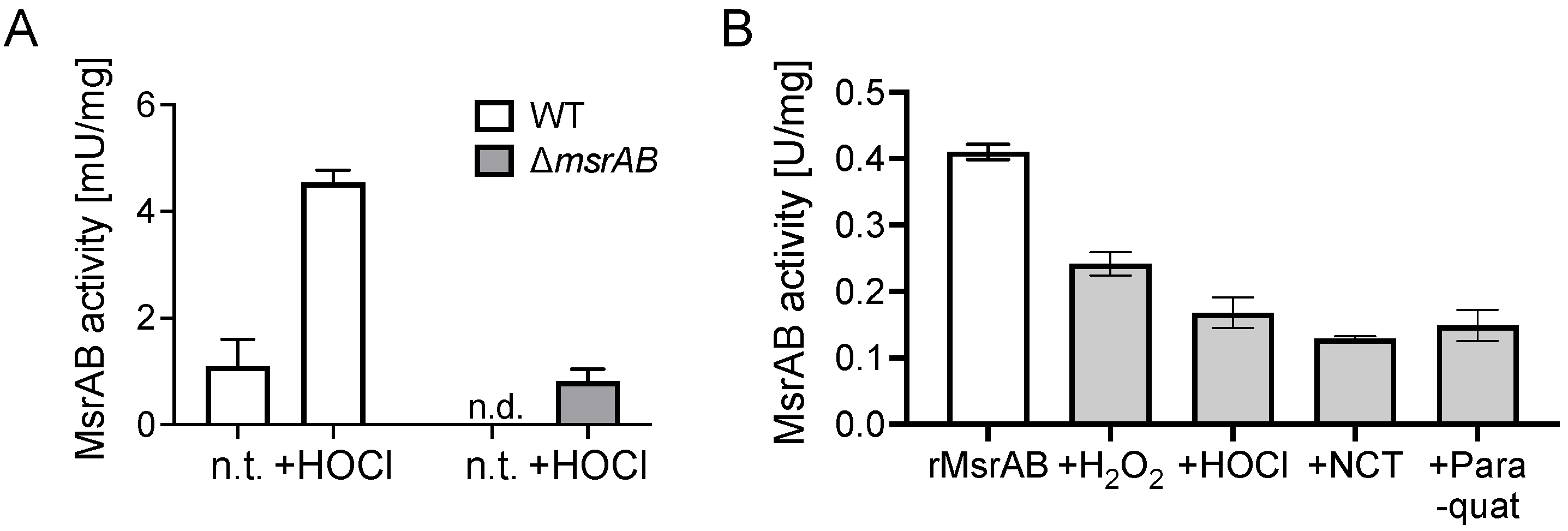
3.6. MsrAB Is Sensitive to Inactivation by Reactive Oxygen and Chlorine Species
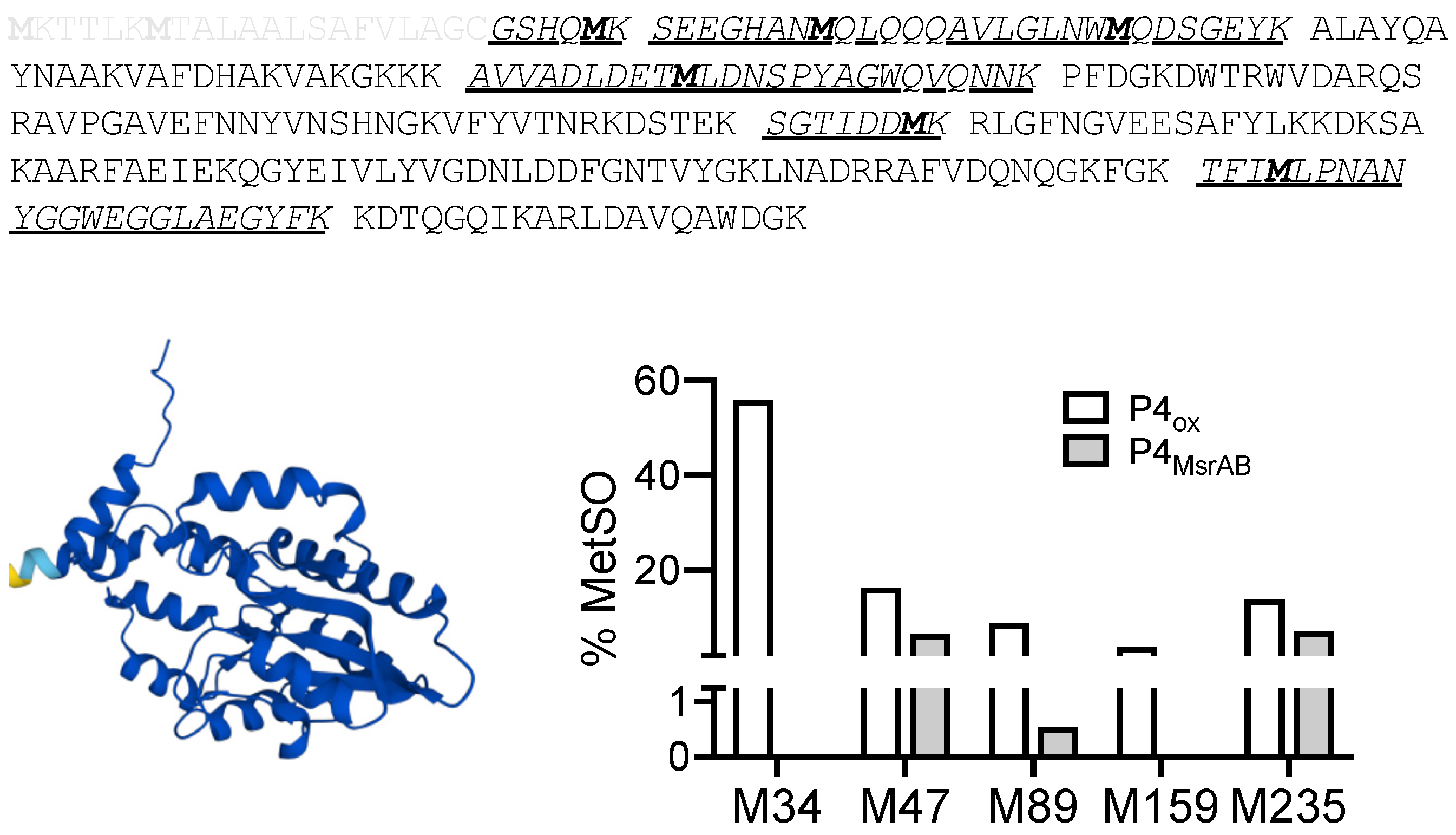
3.7. Identification of Putative H. influenzae MsrAB Target Proteins
3.8. MsrAB Can Efficiently Repair Oxidised Lipoprotein eP4 from H Influenzae
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ezraty, B.; Gennaris, A.; Barras, F.; Collet, J.-F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017, 15, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Kappler, U.; Nasreen, M.; McEwan, A. New insights into the molecular physiology of sulfoxide reduction in bacteria. Adv. Microb. Physiol. 2019, 75, 1–51. [Google Scholar] [CrossRef] [PubMed]
- Aussel, L.; Ezraty, B. Methionine Redox Homeostasis in Protein Quality Control. Front. Mol. Biosci. 2021, 8, 665492. [Google Scholar] [CrossRef]
- Maupin-Furlow, J.A. Methionine Sulfoxide Reductases of Archaea. Antioxidants 2018, 7, 124. [Google Scholar] [CrossRef]
- Lennicke, C.; Rahn, J.; Lichtenfels, R.; Wessjohann, L.A.; Seliger, B. Hydrogen peroxide—Production, fate and role in redox signaling of tumor cells. Cell Commun. Signal. 2015, 13, 39. [Google Scholar] [CrossRef]
- Achilli, C.; Ciana, A.; Minetti, G. The discovery of methionine sulfoxide reductase enzymes: An historical account and future perspectives. BioFactors 2015, 41, 135–152. [Google Scholar] [CrossRef]
- Boschi-Muller, S. Molecular Mechanisms of the Methionine Sulfoxide Reductase System from Neisseria meningitidis. Antioxidants 2018, 7, 131. [Google Scholar] [CrossRef]
- Saleh, M.; Bartual, S.G.; Abdullah, M.R.; Jensch, I.; Asmat, T.M.; Petruschka, L.; Pribyl, T.; Gellert, M.; Lillig, C.H.; Antelmann, H.; et al. Molecular architecture of Streptococcus pneumoniae surface thioredoxin-fold lipoproteins crucial for extracellular oxidative stress resistance and maintenance of virulence. EMBO Mol. Med. 2013, 5, 1852–1870. [Google Scholar] [CrossRef]
- Skaar, E.P.; Tobiason, D.M.; Quick, J.; Judd, R.C.; Weissbach, H.; Etienne, F.; Brot, N.; Seifert, H.S. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc. Natl. Acad. Sci. USA 2002, 99, 10108–10113. [Google Scholar] [CrossRef] [PubMed]
- Scheible, M.; Nguyen, C.T.; Luong, T.T.; Lee, J.H.; Chen, Y.-W.; Chang, C.; Wittchen, M.; Camacho, M.I.; Tiner, B.L.; Wu, C.; et al. The fused methionine sulfoxide reductase MsrAB promotes oxidative stress defense and acterial virulence in Fusobacterium nucleatum. MBio 2022, 13, e03022-21. [Google Scholar] [CrossRef]
- Alamuri, P.; Maier, R.J. Methionine Sulfoxide Reductase in Helicobacter pylori: Interaction with Methionine-Rich Proteins and Stress-Induced Expression. J. Bacteriol. 2006, 188, 5839–5850. [Google Scholar] [CrossRef]
- Alamuri, P.; Maier, R.J. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 2004, 53, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Gennaris, A.; Ezraty, B.; Henry, C.; Agrebi, R.; Vergnes, A.; Oheix, E.; Bos, J.; Leverrier, P.; Espinosa, L.; Szewczyk, J.; et al. Repairing oxidized proteins in the bacterial envelope using respiratory chain electrons. Nature 2015, 528, 409–412. [Google Scholar] [CrossRef]
- Andrieu, C.; Vergnes, A.; Loiseau, L.; Aussel, L.; Ezraty, B. Characterisation of the periplasmic methionine sulfoxide reductase (MsrP) from Salmonella Typhimurium. Free Radic. Biol. Med. 2020, 160, 506–512. [Google Scholar] [CrossRef]
- Tarrago, L.; Grosse, S.; Siponen, M.I.; Lemaire, D.; Alonso, B.; Miotello, G.; Armengaud, J.; Arnoux, P.; Pignol, D.; Sabaty, M. Rhodobacter sphaeroides methionine sulfoxide reductase P reduces R- and S-diastereomers of methionine sulfoxide from a broad-spectrum of protein substrates. Biochem. J. 2018, 475, 3779–3795. [Google Scholar] [CrossRef]
- Nasreen, M.; Fletcher, A.; Hosmer, J.; Zhong, Q.; Essilfie, A.-T.; McEwan, A.G.; Kappler, U. The Alternative Sigma Factor RpoE2 Is Involved in the Stress Response to Hypochlorite and in vivo Survival of Haemophilus influenzae. Front. Microbiol. 2021, 12, 637213. [Google Scholar] [CrossRef]
- Dhouib, R.; Nasreen, M.; Othman, D.S.M.P.; Ellis, D.; Lee, S.; Essilfie, A.-T.; Hansbro, P.M.; McEwan, A.G.; Kappler, U. The DmsABC Sulfoxide Reductase Supports Virulence in Non-typeable Haemophilus influenzae. Front. Microbiol. 2021, 12, 686833. [Google Scholar] [CrossRef]
- Nasreen, M.; Dhouib, R.; Hosmer, J.; Wijesinghe, H.G.S.; Fletcher, A.; Mahawar, M.; Essilfie, A.-T.; Blackall, P.J.; McEwan, A.G.; Kappler, U. Peptide Methionine Sulfoxide Reductase from Haemophilus influenzae Is Required for Protection against HOCl and Affects the Host Response to Infection. ACS Infect. Dis. 2020, 6, 1928–1939. [Google Scholar] [CrossRef] [PubMed]
- Dhouib, R.; Othman, D.S.M.P.; Lin, V.; Lai, X.J.; Wijesinghe, H.G.S.; Essilfie, A.-T.; Davis, A.; Nasreen, M.; Bernhardt, P.V.; Hansbro, P.M.; et al. A Novel, Molybdenum-Containing Methionine Sulfoxide Reductase Supports Survival of Haemophilus influenzae in an in vivo Model of Infection. Front. Microbiol. 2016, 7, 1743. [Google Scholar] [CrossRef]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Current Protoc Mol Biol. In Current Protoc Mol Biol; Janssen, K., Ed.; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2005. [Google Scholar]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Coleman, H.N.; Daines, D.A.; Jarisch, J.; Smith, A.L. Chemically Defined Media for Growth of Haemophilus influenzae Strains. J. Clin. Microbiol. 2003, 41, 4408–4410. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.W. Laboratory Growth and Maintenance of Haemophilus influenzae. Curr. Protoc. Microbiol. 2010, 18, 6D.1.1–6D.1.5. [Google Scholar] [CrossRef] [PubMed]
- Campagnari, A.A.; Gupta, M.R.; Dudas, K.C.; Murphy, T.F.; Apicella, M.A. Antigenic diversity of lipooligosaccharides of nontypable Haemophilus influenzae. Infect. Immun. 1987, 55, 882–887. [Google Scholar] [CrossRef]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.; Smith, J.A.; Struhl, K. Short Protocols in Molecular Biology; Wiley: New York, NY, USA, 1992. [Google Scholar]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Payton, M. Selection of Escherichia coli Expression Systems. Curr. Protoc. Protein Sci. 1995, 5, 5.2.1–5.2.18. [Google Scholar] [CrossRef] [PubMed]
- Markwell, J.P.; Lascelles, J. Membrane-bound, pyridine nucleotide-independent L-lactate dehydrogenase of Rhodopseudomonas sphaeroides. J. Bacteriol. 1978, 133, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.-J.; Ishikawa, K. Identification and characterization of thioredoxin and thioredoxin reductase from Aeropyrum pernix K1. J. Biol. Inorg. Chem. 2002, 269, 5423–5430. [Google Scholar] [CrossRef]
- Holmgren, A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 1979, 254, 9627–9632. [Google Scholar] [CrossRef]
- Spyrou, G.; Enmark, E.; Miranda-Vizuete, A.; Gustafsson, J. Cloning and Expression of a Novel Mammalian Thioredoxin. J. Biol. Chem. 1997, 272, 2936–2941. [Google Scholar] [CrossRef]
- Wu, P.-F.; Zhang, Z.; Guan, X.-L.; Li, Y.-L.; Zeng, J.-H.; Zhang, J.-J.; Long, L.-H.; Hu, Z.-L.; Wang, F.; Chen, J.-G. A specific and rapid colorimetric method to monitor the activity of methionine sulfoxide reductase A. Enzym. Microb. Technol. 2013, 53, 391–397. [Google Scholar] [CrossRef]
- Achilli, C.; Ciana, A.; Minetti, G. Kinetic resolution of phenyl methyl sulfoxides by mammalian methionine sulfoxide reductase A. Tetrahedron Lett. 2017, 58, 4781–4782. [Google Scholar] [CrossRef]
- Carlone, G.M.; Thomas, M.L.; Rumschlag, H.S.; O Sottnek, F. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J. Clin. Microbiol. 1986, 24, 330–332. [Google Scholar] [CrossRef] [PubMed]
- Kappler, U.; Nouwens, A.S. The molybdoproteome of Starkeya novella—Insights into the diversity and functions of molybdenum containing proteins in response to changing growth conditions. Metallomics 2013, 5, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved Prediction of Signal Peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Springer: Berlin, Germany, 2005. [Google Scholar]
- Olry, A.; Boschi-Muller, S.; Marraud, M.; Sanglier-Cianferani, S.; Van Dorsselear, A.; Branlant, G. Characterization of the methionine sulfoxide reductase activities of PILB, a probable virulence factor from Neisseria meningitidis. J. Biol. Chem. 2002, 277, 12016–12022. [Google Scholar] [CrossRef]
- Reynolds, C.M.; Meyer, J.; Poole, L.B. An NADH-Dependent Bacterial Thioredoxin Reductase-like Protein in Conjunction with a Glutaredoxin Homologue Form a Unique Peroxiredoxin (AhpC) Reducing System in Clostridium pasteurianum. Biochemistry 2002, 41, 1990–2001. [Google Scholar] [CrossRef]
- McCarver, A.C.; Lessner, D.J. Molecular characterization of the thioredoxin system from Methanosarcina acetivorans. FEBS J. 2014, 281, 4598–4611. [Google Scholar] [CrossRef]
- Su, Y.-C.; Mukherjee, O.; Singh, B.; Hallgren, O.; Westergren-Thorsson, G.; Hood, D.; Riesbeck, K. Haemophilus influenzaeP4 Interacts With Extracellular Matrix Proteins Promoting Adhesion and Serum Resistance. J. Infect. Dis. 2016, 213, 314–323. [Google Scholar] [CrossRef]
- Kim, Y.K.; Shin, Y.J.; Lee, W.-H.; Kim, H.-Y.; Hwang, K.Y. Structural and kinetic analysis of an MsrA-MsrB fusion protein from Streptococcus pneumoniae. Mol. Microbiol. 2009, 72, 699–709. [Google Scholar] [CrossRef][Green Version]
- Boschi-Muller, S.; Olry, A.; Antoine, M.; Branlant, G. The enzymology and biochemistry of methionine sulfoxide reductases. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2005, 1703, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Hosmer, J.; Nasreen, M.; Dhouib, R.; Essilfie, A.-T.; Schirra, H.J.; Henningham, A.; Fantino, E.; Sly, P.; McEwan, A.G.; Kappler, U. Access to highly specialized growth substrates and production of epithelial immunomodulatory metabolites determine survival of Haemophilus influenzae in human airway epithelial cells. PLoS Pathog. 2022, 18, e1010209. [Google Scholar] [CrossRef] [PubMed]
- Roier, S.; Blume, T.; Klug, L.; Wagner, G.E.; Elhenawy, W.; Zangger, K.; Prassl, R.; Reidl, J.; Daum, G.; Feldman, M.; et al. A basis for vaccine development: Comparative characterization of Haemophilus influenzae outer membrane vesicles. Int. J. Med. Microbiol. 2015, 305, 298–309. [Google Scholar] [CrossRef]
- Morton, D.J.; Smith, A.; VanWagoner, T.M.; Seale, T.W.; Whitby, P.W.; Stull, T.L. Lipoprotein e (P4) of Haemophilus influenzae: Role in heme utilization and pathogenesis. Microbes Infect. 2007, 9, 932–939. [Google Scholar] [CrossRef]
- Kemmer, G.; Reilly, T.J.; Schmidt-Brauns, J.; Zlotnik, G.W.; Green, B.A.; Fiske, M.J.; Herbert, M.; Kraiß, A.; Schlör, S.; Smith, A.; et al. NadN and e (P4) Are Essential for Utilization of NAD and Nicotinamide Mononucleotide but Not Nicotinamide Riboside in Haemophilus influenzae. J. Bacteriol. 2001, 183, 3974–3981. [Google Scholar] [CrossRef] [PubMed]
- Reidl, J.; Schlör, S.; Kraiß, A.; Schmidt-Brauns, J.; Kemmer, G.; Soleva, E. NADP and NAD Utilization in Haemophilus influenzae. Mol. Microbiol. 2002, 35, 1573–1581. [Google Scholar] [CrossRef] [PubMed]
| Plasmid | H. influenzae 2019 Gene Loci | Plasmid Insert Generation | |
|---|---|---|---|
| Primer Name | Primer Sequences | ||
| pProex-Htb MsrAB | C645_RS07025 | Hi_msrAB_pPro_Bam_F | AAAAGGATCCATGAAACTATCAAAAACATTTC |
| Hi_msrAB_pPro_Hind_R | AAAAAAGCTTCTTATTTTTTAATGGATTGAATC | ||
| pProex-Htb-MsrAB-sp | C645_RS07025 | Hi_msrAB-sp_pPro_BamHI_F | AAAAGGATCCATACAAAATTCAACATCATCATC |
| Hi_msrAB_pPro_Hind_R | AAAAAAGCTTCTTATTTTTTAATGGATTGAATC | ||
| pProexHtb Hi-eTrx | C645_RS08405 | Hi_etrx_pPro_Bam_F | AAAAGGATCCCAAACTAATTTGGCAGATGT |
| Hi_etrx_pPro_XbaI_R | AAAACTGCAGCTTTTCATTATTTCCCT | ||
| pProexHtb Hi-TrxR | C645_RS07025 | Hi_trxR_pPro_BamHI F | AAAAGGATCCTCAGATACCAAACACGCAAAAC |
| Hi_trxR_pPro_Eco R | AAAAGAATTCTTAAAAGAGGGGAATTGGTTAG | ||
| pProexHtb-Hi-Trx | C645_RS00695 | Hi_trx_pPro_BamHI_F | AAAAGGATCCAGCGAAGTATTACACATTAATGA |
| Hi_trx_pPro_Eco_R | AAAAGAATTCCAACGATTAAATATGTTGGTTAA | ||
| pET_22_P4 | C645_RS09250 | HiP4_pET_Bam_F | AAAAGGATCCGGGTTCACACCAAATGAAATCAGAA |
| HiP4_pET_SacI_R | AAAAGAGCTCGCTTTACCATCCCAAGCTTGTACTGC | ||
| Substrate | Vmax_app (U/mg) | kcat_app (s−1) | KM _app (mM) | kcat/KM (s−1 M−1) |
|---|---|---|---|---|
| S/R-MPTS | 0.34 ± 0.01 | 0.204 ± 0.06 | 2.96 ± 0.44 | 68.9 |
| S-MPTS | 0.52 ± 0.02 | 0.312 ± 0.12 | 3.62 ± 0.55 | 86.1 |
| R-MPTS | 0.26 ± 0.01 | 0.156 ± 0.06 | 2.31 ± 0.35 | 67.5 |
| % Met Oxidation | ||||
|---|---|---|---|---|
| Accession | Protein Name | Peptides | WT | ΔmsrAB |
| Transport Proteins | ||||
| WP_005651801.1 | ABC transporter substrate-binding protein OppA | VAIAAASmWK | 0 | 12.23 ± 4.45 |
| AmAESYAATDAEGR | 0 | 5.43 ± 0.75 | ||
| WP_005655633.1 | galactose ABC transporter substrate-binding protein MglA | LLmNDSQNAQSIQNDQVDVLLSK | 0 | 29.16 ± 5.89 |
| YDDNFmSLMR | 0 | 20.83 ± 17.67 | ||
| WP_005657776.1 | C4-dicarboxylate ABC transporter substrate-binding protein DctP-like | AADDSMmYHK | 42.22 ± 22.68 | 71.1 ± 7.69 |
| mIAETTQEAK | 6.94 ± 1.49 | 22.5 ± 3.53 | ||
| WP_005688477.1 | putrescine/spermidine ABC transporter substrate-binding protein PotD | APLNmVFPK | 0 * | 10.1 ± 3.36 |
| WP_012054840.1 | sialic acid-binding protein SiaP | FGmNAGTSSNEYK | 0 * | 36.1 ± 12.72 |
| WP_046067550.1 | methionine ABC transporter substrate-binding protein MetQ | VGVmSGPEHQVAEIAAK | 0 * | 13.9 ± 10.01 |
| WP_005650782.1 | glycerol-3-phosphate transporter GlpT | FVMAGmSDR | 0 * | 50.2 ± 46.58 |
| WP_005653411.1 | preprotein translocase subunit SecD | NmLPADSEVKYDR | 0 * | 75 ± 35.35 |
| Enzymes and Virulence Factors | ||||
| WP_005631652.1 | NADP transhydrogenase subunit alpha PntA | VmSEEFNRR | 0 | 100 ± 0 |
| mQNPELMK | 0 | 26.78 ± 2.52 | ||
| WP_005657875.1 | alpha-hydroxy-acid oxidizing enzyme LldD | DmHSGMSGPYK | 9.72 ± 8.67 | 33.2 ± 14.36 |
| MLALGADATmLGR | 0 * | 33.3 ± 0 | ||
| WP_005687981.1 | thiamine biosynthesis lipoprotein ApbE | TmGTTYHVK | 0 | 41.66 ± 11.78 |
| WP_005649107.1 | cytochrome c NapC | LEmAQNEWAR | 0 | 16.7 ± 28.86 |
| WP_046067759.1 | membrane protein OmpA | ANLKPQAQATLDSIYGEmSQVK | 26.13 ± 1.61 | 41.66 ± 11.78 |
| WP_046067581.1 | opacity-associated protein OapA | ATAPVQPmKK | 0 * | 83.33 ± 23.57 |
| WP_005647222.1 | Outer membrane protein SlyB | mSQVNGAELVIK | 0 | 33.34 ± 0 |
| WP_005661229.1 | glycerophosphoryl diester phosphodiesterase GlpQ/Protein D | IKTELLPQmGMDLK | 0 | 46.7 ± 17.63 |
| ALAFAQQADYLEQDLAmTK | 0 | 32.5 ± 10.61 | ||
| WP_005661232.1 | 5′-nucleotidase, lipoprotein e P4 family | DSTEKAGTIDDmKR | 0 | 50 ± 0 |
| WP_046067826.1 | Histone | ASEmKEAASEKASEMK | 0 | 93.3 ± 11.54 |
| EAVSEKASEmKEAASEK | 0 | 100 ± 0 | ||
| EAASEKASEmKEAASEK | 0 | 61.1 ± 34.69 | ||
| EAASEKTSEmKEAVSEK | 0 | 50 ± 0 | ||
| DAAANTmTEVK | 0 | 21.36 ± 10.91 | ||
| Proteins of unknown function | ||||
| WP_005656625.1 | hypothetical protein | KPLNAEMAmTR | 0 | 20 ± 0 |
| WP_005668839.1 | hypothetical protein | SSAQMAEmQTLPTITDK | 0 | 23 ± 23.38 |
| WP_005655950.1 | hypothetical protein | NGmVEVQKNEDGTPK | 0 | 61.1 ± 34.69 |
| WP_005689200.1 | membrane protein | NGEAYLmPK | 47.77 ± 27.16 | 55.6 ± 9.62 |
| WP_005658466.1 | membrane protein | IVAPmQR | 0 | 50.0 ± 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasreen, M.; Nair, R.P.; McEwan, A.G.; Kappler, U. The Peptide Methionine Sulfoxide Reductase (MsrAB) of Haemophilus influenzae Repairs Oxidatively Damaged Outer Membrane and Periplasmic Proteins Involved in Nutrient Acquisition and Virulence. Antioxidants 2022, 11, 1557. https://doi.org/10.3390/antiox11081557
Nasreen M, Nair RP, McEwan AG, Kappler U. The Peptide Methionine Sulfoxide Reductase (MsrAB) of Haemophilus influenzae Repairs Oxidatively Damaged Outer Membrane and Periplasmic Proteins Involved in Nutrient Acquisition and Virulence. Antioxidants. 2022; 11(8):1557. https://doi.org/10.3390/antiox11081557
Chicago/Turabian StyleNasreen, Marufa, Remya Purushothaman Nair, Alastair G. McEwan, and Ulrike Kappler. 2022. "The Peptide Methionine Sulfoxide Reductase (MsrAB) of Haemophilus influenzae Repairs Oxidatively Damaged Outer Membrane and Periplasmic Proteins Involved in Nutrient Acquisition and Virulence" Antioxidants 11, no. 8: 1557. https://doi.org/10.3390/antiox11081557
APA StyleNasreen, M., Nair, R. P., McEwan, A. G., & Kappler, U. (2022). The Peptide Methionine Sulfoxide Reductase (MsrAB) of Haemophilus influenzae Repairs Oxidatively Damaged Outer Membrane and Periplasmic Proteins Involved in Nutrient Acquisition and Virulence. Antioxidants, 11(8), 1557. https://doi.org/10.3390/antiox11081557







