PPARδ Activation Mitigates 6-OHDA-Induced Neuronal Damage by Regulating Intracellular Iron Levels
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Gene Silencing
2.4. Cytotoxicity Assay
2.5. Measurement of Intracellular Iron Levels
2.6. Western Blot Analysis
2.7. Real-Time PCR Analysis
2.8. Measurement of Intracellular ROS
2.9. Measurement of Lipid Peroxidation
2.10. Statistical Analysis
3. Results
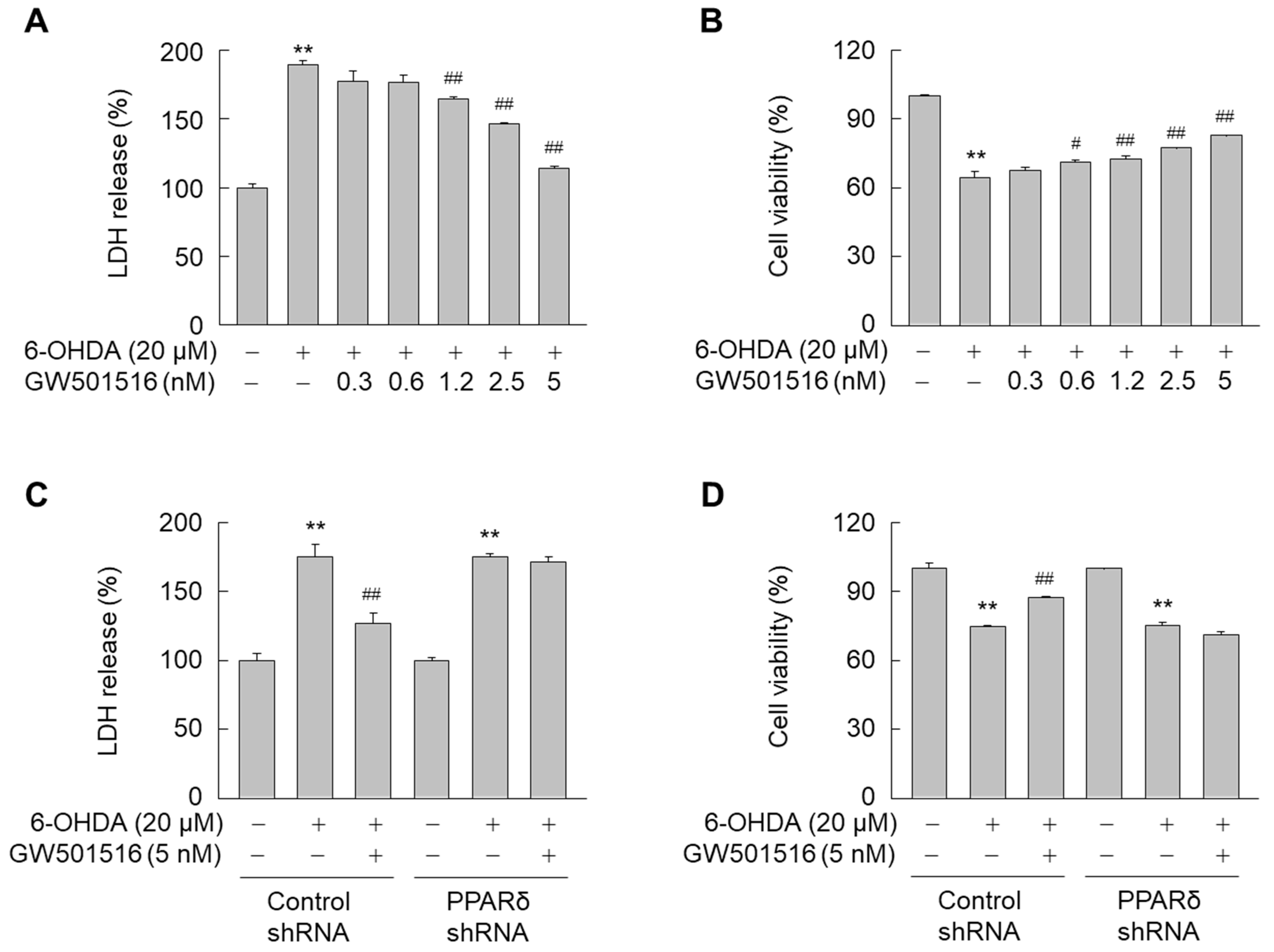
3.1. GW501516 Activation of PPARδ Attenuates 6-OHDA-Triggered Cellular Damage in SH-SY5Y Cells
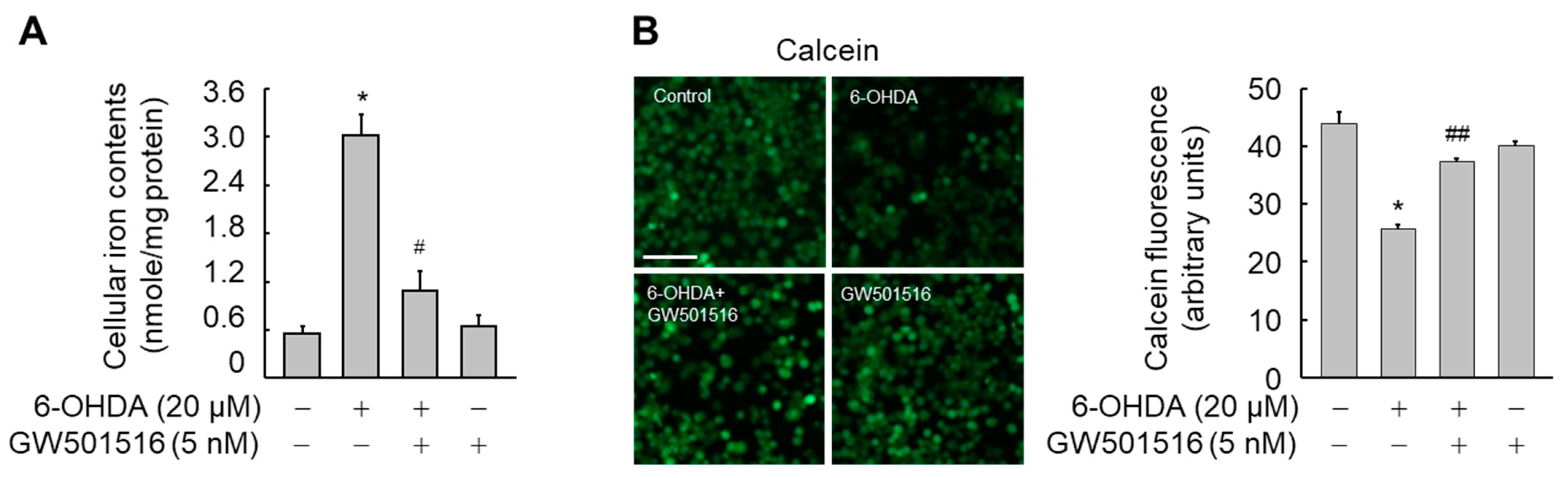
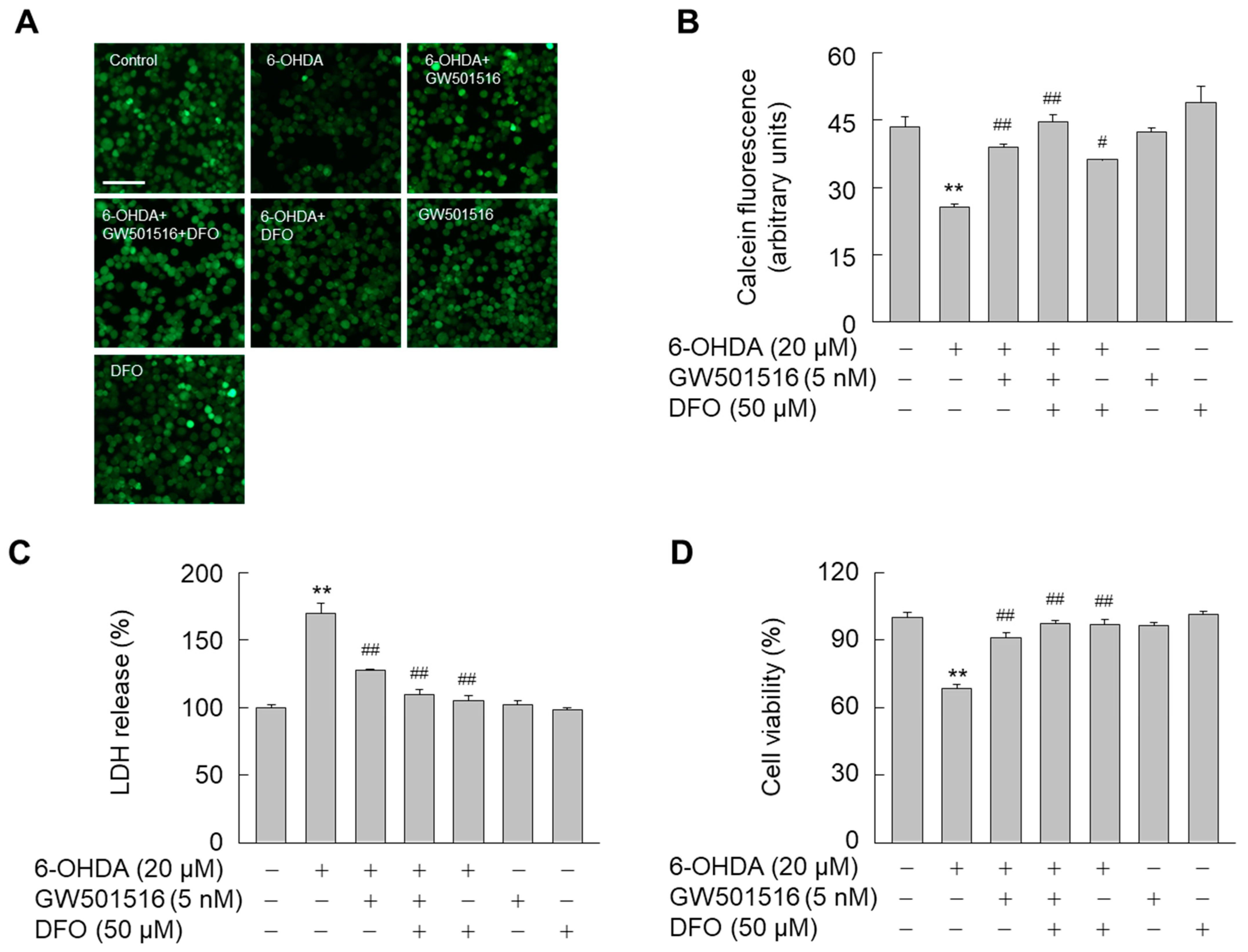
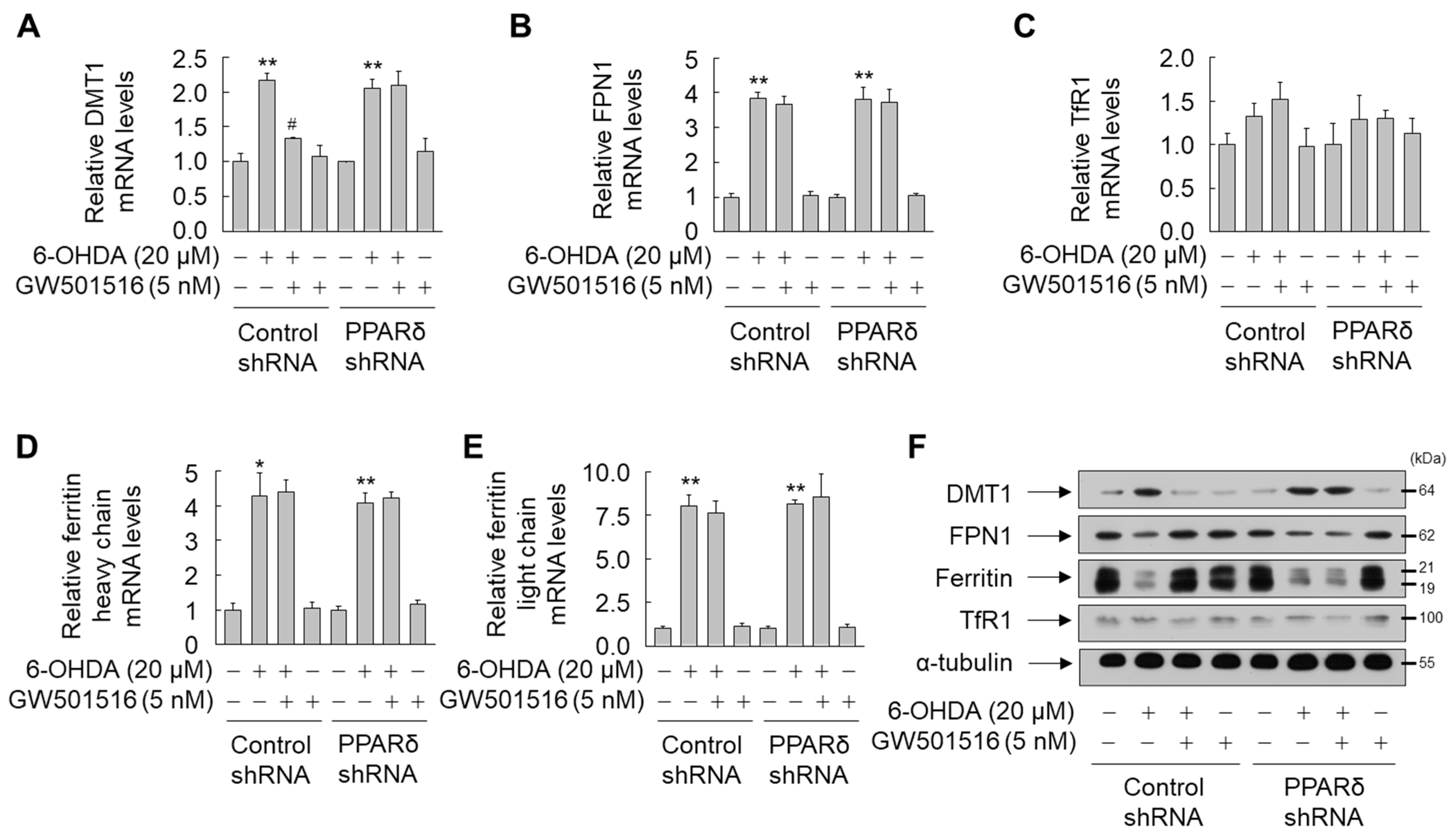
3.2. GW501516 Activation of PPARδ Decreases 6-OHDA-Induced Iron Accumulation
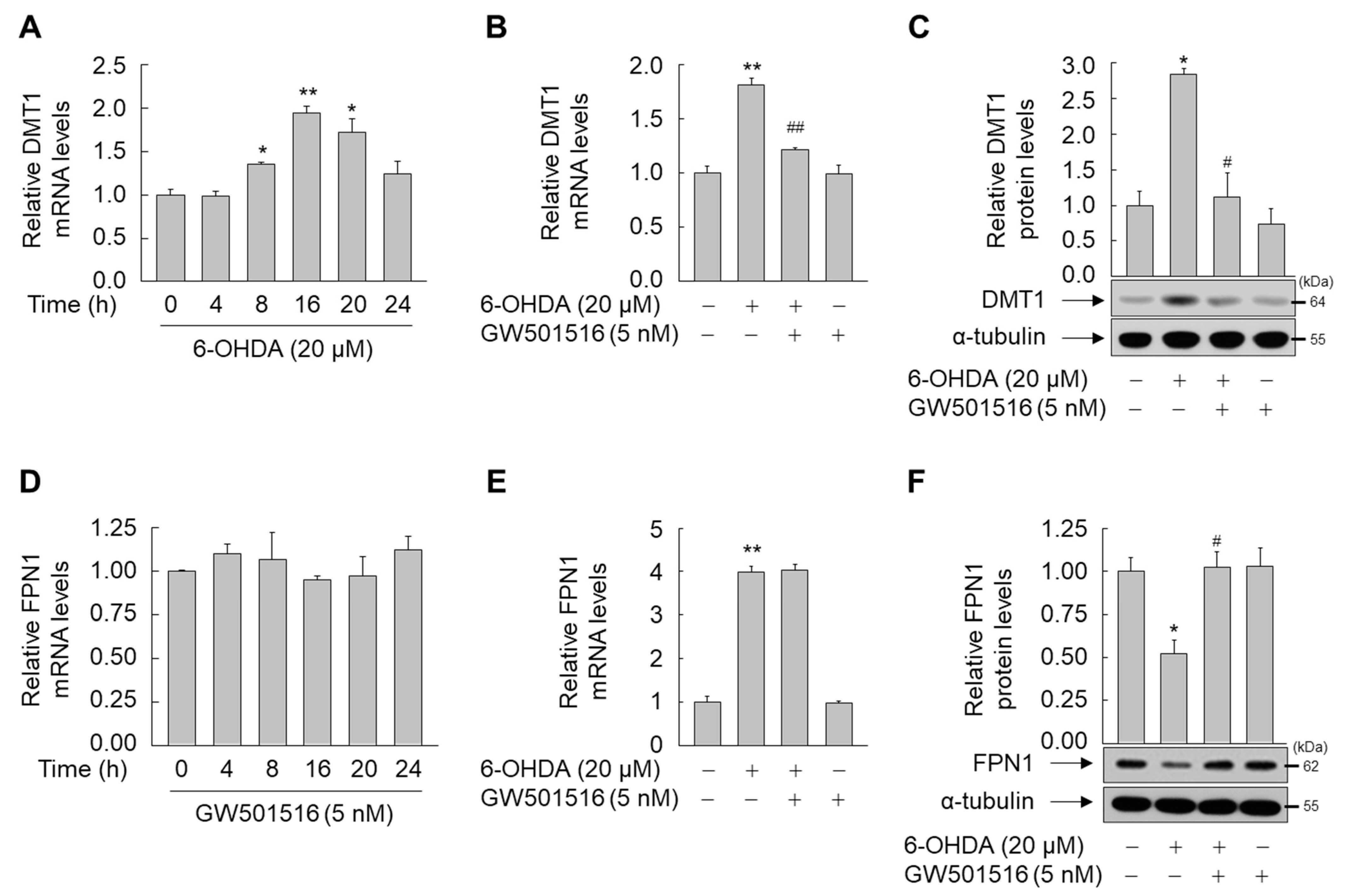
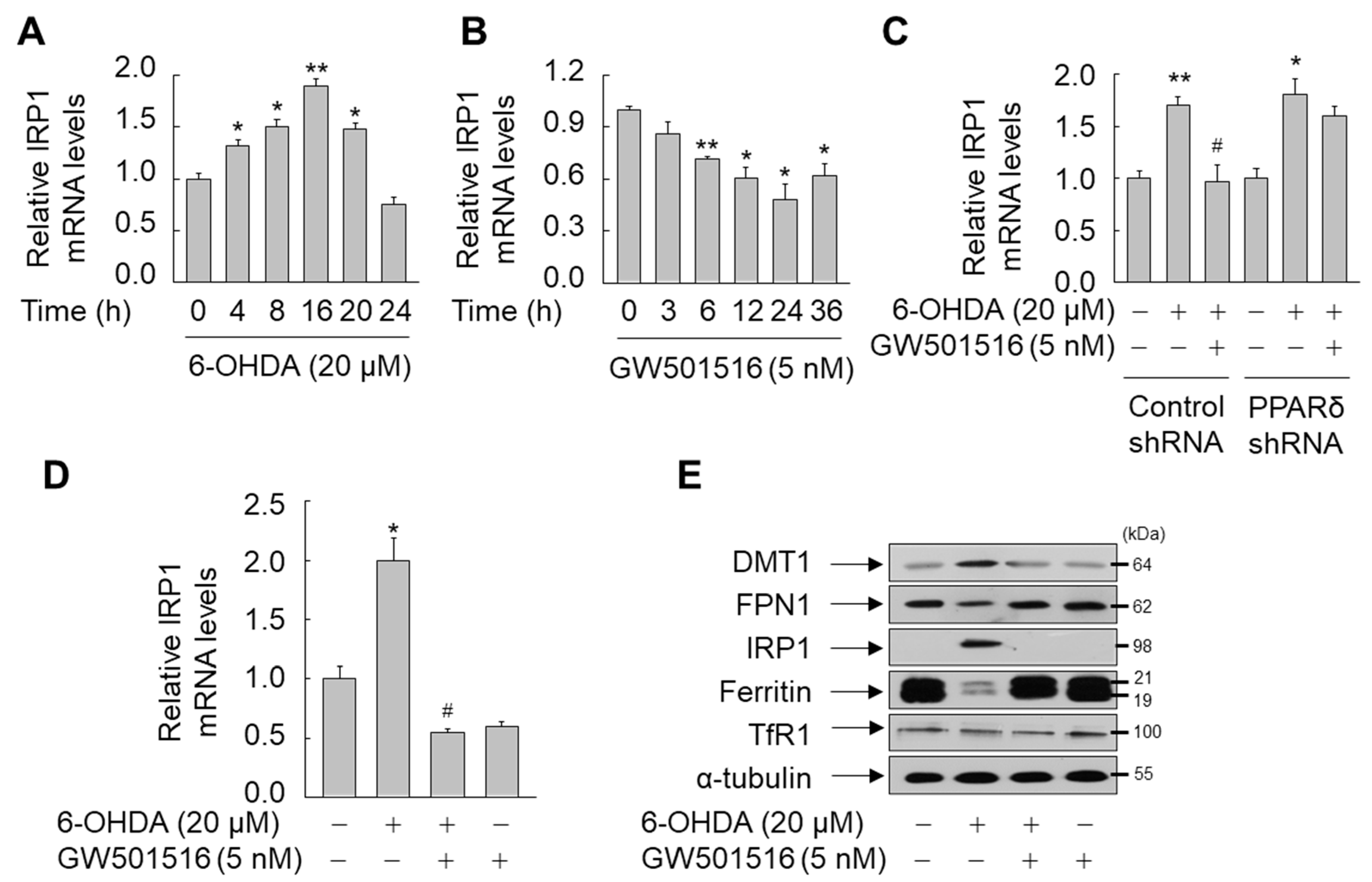
3.3. GW501516 Activation of PPARδ Suppresses IRP1 Expression in SH-SY5Y Cells
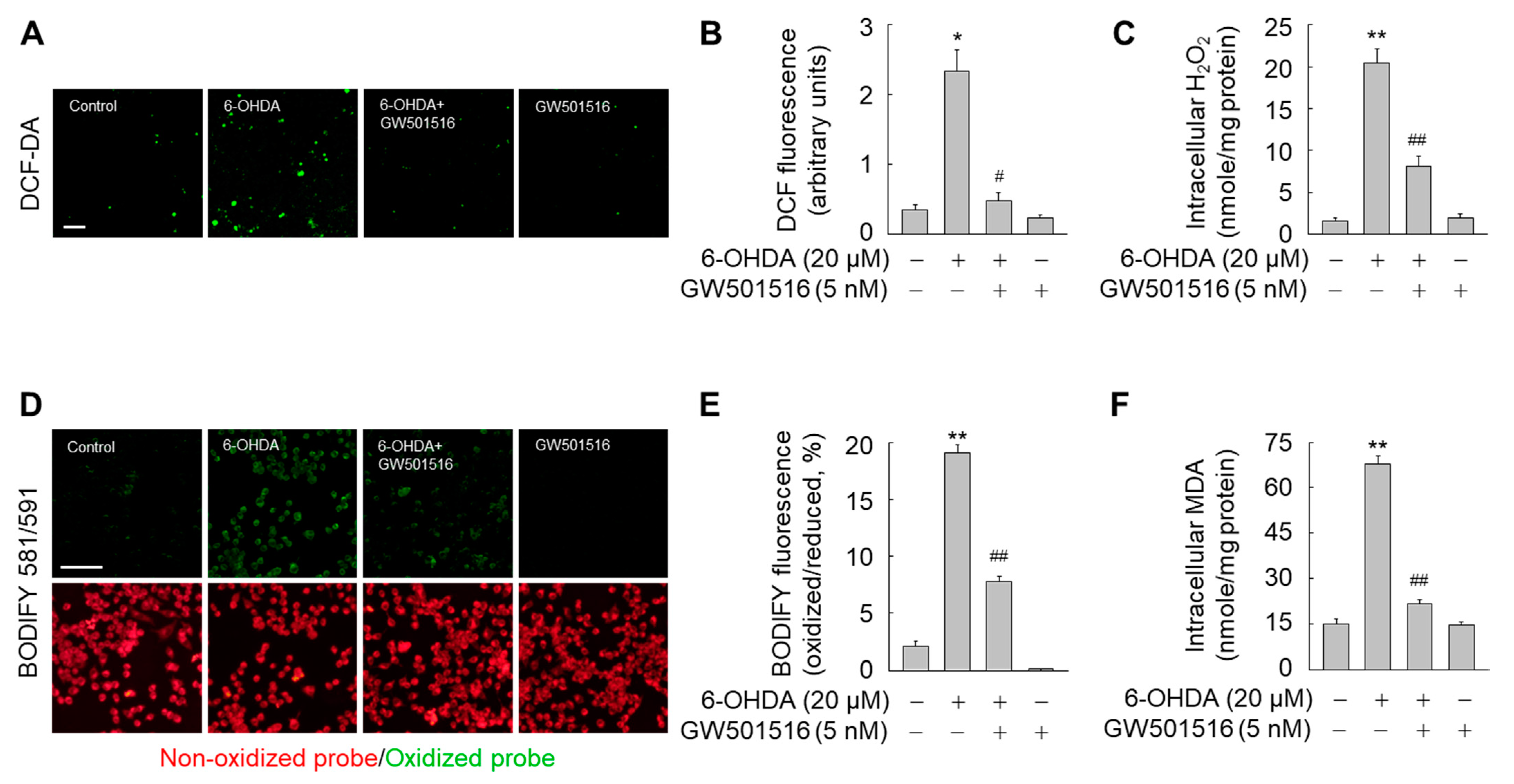
3.4. GW501516 Activation of PPARδ Decreases 6-OHDA-Triggered Accumulation of ROS and Lipid Peroxides
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maiti, P.; Manna, J.; Dunbar, G.L. Current understanding of the molecular mechanisms in Parkinson’s disease: Targets for potential treatments. Transl. Neurodegener. 2017, 6, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.H.; Katzenschlager, R.; Lim, S.Y.; Barton, B.; de Bie, R.M.A.; Seppi, K.; Coelho, M.; Sampaio, C.; Movement Disorder Society Evidence-Based Medicine Committee. International Parkinson and movement disorder society evidence-based medicine review: Update on treatments for the motor symptoms of Parkinson’s disease. Mov. Disord. 2018, 33, 1248–1266. [Google Scholar] [CrossRef] [PubMed]
- Olanow, C.W.; Agid, Y.; Mizuno, Y.; Albanese, A.; Bonuccelli, U.; Damier, P.; De Yebenes, J.; Gershanik, O.; Guttman, M.; Grandas, F.; et al. Levodopa in the treatment of Parkinson’s disease: Current controversies. Mov. Disord. 2004, 19, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Ungerstedt, U. 6-Hydroxy-dopamine induced degeneration of central monoamine neurons. Eur. J. Pharmacol. 1968, 5, 107–110. [Google Scholar] [CrossRef]
- Luthman, J.; Fredriksson, A.; Sundström, E.; Jonsson, G.; Archer, T. Selective lesion of central dopamine or noradrenaline neuron systems in the neonatal rat: Motor behavior and monoamine alterations at adult stage. Behav. Brain Res. 1989, 33, 267–277. [Google Scholar] [CrossRef]
- Freinbichler, W.; Misini, B.; Colivicchi, M.A.; Linert, W.; Tipton, K.F.; Della Corte, L. The application of bathophenanthroline for the determination of free iron in parallel with hROS in microdialysis samples. J. Neurosci. Methods 2020, 331, 108530. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Guiney, S.J.; Adlard, P.A.; Bush, A.I.; Finkelstein, D.I.; Ayton, S. Ferroptosis and cell death mechanisms in Parkinson’s disease. Neurochem. Int. 2017, 104, 34–48. [Google Scholar] [CrossRef]
- Wan, J.; Ren, H.; Wang, J. Iron toxicity, lipid peroxidation and ferroptosis after intracerebral haemorrhage. Stroke Vasc. Neurol. 2019, 4, 93–95. [Google Scholar] [CrossRef]
- Xie, Y.; Hou, W.; Song, X.; Yu, Y.; Huang, J.; Sun, X.; Kang, R.; Tang, D. Ferroptosis: Process and function. Cell Death Differ. 2016, 23, 369–379. [Google Scholar] [CrossRef]
- Masaldan, S.; Bush, A.I.; Devos, D.; Rolland, A.S.; Moreau, C. Striking while the iron is hot: Iron metabolism and ferroptosis in neurodegeneration. Free Radic. Biol. Med. 2019, 133, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Wang, H.; Han, D.; Xie, E.; Yang, X.; Wei, J.; Gu, S.; Gao, F.; Zhu, N.; Yin, X.; et al. Ferroptosis as a target for protection against cardiomyopathy. Proc. Natl. Acad. Sci. USA 2019, 116, 2672–2680. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Feng, D.; Wang, Z.; Zhao, Y.; Sun, R.; Tian, D.; Liu, D.; Zhang, F.; Ning, S.; Yao, J.; et al. Ischemia-induced ACSL4 activation contributes to ferroptosis-mediated tissue injury in intestinal ischemia/reperfusion. Cell Death Differ. 2019, 26, 2284–2299. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.; Gao, X.; Xu, H.; Cui, Y.; Zhang, Y.; Gou, X. The Emerging Roles of Ferroptosis in Huntington’s Disease. Neuromol. Med. 2019, 21, 110–119. [Google Scholar] [CrossRef]
- Dexter, D.T.; Wells, F.R.; Lees, A.J.; Agid, F.; Agid, Y.; Jenner, P.; Marsden, C.D. Increased nigral iron content and alterations in other metal ions occurring in brain in Parkinson’s disease. J. Neurochem. 1989, 52, 1830–1836. [Google Scholar] [CrossRef]
- Takanashi, M.; Mochizuki, H.; Yokomizo, K.; Hattori, N.; Mori, H.; Yamamura, Y.; Mizuno, Y. Iron accumulation in the substantia nigra of autosomal recessive juvenile parkinsonism (ARJP). Parkinsonism Relat. Disord. 2001, 7, 311–314. [Google Scholar] [CrossRef]
- Salazar, J.; Mena, N.; Hunot, S.; Prigent, A.; Alvarez-Fischer, D.; Arredondo, M.; Duyckaerts, C.; Sazdovitch, V.; Zhao, L.; Garrick, L.M.; et al. Divalent metal transporter 1 (DMT1) contributes to neurodegeneration in animal models of Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2008, 105, 18578–18583. [Google Scholar] [CrossRef]
- Song, N.; Wang, J.; Jiang, H.; Xie, J. Ferroportin 1 but not hephaestin contributes to iron accumulation in a cell model of Parkinson’s disease. Free Radic. Biol. Med. 2010, 48, 332–341. [Google Scholar] [CrossRef]
- Zhang, Z.; Hou, L.; Song, J.L.; Song, N.; Sun, Y.J.; Lin, X.; Wang, X.L.; Zhang, F.Z.; Ge, Y.L. Pro-inflammatory cytokine-mediated ferroportin down-regulation contributes to the nigral iron accumulation in lipopolysaccharide-induced Parkinsonian models. Neuroscience 2014, 257, 20–30. [Google Scholar] [CrossRef][Green Version]
- Zhou, Z.D.; Tan, E.K. Iron regulatory protein (IRP)-iron responsive element (IRE) signaling pathway in human neurodegenerative diseases. Mol. Neurodegener. 2017, 12, 75. [Google Scholar] [CrossRef]
- Cairo, G.; Recalcati, S. Iron-regulatory proteins: Molecular biology and pathophysiological implications. Expert Rev. Mol. Med. 2007, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Giordano Attianese, G.M.; Desvergne, B. Integrative and systemic approaches for evaluating PPARβ/δ (PPARD) function. Nucl. Recept. Signal. 2015, 13, e001. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.B.; Most, D.; Blednov, Y.A.; Harris, R.A. PPAR agonists regulate brain gene expression: Relationship to their effects on ethanol consumption. Neuropharmacology 2014, 86, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.J.; Lee, E.J.; Park, J.S.; Kim, S.N.; Park, E.M.; Kim, H.S. Anti-inflammatory mechanism of galangin in lipopolysaccharide-stimulated microglia: Critical role of PPAR-γ signaling pathway. Biochem. Pharmacol. 2017, 144, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Moosecker, S.; Pissioti, A.; Leidmaa, E.; Harb, M.R.; Dioli, C.; Gassen, N.C.; Yu, S.; Gazea, M.; Catania, C.; Anderzhanova, E.; et al. Brain Expression, Physiological Regulation and Role in Motivation and Associative Learning of Peroxisome Proliferator-activated Receptor γ. Neuroscience 2021, 479, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Woods, J.W.; Tanen, M.; Figueroa, D.J.; Biswas, C.; Zycband, E.; Moller, D.E.; Austin, C.P.; Berger, J.P. Localization of PPARdelta in murine central nervous system: Expression in oligodendrocytes and neurons. Brain Res. 2003, 975, 10–21. [Google Scholar] [CrossRef]
- Iwashita, A.; Muramatsu, Y.; Yamazaki, T.; Muramoto, M.; Kita, Y.; Yamazaki, S.; Mihara, K.; Moriguchi, A.; Matsuoka, N. Neuroprotective efficacy of the peroxisome proliferator-activated receptor delta-selective agonists in vitro and in vivo. J. Pharmacol. Exp. Ther. 2007, 320, 1087–1096. [Google Scholar] [CrossRef]
- Chen, L.; Xue, L.; Zheng, J.; Tian, X.; Zhang, Y.; Tong, Q. PPARß/δ agonist alleviates NLRP3 inflammasome-mediated neuroinflammation in the MPTP mouse model of Parkinson’s disease. Behav. Brain Res. 2019, 356, 483–489. [Google Scholar] [CrossRef]
- Jin, H.; Ham, S.A.; Kim, M.Y.; Woo, I.S.; Kang, E.S.; Hwang, J.S.; Lee, K.W.; Kim, H.J.; Roh, G.S.; Lim, D.S.; et al. Activation of peroxisome proliferator-activated receptor-δ attenuates glutamate-induced neurotoxicity in HT22 mouse hippocampal cells. J. Neurosci. Res. 2012, 90, 1646–1653. [Google Scholar] [CrossRef]
- Lee, W.J.; Ham, S.A.; Yoo, H.; Hwang, J.S.; Yoo, T.; Paek, K.S.; Lim, D.S.; Han, S.G.; Lee, C.H.; Hong, K.; et al. Activation of PPARδ attenuates neurotoxicity by inhibiting lipopolysaccharide-triggered glutamate release in BV-2 microglial cells. J. Cell. Biochem. 2018, 119, 5609–5619. [Google Scholar] [CrossRef]
- Venkatesh, D.; O’Brien, N.A.; Zandkarimi, F.; Tong, D.R.; Stokes, M.E.; Dunn, D.E.; Kengmana, E.S.; Aron, A.T.; Klein, A.M.; Csuka, J.M.; et al. MDM2 and MDMX promote ferroptosis by PPARα-mediated lipid remodeling. Genes Dev. 2020, 34, 526–543. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.S.; Kim, E.; Lee, H.G.; Lee, W.J.; Won, J.P.; Hur, J.; Fujii, J.; Seo, H.G. Peroxisome proliferator-activated receptor δ rescues xCT-deficient cells from ferroptosis by targeting peroxisomes. Biomed. Pharmacother. 2021, 143, 112223. [Google Scholar] [CrossRef] [PubMed]
- Epsztejn, S.; Kakhlon, O.; Glickstein, H.; Breuer, W.; Cabantchik, I. Fluorescence analysis of the labile iron pool of mammalian cells. Anal. Biochem. 1997, 248, 31–40. [Google Scholar] [CrossRef]
- Abbasi, U.; Abbina, S.; Gill, A.; Bhagat, V.; Kizhakkedathu, J.N. A facile colorimetric method for the quantification of labile iron pool and total iron in cells and tissue specimens. Sci. Rep. 2021, 11, 6008. [Google Scholar] [CrossRef] [PubMed]
- Wojtala, A.; Bonora, M.; Malinska, D.; Pinton, P.; Duszynski, J.; Wieckowski, M.R. Methods to Monitor ROS Production by Fluorescence Microscopy and Fluorometry. Methods Enzymol. 2014, 542, 243–262. [Google Scholar] [CrossRef]
- Drummen, G.P.; van Liebergen, L.C.; Op den Kamp, J.A.; Post, J.A. C11-BODIPY(581/591), an oxidation-sensitive fluorescent lipid peroxidation probe: (micro)spectroscopic characterization and validation of methodology. Free Radic. Biol. Med. 2002, 33, 473–490. [Google Scholar] [CrossRef]
- De Winter, J.C.F. Using the Student’s t-test with extremely small sample sizes. Pract. Assess. Res. Eval. 2013, 18, 10. [Google Scholar] [CrossRef]
- Volz, K. The functional duality of iron regulatory protein 1. Curr. Opin. Struct. Biol. 2008, 18, 106–111. [Google Scholar] [CrossRef]
- Meyron-Holtz, E.G.; Ghosh, M.C.; Iwai, K.; LaVaute, T.; Brazzolotto, X.; Berger, U.V.; Land, W.; Ollivierre-Wilson, H.; Grinberg, A.; Love, P.; et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004, 23, 386–395. [Google Scholar] [CrossRef]
- Cui, R.; Choi, S.; Kim, T.H.; Lee, H.J.; Lee, S.J.; Kang, Y.; Jeon, J.Y.; Kim, H.J.; Lee, K. Iron overload by transferrin receptor protein 1 regulation plays an important role in palmitate-induced insulin resistance in human skeletal muscle cells. FASEB J. 2019, 33, 1771–1786. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Doll, S.; Freitas, F.P.; Shah, R.; Aldrovandi, M.; da Silva, M.C.; Ingold, I.; Goya Grocin, A.; Xavier da Silva, T.N.; Panzilius, E.; Scheel, C.H.; et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 2019, 575, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.; Farioli-Vecchioli, S.; Cerù, M.P. Immunolocalization of peroxisome proliferator-activated receptors and retinoid X receptors in the adult rat CNS. Neuroscience 2004, 123, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Xie, Y.; Kang, R.; Hou, W.; Sun, X.; Epperly, M.W.; Greenberger, J.S.; Tang, D. FANCD2 protects against bone marrow injury from ferroptosis. Biochem. Biophys. Res. Commun. 2016, 480, 443–449. [Google Scholar] [CrossRef]
- Beck, G.; Shinzawa, K.; Hayakawa, H.; Baba, K.; Yasuda, T.; Sumi-Akamaru, H.; Tsujimoto, Y.; Mochizuki, H. Deficiency of Calcium-Independent Phospholipase A2 Beta Induces Brain Iron Accumulation through Upregulation of Divalent Metal Transporter 1. PLoS ONE 2015, 10, e0141629. [Google Scholar] [CrossRef]
- Wang, W.; Song, N.; Zhang, H.; Xie, J.; Wang, J. 6-Hydroxydopamine upregulates iron regulatory protein 1 by activating certain protein kinase C isoforms in the dopaminergic MES23.5 cell line. Int. J. Biochem. Cell Biol. 2012, 44, 1987–1992. [Google Scholar] [CrossRef]
- Das, N.R.; Gangwal, R.P.; Damre, M.V.; Sangamwar, A.T.; Sharma, S.S. A PPAR-β/δ agonist is neuroprotective and decreases cognitive impairment in a rodent model of Parkinson’s disease. Curr. Neurovasc. Res. 2014, 11, 114–124. [Google Scholar] [CrossRef]
- Bosco, D.A.; Fowler, D.M.; Zhang, Q.; Nieva, J.; Powers, E.T.; Wentworth, P., Jr.; Lerner, R.A.; Kelly, J.W. Elevated levels of oxidized cholesterol metabolites in Lewy body disease brains accelerate alpha-synuclein fibrilization. Nat. Chem. Biol. 2006, 2, 249–253. [Google Scholar] [CrossRef]
- Medeiros, M.S.; Schumacher-Schuh, A.; Cardoso, A.M.; Bochi, G.V.; Baldissarelli, J.; Kegler, A.; Santana, D.; Chaves, C.M.; Schetinger, M.R.; Moresco, R.N.; et al. Iron and Oxidative Stress in Parkinson’s Disease: An Observational Study of Injury Biomarkers. PLoS ONE 2016, 11, e0146129. [Google Scholar] [CrossRef]
- Do Van, B.; Gouel, F.; Jonneaux, A.; Timmerman, K.; Gelé, P.; Pétrault, M.; Bastide, M.; Laloux, C.; Moreau, C.; Bordet, R.; et al. Ferroptosis, a newly characterized form of cell death in Parkinson’s disease that is regulated by PKC. Neurobiol. Dis. 2016, 94, 169–178. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, P.; Zhai, B.; Zhang, M.; Xiang, Y.; Fang, J.; Xu, S.; Gao, Y.; Chen, X.; Sui, X.; et al. The emerging role of ferroptosis in inflammation. Biomed. Pharmacother. 2020, 127, 110108. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Lu, J.; Hao, X.; Li, H.; Zhang, G.; Liu, X.; Li, X.; Zhao, C.; Kuang, W.; Chen, D.; et al. FTH1 Inhibits Ferroptosis Through Ferritinophagy in the 6-OHDA Model of Parkinson’s Disease. Neurotherapeutics 2020, 17, 1796–1812. [Google Scholar] [CrossRef] [PubMed]
- Beal, M.F.; Oakes, D.; Shoulson, I.; Henchcliffe, C.; Galpern, W.R.; Haas, R.; Juncos, J.L.; Nutt, J.G.; Voss, T.S.; Ravina, B.; et al. A randomized clinical trial of high-dosage coenzyme Q10 in early Parkinson disease: No evidence of benefit. JAMA Neurol. 2014, 71, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Parkinson Study Group. Mortality in DATATOP: A multicenter trial in early Parkinson’s disease. Ann. Neurol. 1998, 43, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Writing Group for the NINDS Exploratory Trials in Parkinson Disease (NET-PD) Investigators; Kieburtz, K.; Tilley, B.C.; Elm, J.J.; Babcock, D.; Hauser, R.; Ross, G.W.; Augustine, A.H.; Augustine, E.U.; Aminoff, M.J.; et al. Effect of creatine monohydrate on clinical progression in patients with Parkinson disease: A randomized clinical trial. JAMA 2015, 313, 584–593. [Google Scholar] [CrossRef]
- Devos, D.; Moreau, C.; Devedjian, J.C.; Kluza, J.; Petrault, M.; Laloux, C.; Jonneaux, A.; Ryckewaert, G.; Garçon, G.; Rouaix, N.; et al. Targeting chelatable iron as a therapeutic modality in Parkinson’s disease. Antioxid. Redox Signal. 2014, 21, 195–210. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, W.J.; Lee, H.G.; Hur, J.; Lee, G.H.; Won, J.P.; Kim, E.; Hwang, J.S.; Seo, H.G. PPARδ Activation Mitigates 6-OHDA-Induced Neuronal Damage by Regulating Intracellular Iron Levels. Antioxidants 2022, 11, 810. https://doi.org/10.3390/antiox11050810
Lee WJ, Lee HG, Hur J, Lee GH, Won JP, Kim E, Hwang JS, Seo HG. PPARδ Activation Mitigates 6-OHDA-Induced Neuronal Damage by Regulating Intracellular Iron Levels. Antioxidants. 2022; 11(5):810. https://doi.org/10.3390/antiox11050810
Chicago/Turabian StyleLee, Won Jin, Hyuk Gyoon Lee, Jinwoo Hur, Gyeong Hee Lee, Jun Pil Won, Eunsu Kim, Jung Seok Hwang, and Han Geuk Seo. 2022. "PPARδ Activation Mitigates 6-OHDA-Induced Neuronal Damage by Regulating Intracellular Iron Levels" Antioxidants 11, no. 5: 810. https://doi.org/10.3390/antiox11050810
APA StyleLee, W. J., Lee, H. G., Hur, J., Lee, G. H., Won, J. P., Kim, E., Hwang, J. S., & Seo, H. G. (2022). PPARδ Activation Mitigates 6-OHDA-Induced Neuronal Damage by Regulating Intracellular Iron Levels. Antioxidants, 11(5), 810. https://doi.org/10.3390/antiox11050810






