Active Transposition of Insertion Sequences in Prokaryotes: Insights from the Response of Deinococcus geothermalis to Oxidative Stress
Abstract
1. Introduction
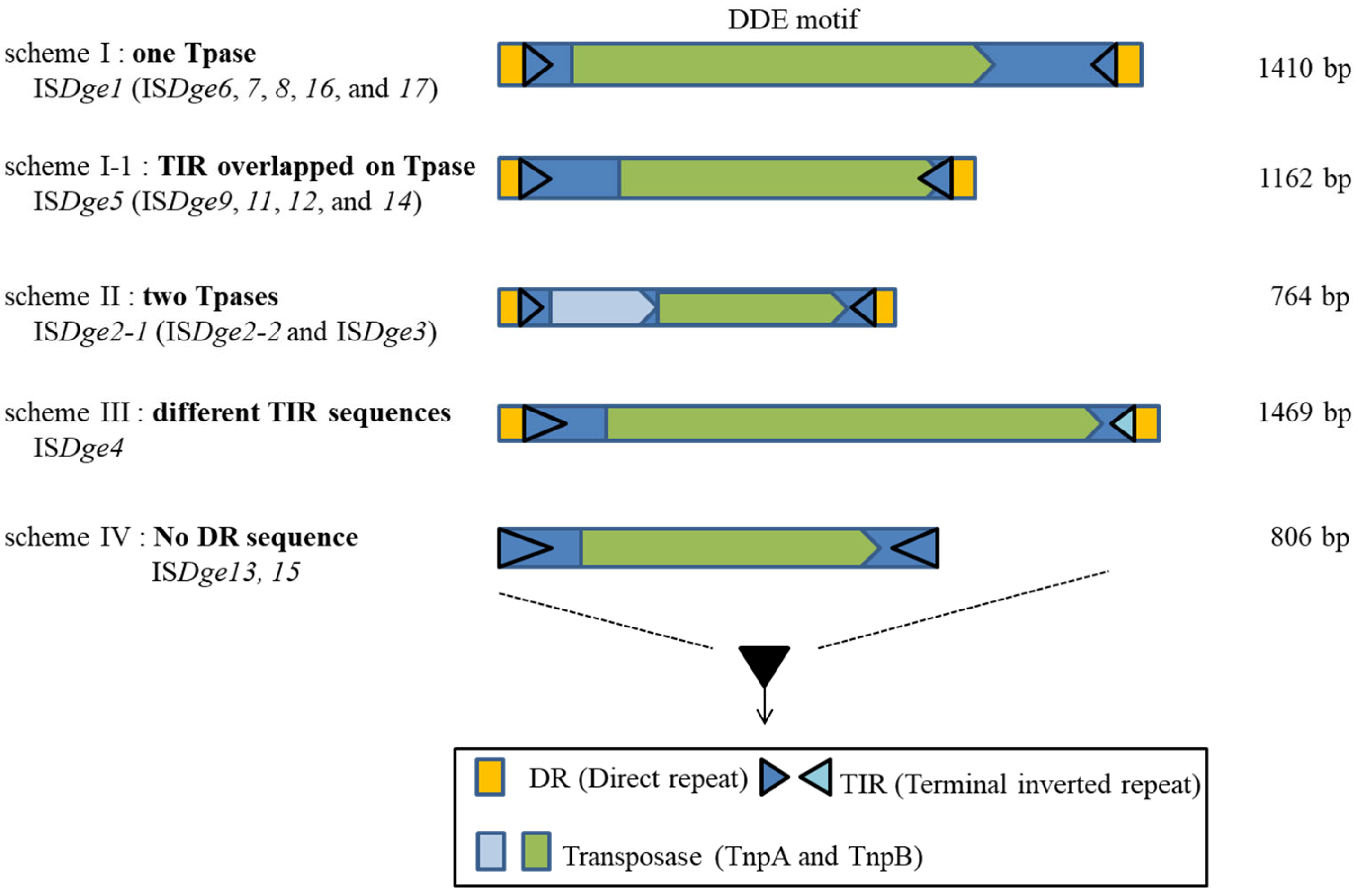
2. Structural Properties of IS Elements and Functional Aspects
3. IS Transpositional Procedures and Trigger Factors
3.1. IS Transposition in Deinococcus-Thermus
3.2. IS Transposition in Gram-Negative Bacteria
3.3. IS Transposition in Gram-Positive Bacteria
3.4. IS Transposition in Archaea
3.5. Use of Transposon Mutagenesis
4. Inductive Signals of Active Transposition
4.1. Host Factors
4.2. Nutrition and Temperature
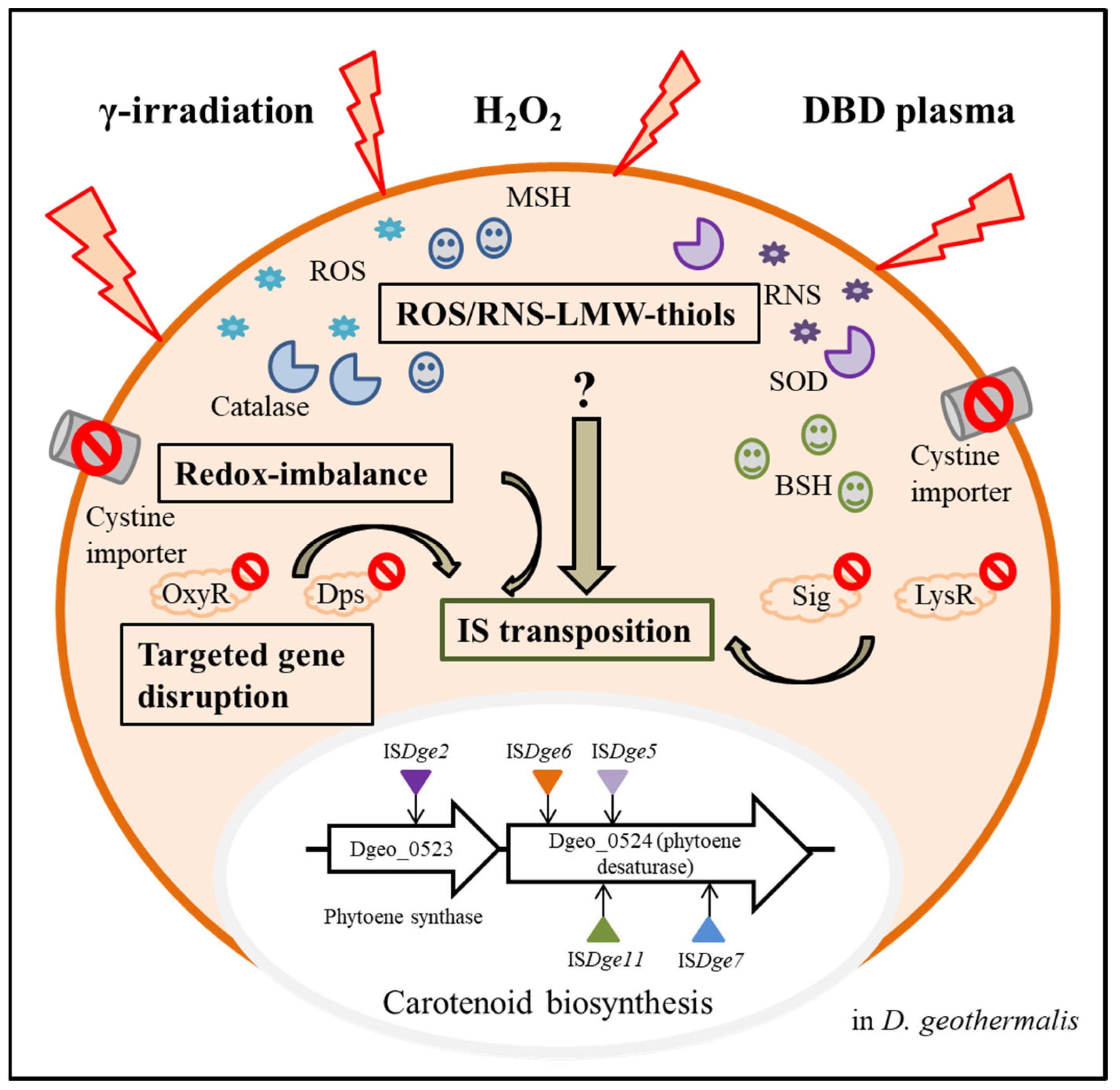
4.3. Gamma-Irradiation and Dielectric Bilayer Discharge Plasma
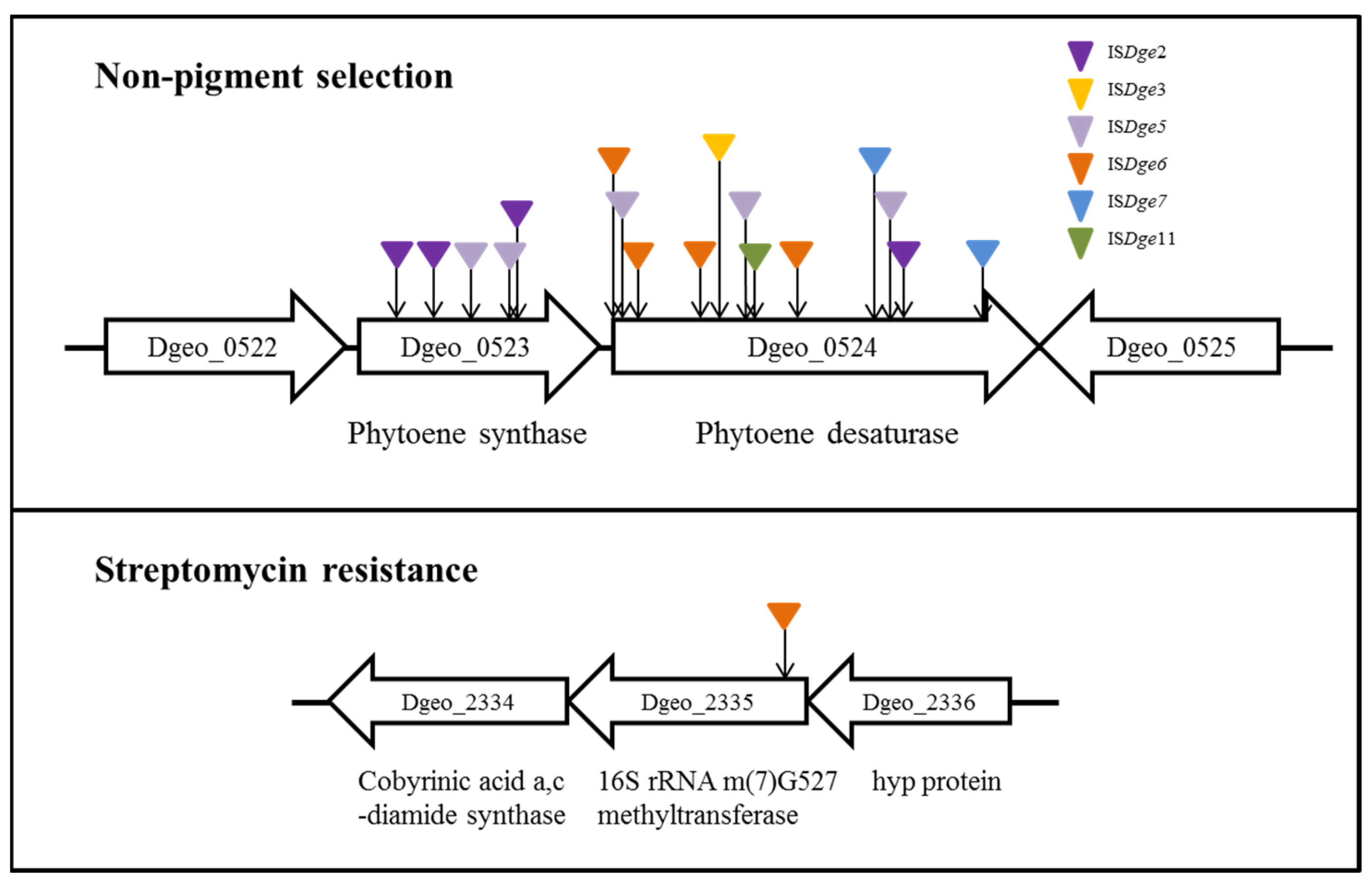
4.4. Redox Imbalance
4.5. Antibiotics
4.6. Metals
5. Redox-Switched Regulators and Redox Signalling
6. The Evolution of Prokaryotic Genomes via IS Elements
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviation
| IS | insertion sequence |
| Tpase (Tnp) | transposase |
| InsA; InsB’ | two ORFs in IS element and in some cases both genes were produced a fused protein (InsAB’) or InsA regulated InsB’ expression |
| TIR | terminal inverted repeat |
| DR | direct repeat |
| Dps | DNA-protection protein from starved cell |
| H-NS | histone-like nucleoid structuring protein |
| LysR | a broad transcriptional regulator family |
| DBD | dielectric bilayer discharge |
| DdrO-IrrE | Deinococcus unique regulatory system for RDR regulon |
| RDR | radiation-desiccation responded regulon |
| ROS | reactive oxygen species |
| SIDD | stress-induced DNA destabilisation region |
References
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Gourbeyre, E.; Varani, A.; Ton-Hoang, B.; Chandler, M. Everyman’s guide to bacterial insertion sequences. Microbiol. Spectrum 2015, 3, 555–590. [Google Scholar] [CrossRef] [PubMed]
- Blesa, A.; Sanchez, M.; Sacristan-Horcajada, E.; Fuente, S.G.; Peiro, R.; Berenguer, J. Into the Thermus mobilome: Presence, diversity and recent activities of insertion sequences across Thermus spp. Microorganisms 2019, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Fayad, N.; Awad, M.K.; Mahillon, J. Diversity of Bacillus cereus sensu lato mobilome. BMC Genom. 2019, 20, 436. [Google Scholar] [CrossRef]
- Durrant, M.G.; Li, M.M.; Siranosian, B.A.; Montgomery, S.B.; Bhatt, A.S. A bioinformatic analysis of integrative mobile genetic elements highlights their role in bacterial adaptation. Cell Host Microbe 2020, 27, 140–153. [Google Scholar] [CrossRef]
- Lee, C.; Bae, M.K.; Choi, N.; Lee, S.J.; Lee, S.-J. Genome plasticity by insertion sequences learned from a case of radiation-resistant bacterium Deinococcus geothermalis. Bioinformat. Biol. Insights 2021, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hawkey, J.; Hamidian, M.; Wick, R.R.; Edwards, D.J.; Billman-Jacobe, H.; Hall, R.M.; Holt, K.E. ISMapper: Identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genom. 2015, 16, 667. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Gauthier, D.T.; Ranjan, D.; Zubair, M. ISQuest: Finding insertion sequences in prokaryotics sequence fragment data. Bioinformatics 2015, 31, 3406–3412. [Google Scholar] [CrossRef]
- Robinson, D.G.; Lee, M.-C.; Marx, C.J. Oasis: An automatic program for global investigation of bacterial and archaeal insertion sequences. Nucleic Acid Res. 2012, 40, e174. [Google Scholar] [CrossRef] [PubMed]
- Hickman, A.B.; Dyda, F. Mechanisms of DNA transposition. Microbiol. Spectrum 2015, 3, 3.2.12. [Google Scholar] [CrossRef] [PubMed]
- Hickman, A.B.; Dyda, F. DNA transposition at work. Chem. Rev. 2016, 116, 12758–12784. [Google Scholar] [CrossRef] [PubMed]
- Machida, C.; Machida, Y. Regulation of transposition by the insA gene product. J. Mol. Biol. 1989, 208, 567–574. [Google Scholar] [CrossRef]
- Ton-Hoang, B.; Turlan, C.; Chandler, M. Functional domains of the IS1 transposase: Analysis in vivo and in vitro. Mol. Microbiol. 2004, 53, 1529–1543. [Google Scholar] [CrossRef] [PubMed]
- Mahillon, J.; Chandler, M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998, 62, 725–774. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef] [PubMed]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; Van Houdt, R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef] [PubMed]
- Barabas, O.; Ronning, D.R.; Guynet, C.; Hickman, A.B.; Ton-Hoang, B.; Chandler, M.; Dyda, F. Mechanism of IS200/IS605 family DNA transposases: Activation and transposon-directed target site selection. Cell 2008, 132, 208–220. [Google Scholar] [CrossRef]
- Hickman, A.B.; James, J.A.; Barabas, O.; Pasternak, C.; Ton-Hoang, B.; Chandler, M.; Sommer, S.; Dyda, F. DNA recognition and the precleavage state during single-stranded DNA transposition in D. radiodurans. EMBO J. 2010, 29, 3840–3852. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Corneloup, A.; Guynet, C.; Lavatine, L.; Caumont-Sarcos, A.; Siguier, P.; Marty, B.; Dyda, F.; Chandler, M.; Ton Hoang, B. The IS200/IS605 family and “peel and paste” single-strand transposition mechanism. Microbiol. Spectrum 2015, 3, 3.4.02. [Google Scholar] [CrossRef] [PubMed]
- Lavatine, L.; He, S.; Caumont-Sarcos, A.; Guynet, C.; Marty, B.; Chandler, M.; Ton-Hoang, B. Single strand transposition at the host replication fork. Nucleic Acids Res. 2016, 44, 7866–7883. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Known knowns, known unknowns and unknown unknowns in prokaryotic transposition. Curr. Opin. Microbiol. 2017, 38, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Harmer, C.J.; Hall, R.M. An analysis of the IS6/IS26 family of insertion sequences: Is it a single family? Microb. Genom. 2019, 5, e000291. [Google Scholar] [CrossRef] [PubMed]
- Varani, A.; He, S.; Siguier, P.; Ross, K.; Chandler, M. The IS6 family, a clinically important group of insertion sequences including IS26. Mob. DNA 2021, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Choi, N.; Bae, M.K.; Choo, K.; Lee, S.-J. Transposition of insertion sequences was triggered by oxidative stress in radiation-resistant bacterium Deinococcus geothermalis. Microorganisms 2019, 7, 446. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Choo, K.; Lee, S.-J. Active transposition of insertion sequences by oxidative stress in Deinococcus geothermalis. Front. Microbiol. 2020, 11, 558747. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Lee, C.; Shin, E.; Lee, S.-J. Influence of redox imbalances on the transposition of insertion sequences in Deinococcus geothermalis. Antioxidants 2021, 10, 1623. [Google Scholar] [CrossRef]
- Pasternak, C.; Dulermo, R.; Ton-Hoang, B.; Debuchy, R.; Siguier, P.; Coste, G.; Chandler, M.; Sommer, S. ISDra2 transpositionin Deinococcus radiodurans is downregulated by TnpB. Mol. Microbiol. 2013, 88, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Quentin, Y.; Siguier, P.; Chandler, M.; Fichant, G. Single-strand DNA processing: Phylogenomics and sequence diversity of a superfamily of potential prokaryotic HuH endonuclease. BMC Genom. 2018, 19, 475. [Google Scholar] [CrossRef] [PubMed]
- Gregory, S.T.; Dahlberg, A.E. Transposition of an insertion sequence, ISTth7, in the genome of the extreme thermophile Thermus thermophilus HB8. FEMS Lett. 2008, 289, 187–192. [Google Scholar] [CrossRef]
- Mennecier, S.; Servant, P.; Coste, G.; Bailone, A.; Sommer, S. Mutagenesis via IS transposition in Deinococcus radiodurans. Mol. Microbiol. 2006, 59, 317–325. [Google Scholar] [CrossRef]
- Lee, C.; Bae, M.K.; Lee, S.-J. An antioxidant defense system in radiation-resistant bacterium Deinococcus geothermalis against oxidative stress. In Antioxidants: Benefits, Sources, Mechanisms of Action; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Chandler, M.; Fayet, O.; Rousseau, P.; Ton-Hoang, B.; Duval-Valentin, G. Copy-out-paste-in transposition of IS911: A major transposition pathway. Miocrobiol. Spectrum 2015, 3, 3.4.01. [Google Scholar]
- Haren, L.; Betermier, M.; Polard, P.; Chandler, M. IS911-mediated intramolecular transposition is naturally temperature sensitive. Mol. Microbiol. 1997, 25, 531–540. [Google Scholar] [CrossRef]
- Ohtsubo, Y.; Genka, H.; Komatsu, H.; Nagata, Y.; Tsuda, M. High-temperature-induced transposition of insertion elements in Burkholderia multivorans ATCC17616. Appl. Environ. Microbiol. 2005, 71, 1822–1828. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Christie-Oleza, J.A.; Lanfranconi, M.P.; Nogales, B.; Lalucat, J.; Bosch, R. Conjugative interaction induces transposition of ISPst9 in Pseudomonas stutzeri AN10. J. Bacteriol. 2009, 191, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Drevinek, P.; Baldwin, A.; Lindenburg, L.; Joshi, L.T.; Marchbank, A.; Vosahlikova, S.; Dowson, C.G.; Mahenthiralingam, E. Oxidative stress of Burkholderia cenocepacia induces insertion sequence-mediated genomic rearrangements that interfere with macrorestriction-based genotyping. J. Clin. Microbiol. 2010, 48, 34–40. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Suzuki, H.; Taketani, T.; Tanabiki, M.; Ohara, M.; Kobayashi, J.; Ohshiro, T. Frequent transposition of multiple insertion sequences in Geobacillus kaustophilus HTA426. Front. Microbiol. 2021, 12, 650461. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Sekine, Y.; Chibazakura, T.; Yoshikawa, H. Development of an intermolecular transposition assay system in Bacillus subtilis 168 using IS4Bsu1 from Bacillus subtilis (natto). Microbiology 2007, 153, 2553–2559. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Filée, J.; Siguier, P.; Chandler, M. Insertion sequence diversity in archaea. Micribiol. Mol. Biol. Rev. 2007, 71, 121–157. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, F.; Blaseio, U. Transposition burst of the ISH27 insertion element family in Halobacterium halobium. Nucleic Acids Res. 1990, 18, 6921–6925. [Google Scholar] [CrossRef] [PubMed]
- Zivanovic, Y.; Lopez, P.; Philippe, H.; Forterre, P. Pyrococcus genome comparison evidences chromosome shuffling-driven evolution. Nucleic Acids Res. 2002, 30, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Sakanaka, M.; Fukiya, S.; Kobayashi, R.; Abe, A.; Hirayama, Y.; Kano, Y.; Yokota, A. Isolation and transposition properties of ISBlo11, an active insertion sequence belonging to the IS3 family, from Bifidobacterium longum 105-A. FEMS Lett. 2015, 362, fnv032. [Google Scholar] [CrossRef] [PubMed]
- Sakanaka, M.; Nakakawaji, S.; Nakajima, S.; Fukiya, S.; Abe, A.; Saburi, W.; Mori, H.; Yokota, A. A transposon mutagenesis system for Bifidobacterium longum subsp. longum based on an IS3 family insertion sequence, ISBlo11. Appl. Environ. Microbiol. 2018, 84, e00824-18. [Google Scholar]
- Nagy, Z.; Chandler, M. Regulation of transposition in bacteria. Res. Microbiol. 2004, 155, 387–398. [Google Scholar] [CrossRef]
- Swingle, B.; O’Carroll, M.; Haniford, D.; Derbyshire, K.M. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. Mol. Microbiol. 2004, 52, 1055–1067. [Google Scholar] [CrossRef] [PubMed]
- Shiga, Y.; Sekina, Y.; Kano, Y.; Ohtsubo, E. Involvment of H-NS in transpositional recombination mediated by IS1. J. Bacteriol. 2001, 183, 2476–2484. [Google Scholar] [CrossRef]
- Humayun, M.Z.; Zhang, Z.; Butcher, A.M.; Moshayedi, A.; Saier, M.H., Jr. Hopping into a host seat: Role of DNA structural features on IS5-mediated gene activation and inactivation under stress. PLoS ONE 2017, 12, e0180156. [Google Scholar] [CrossRef]
- Rouquette, C.; Serre, M.-C.; Lane, D. Protective role for H-NS protein in IS1 transposition. J. Bacteriol. 2004, 186, 2091–2098. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.; Mahillon, J. Insertion sequences revisited. In Mobile DNA; ASM Press: Washington, DC, USA, 2002; Volume II, pp. 305–366. [Google Scholar]
- Coros, A.M.; Twiss, E.; Tavakoli, N.P.; Derbyshire, K.M. Genetic Evidence that GTP Is Required for Transposition of IS903 and Tn552 in Escherichia coli. J. Bacteriol. 2005, 187, 4598–4606. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Twiss, E.; Coros, A.M.; Tavakoli, N.P.; Derbyshire, K.M. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Mol. Microbiol. 2005, 57, 1593–1607. [Google Scholar] [CrossRef]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef]
- Sobrero, P.; Valverde, C. The bacterial protein Hfq: Much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 2012, 38, 276–299. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.J.; Trussler, R.S.; Haniford, D.B. Hfq binds directly to the ribosome-binding site of IS10 transposase mRNA to inhibit translation. Mol. Microbiol. 2015, 96, 633–650. [Google Scholar] [CrossRef] [PubMed]
- Zerbib, D.; Polard, P.; Escoubas, J.M.; Galas, D.; Chandler, M. The regulatory role of the IS1-encoded InsA protein in transposition. Mol. Microbiol. 1990, 4, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.T.; Hwang, J.H.; Lee, L.C.; Lee, C.H.; Li, P.L.; Hsieh, Y.C. Functional analysis of the 14 kDa protein of insertion sequence 2. J. Mol. Biol. 1994, 236, 503–513. [Google Scholar] [CrossRef]
- Kleckner, N.; Chalmers, R.M.; Kwon, D.; Sakai, J.; Bolland, S. Tn10 and IS10 transposition and chromosome rearrangements: Mechanism and regulation in vivo and in vitro. In Transposable Elements; Springer: Berlin/Heidelberg, Germany, 1996; pp. 49–82. [Google Scholar]
- Escoubas, J.M.; Prere, M.F.; Fayet, O.; Salvignol, I.; Galas, D.; Zerbib, D.; Chandler, M. Translational control of transposition activity of the bacterial insertion sequence IS1. EMBO J. 1991, 10, 705–712. [Google Scholar] [CrossRef]
- Beuzon, C.R.; Marques, S.; Casadesus, J. Repression of IS200 transposase synthesis by RNA secondary structures. Nucleic Acids Res. 1999, 27, 3690–3695. [Google Scholar] [CrossRef][Green Version]
- Roberts, D.; Hoopes, B.C.; McClure, W.R.; Kleckner, N. IS10 transposition is regulated by DNA adenine methylation. Cell 1990, 43, 117–130. [Google Scholar] [CrossRef]
- Derbyshire, K.M.; Kramer, M.; Grindley, N.D. Role of instability in the cis action of the insertion sequence IS903 transposase. Proc. Natl. Acad. Sci. USA 1990, 87, 4048–4052. [Google Scholar] [CrossRef]
- Olasz, F.; Kiss, J.; Konig, P.; Buzas, Z.; Stalder, R.; Arber, W. Target specificity of insertion element IS30. Mol. Microbiol. 1998, 28, 691–704. [Google Scholar] [CrossRef]
- Kiss, J.; Nagy, Z.; Toth, G.; Kiss, G.B.; Jakab, J.; Chandler, M.; Olasz, F. Transposition and target specificity of the typical IS30 family element IS1655 from Neisseria meningitidis. Mol. Microbiol. 2007, 63, 1731–1747. [Google Scholar] [CrossRef]
- Kharat, A.; Coursange, E.; Noirclerc-Savoye, M.; Lacoste, J.; Blot, M. IS1 transposition is enhanced by translation errors and by bacterial growth at extreme glucose levels. Acta Biochim. Pol. 2006, 53, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Reif, H.J.; Saedler, H. IS1 is involved in deletion formation in the gal region of E. coli K12. Mol. Gen. Genet. 1975, 137, 17–28. [Google Scholar] [CrossRef]
- Pasternak, C.; Ton-Hoang, B.; Coste, G.; Bailone, A.; Chandler, M.; Sommer, S. Irradiation-induced Deinococcus radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet. 2010, 6, e1000799. [Google Scholar] [CrossRef] [PubMed]
- Narumi, I.; Cherdchu, K.; Kitayama, S.; Watanabe, H. The Deinococcus radiodurans uvrA gene: Identification of mutation sites in two mitomycin-sensitive strains and the first discovery of insertion sequence element from deinobacteria. Gene 1997, 198, 115–126. [Google Scholar] [CrossRef]
- Demirci, H.; Murphy, I.V.F.; Murphy, E.; Gregory, S.T.; Dahlberg, A.E.; Jogl, G. A structural basis for Sm-induced misreading of the genetic code. Nat. Commun. 2013, 4, 1355. [Google Scholar] [CrossRef] [PubMed]
- Carr, J.F.; Hamburg, D.-M.; Gregory, S.T.; Limbach, P.A.; Dahlberg, A.E. Effects of streptomycin resistance mutations on posttranslational modification of ribosomal protein S12. J. Bacteriol. 2006, 188, 2020–2023. [Google Scholar] [CrossRef]
- Okamoto, S.; Tamaru, A.; Nakajima, C.; Nishimura, K.; Tanaka, Y.; Tokuyama, S.; Suzuki, Y.; Ochi, K. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 2007, 63, 1096–1106. [Google Scholar] [CrossRef]
- Paine, T.F.; Finland, M. Streptomycin-sensitive, -dependent, and -resistant bacteria. Science 1948, 107, 143–144. [Google Scholar] [CrossRef]
- Gregory, S.T.; Cate, J.H.; Dahlberg, A.E. Streptomycin-resistant and streptomycin-dependent mutants of the extreme thermophile Thermus thermophilus. J. Mol. Biol. 2001, 309, 333–338. [Google Scholar] [CrossRef]
- Mijnendonckx, K.; Provoost, A.; Monsieurs, P.; Leys, N.; Mergeay, M.; Mahillon, J.; Van Houdt, R. Insertion sequence elements in Cupriavidus metallidurans CH34: Distribution and role in adaptation. Plasmid 2011, 65, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Vandecraen, J.; Monsieurs, P.; Mergeay, M.; Leys, N.; Aertsen, A.; Van Houdt, R. Zinc-induced transposition of insertion sequence elements contributes to increased adaptability of Cupriavidus metallidurans. Front. Microbiol. 2016, 7, 359. [Google Scholar] [CrossRef]
- Eugénie, N.; Zivanovic, Y.; Lelandais, G.; Coste, G.; Bouthier de la Tour, C.; Bentchikou, E.; Servant, P.; Confalonieri, F. Characterization of the radiation desiccation response regulon of the radioresistant bacterium Deinococus radiodurans by integrative genomic analyses. Cells 2021, 10, 2536. [Google Scholar] [CrossRef] [PubMed]
- Ludanyi, M.; Blanchard, L.; Dulermo, R.; Brandelet, G.; Bellanger, L.; Pignol, D.; Lemaire, D.; de Groot, A. Radiation response in Deinococcus deserti: IrrE is a metalloprotease that cleaves repressor protein DdrO. Mol. Microbiol. 2014, 94, 434–449. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, L.; Guerin, P.; Roche, D.; Cruveiller, S.; Pignol, D.; Vallenet, D.; Armengaud, J.; de Groot, A. Conservation and diversity of the IrrE/DdrO-controlled radiation response in radiation-resistant Deinococcus bacteria. MicrobiologyOpen 2017, 6, e477. [Google Scholar] [CrossRef] [PubMed]
- Magerand, R.; Rey, P.; Blanchard, L.; de Groot, A. Redox signaling through zinc activates the radiation response in Deinococcus bacteria. Sci. Rep. 2021, 11, 4528. [Google Scholar] [CrossRef] [PubMed]
- Antelmann, H.; Helmann, J.D. Thiol-based redox switches and gene regulation. Antioxid. Redox Signal. 2011, 14, 1049–1063. [Google Scholar] [CrossRef]
- Hillion, M.; Antelmann, H. Thiol-based redox switches in prokaryotes. Biol. Chem. 2015, 396, 415–444. [Google Scholar] [CrossRef]
- Sevilla, E.; Bes, M.T.; González, A.; Peleato, M.L.; Fillat, M.F. Redox-based transcriptional regulation in prokaryotes: Revisiting model mechanisms. Antioxid. Redox Signal. 2019, 30, 1651–1696. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Jung, J.-H.; Blanchard, L.; de Groot, A. Conservation and diversity of radiation and oxidative stress resistance mechanisms in Deinococcus species. FEMS Microbiol. Rev. 2019, 43, 19–52. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhou, Z.; Chen, X.; Zhang, W.; Lin, M.; Chen, M. Comparative proteomics analysis reveals new features of the oxidative stress response in the polyextremophilic bacterium Deinococcus radiodurans. Microorganisms 2020, 8, 451. [Google Scholar] [CrossRef]
- Orsi, R.H.; Bowen, B.M.; Wiedmann, M. Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC Genom. 2010, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Fahrner, K.A.; Berg, H.C. Mutations that stimulate flhDC expression in Escherichia coli K-12. J. Bacteriol. 2015, 197, 3087–3096. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Kukita, C.; Humayun, M.Z.; Saier, M.H., Jr. Environmental-directed activation of the Escherichia coli flhDC operon by transposons. Microbiology 2017, 163, 554–569. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yen, M.R.; Saier, M.H., Jr. Precise excision of IS5 from the intergenic region between the fucPIK and the fucAO operons and mutational control of fucPIK operon expression in Escherichia coli. J. Bacteriol. 2010, 192, 2013–2019. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, E.; Ye, Q.; Lee, S.-J. Active Transposition of Insertion Sequences in Prokaryotes: Insights from the Response of Deinococcus geothermalis to Oxidative Stress. Antioxidants 2022, 11, 481. https://doi.org/10.3390/antiox11030481
Shin E, Ye Q, Lee S-J. Active Transposition of Insertion Sequences in Prokaryotes: Insights from the Response of Deinococcus geothermalis to Oxidative Stress. Antioxidants. 2022; 11(3):481. https://doi.org/10.3390/antiox11030481
Chicago/Turabian StyleShin, Eunjung, Qianying Ye, and Sung-Jae Lee. 2022. "Active Transposition of Insertion Sequences in Prokaryotes: Insights from the Response of Deinococcus geothermalis to Oxidative Stress" Antioxidants 11, no. 3: 481. https://doi.org/10.3390/antiox11030481
APA StyleShin, E., Ye, Q., & Lee, S.-J. (2022). Active Transposition of Insertion Sequences in Prokaryotes: Insights from the Response of Deinococcus geothermalis to Oxidative Stress. Antioxidants, 11(3), 481. https://doi.org/10.3390/antiox11030481





