Development of an In Vitro Model of SARS-CoV-Induced Acute Lung Injury for Studying New Therapeutic Approaches
Abstract
1. Introduction
2. Materials and Methods
2.1. A549 Cultivation
2.2. Blood Sampling and Peripheral Blood Mononuclear Cells (PBMC) Isolation
2.3. Real-Time Cell Proliferation Monitoring
2.4. MSC Isolation and Culturing
2.5. Isolation of Extracellular Vesicles
2.6. Inflammation Modeling In Vitro
2.7. Cell Viability Estimation
2.8. Cytokine Multiplex Assay
2.9. ROS and Lipid Peroxidation Detection
2.10. Western Blot Analysis
2.11. Statistics
3. Results
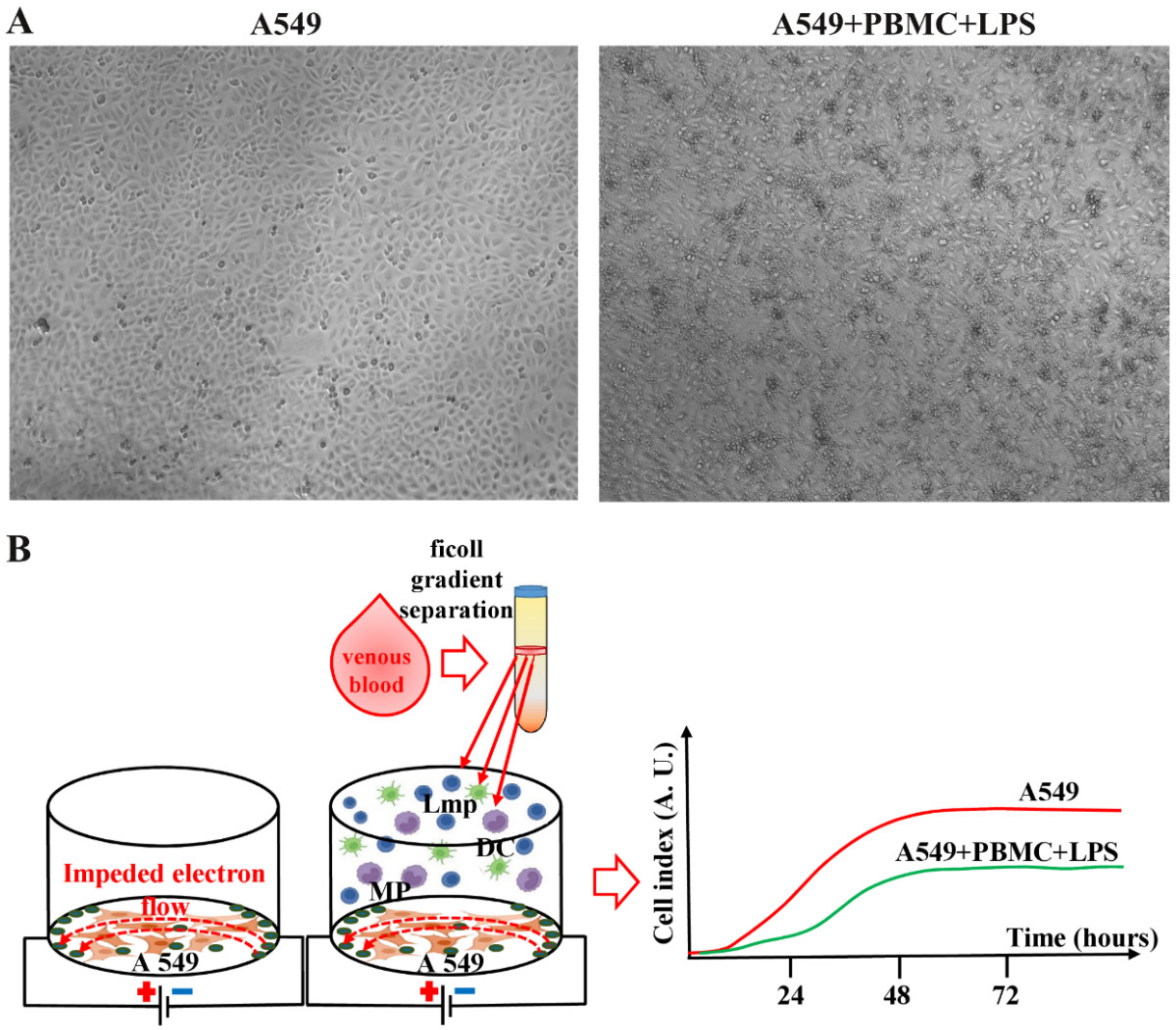
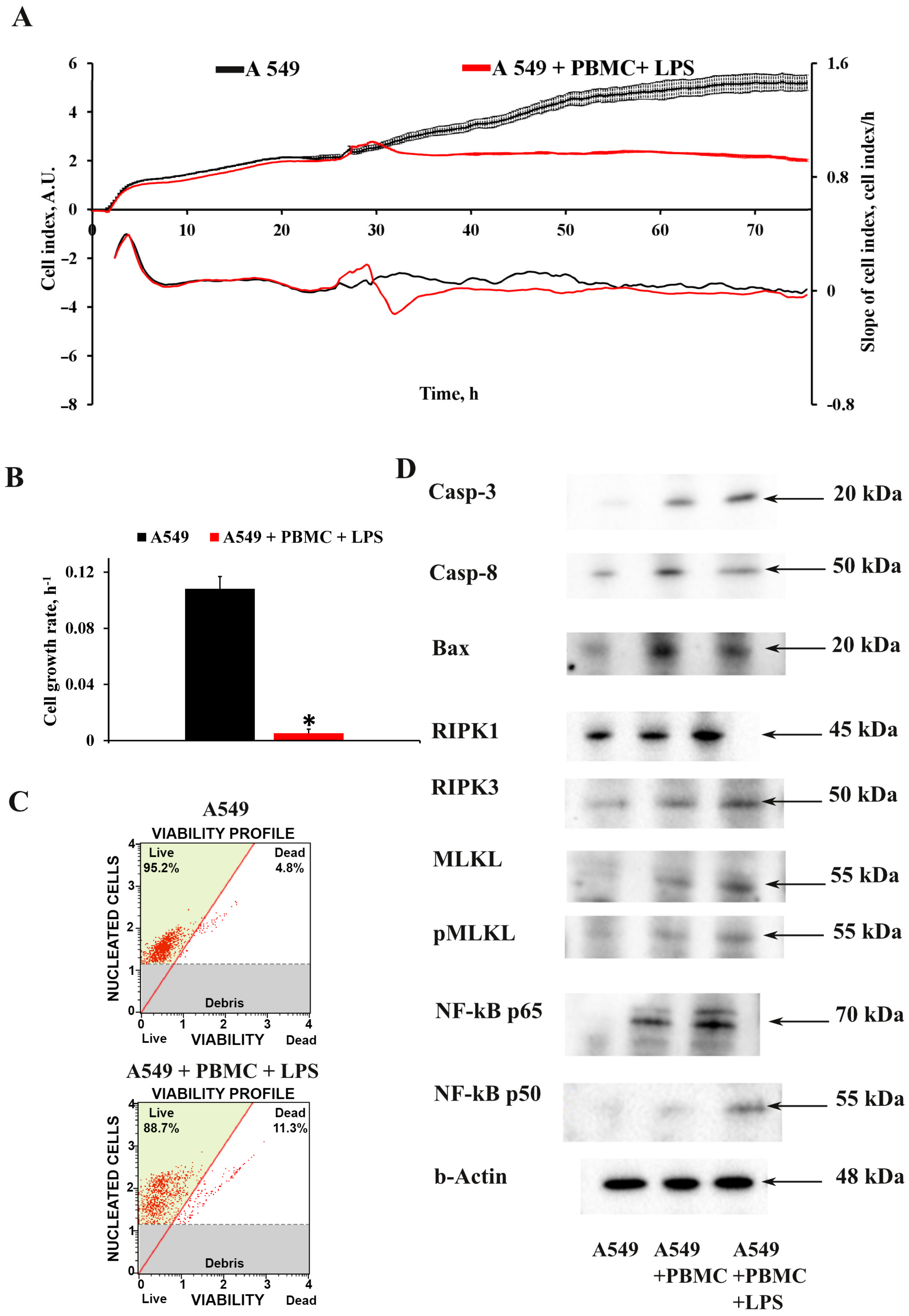
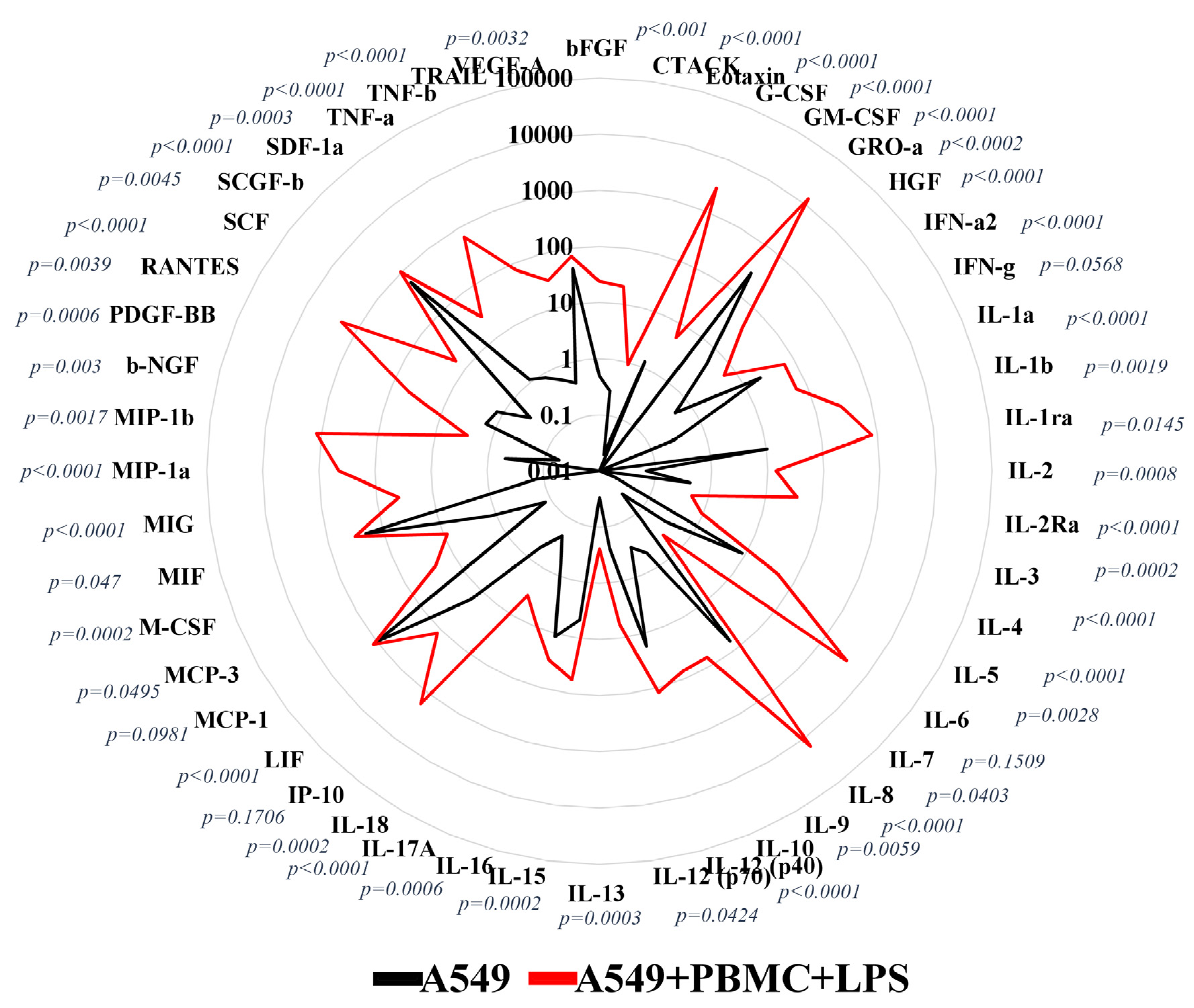
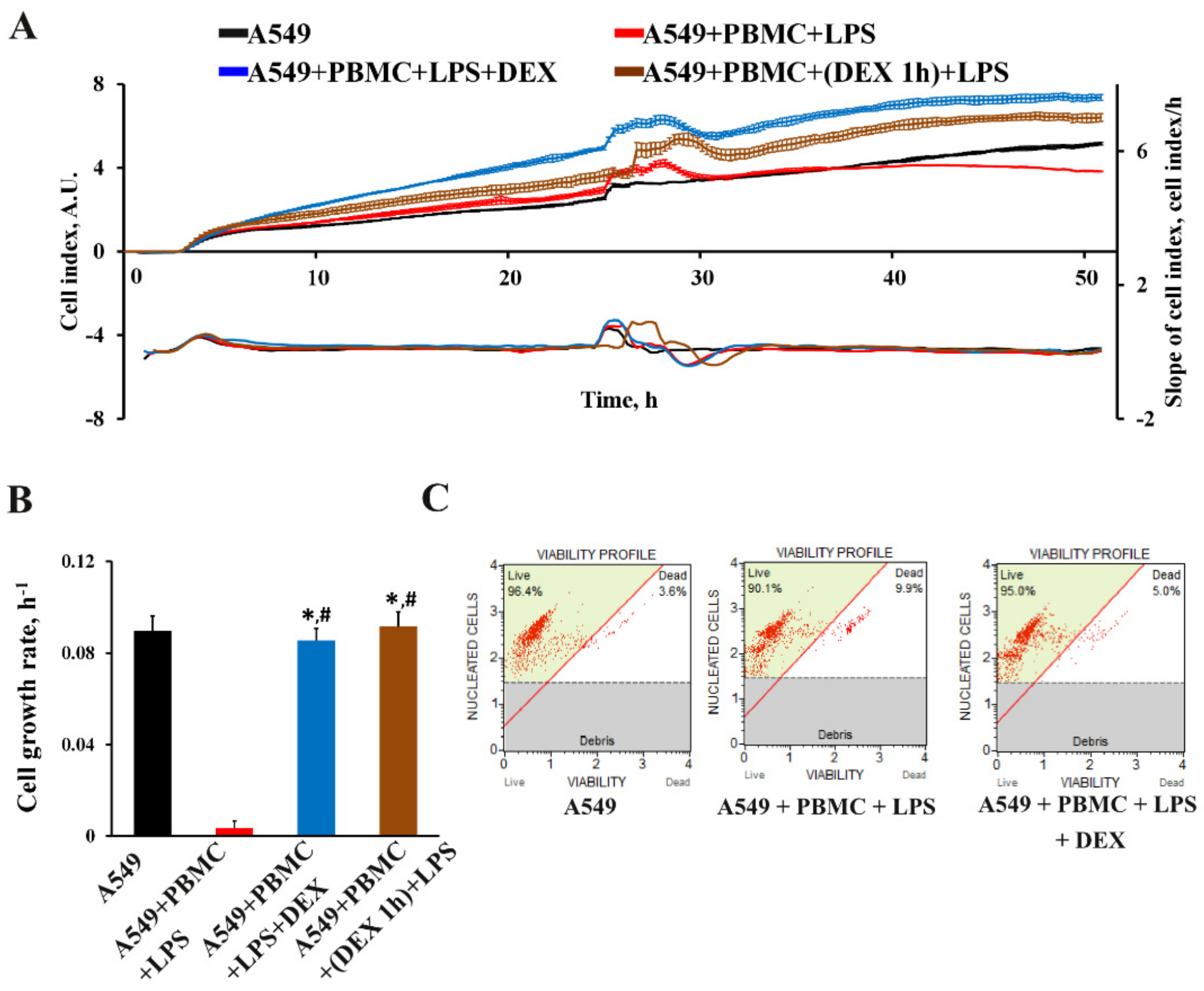
3.1. Inflammation Modeling
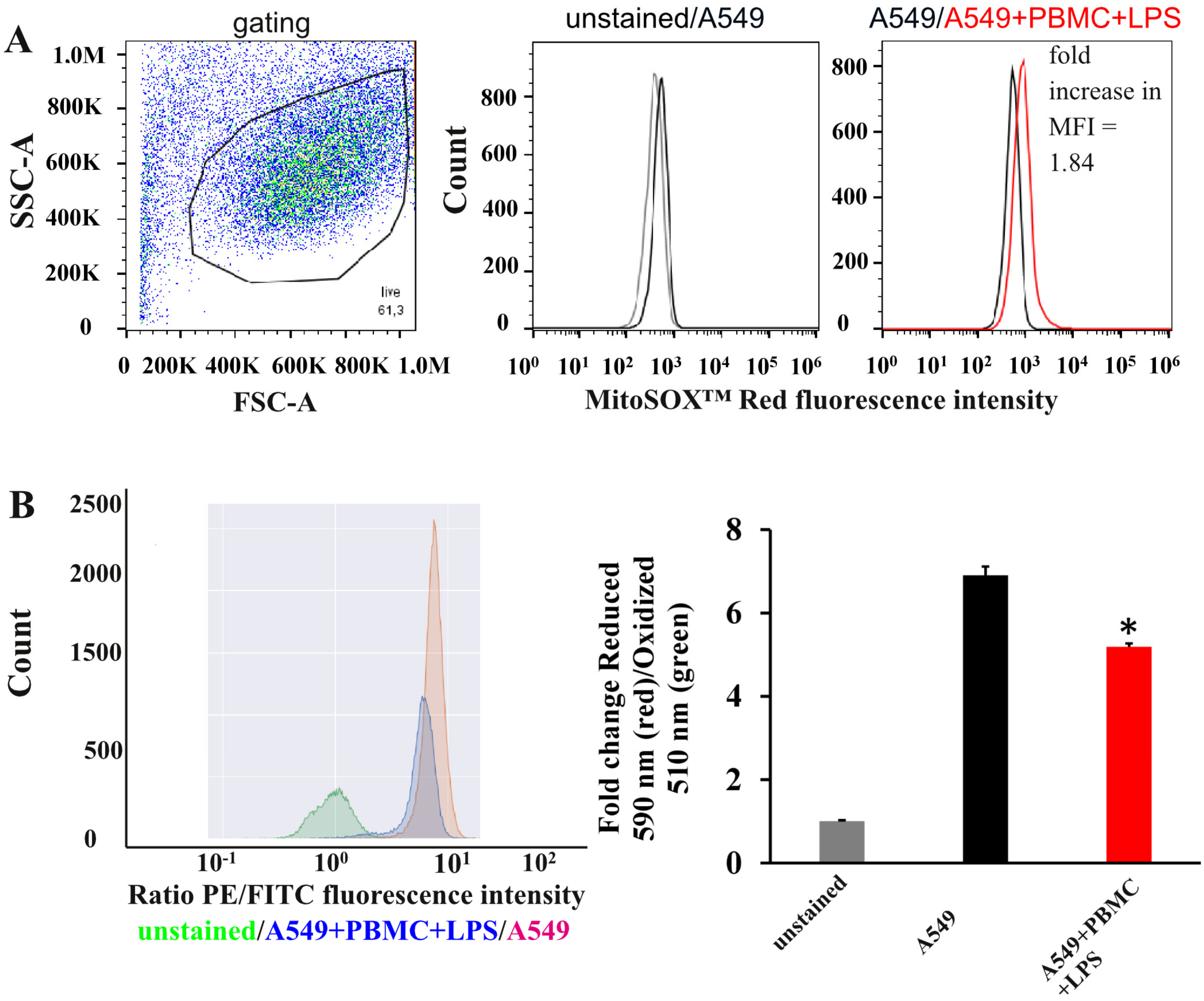
3.2. Oxidative Stress in A549 Cells under Inflammation Conditions
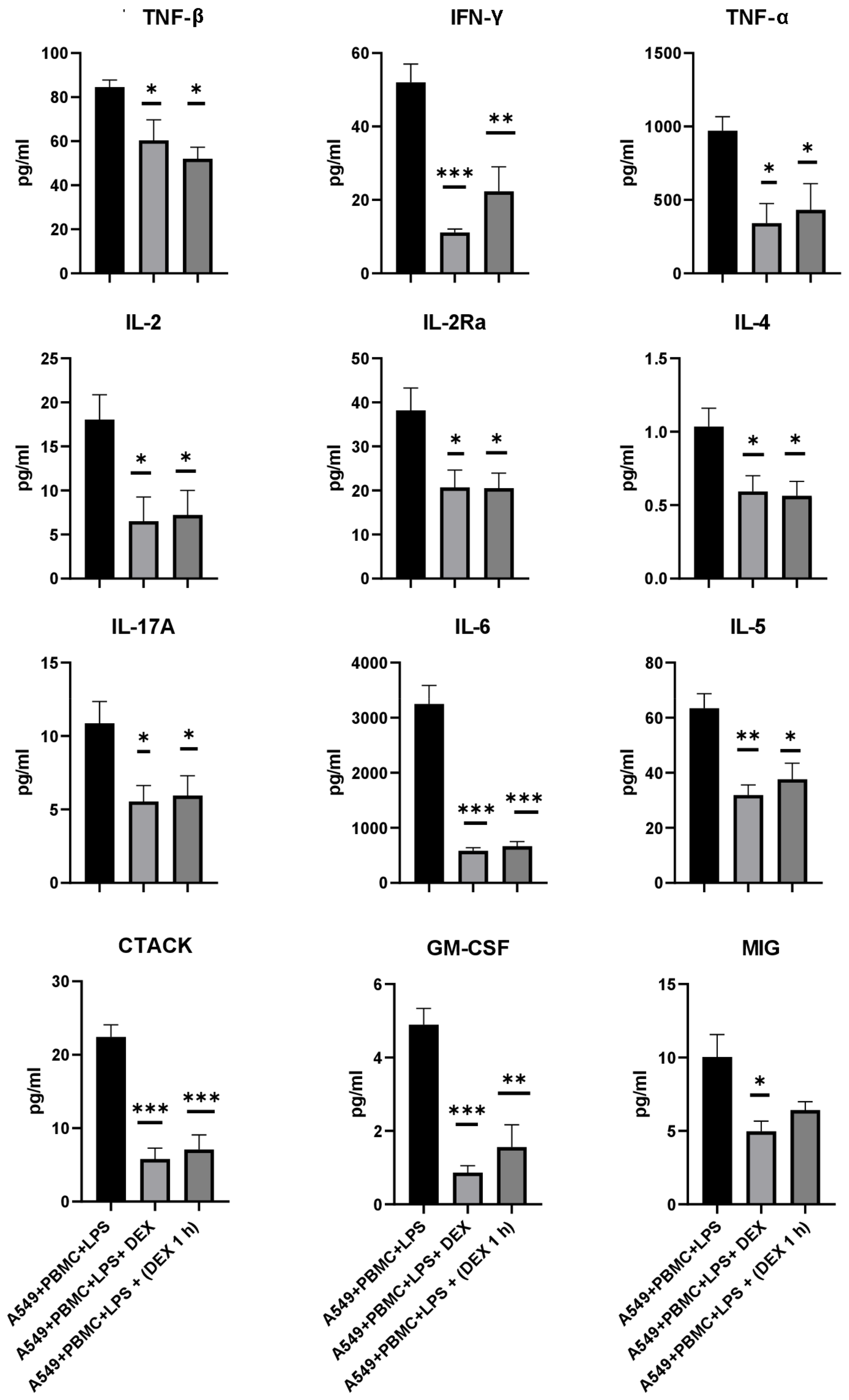
3.3. The Effect of Dexamethasone on the Cytokine Profile and Cell Index of A549 Cells
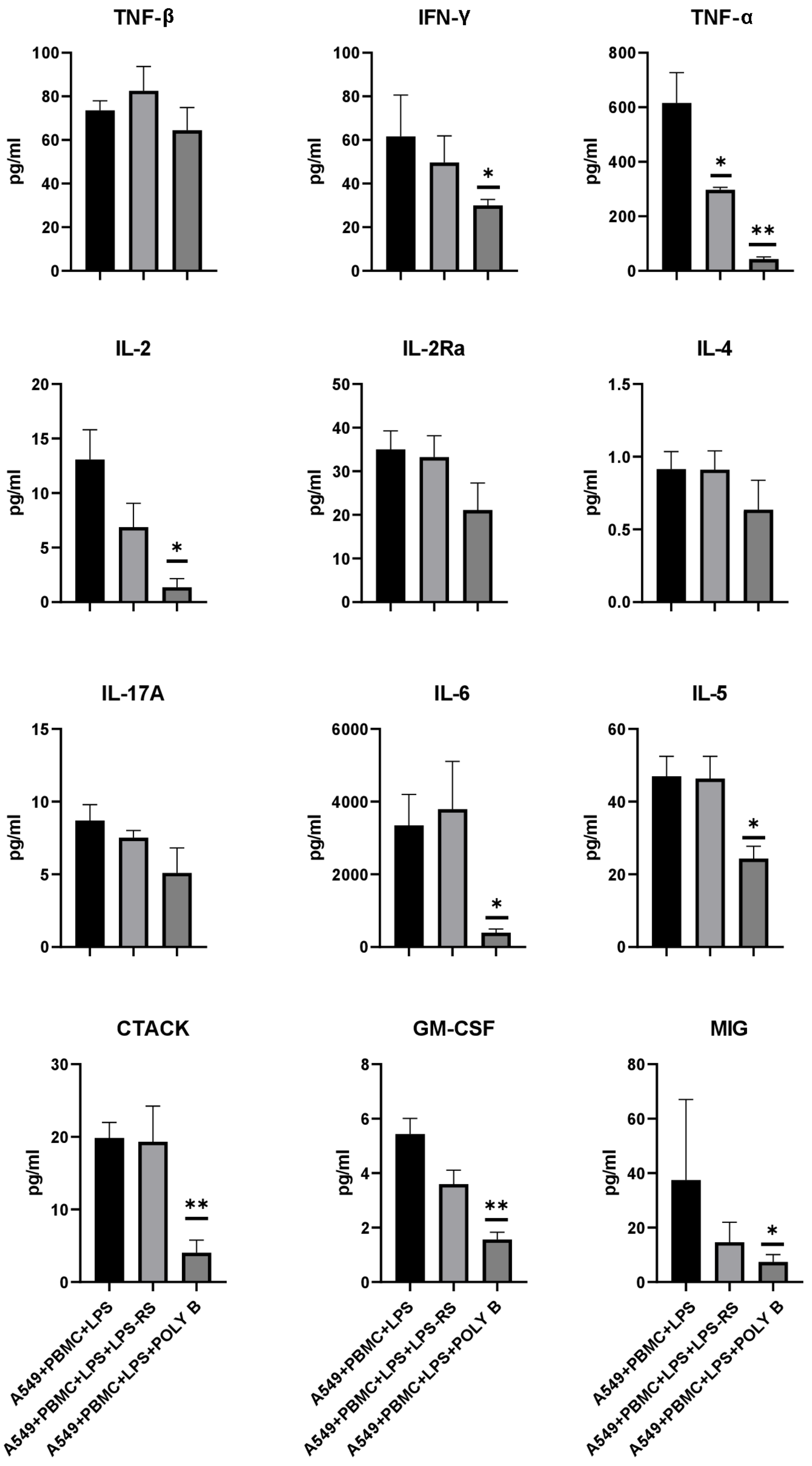
3.4. Effect of TLR4 Inhibitors on Cytokine/Chemokine Concentrations and Proliferation of A549 Cells in the Model System
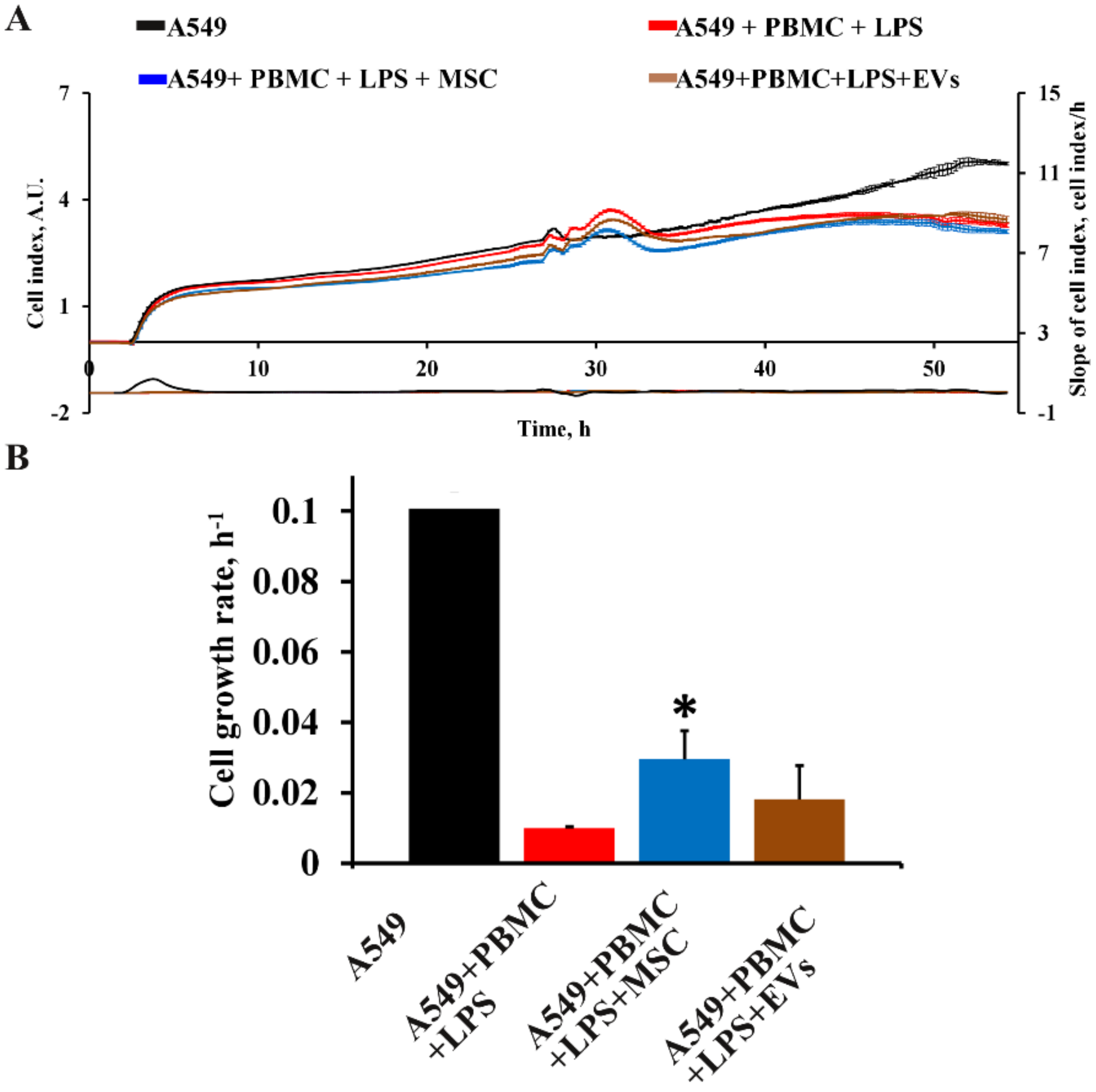
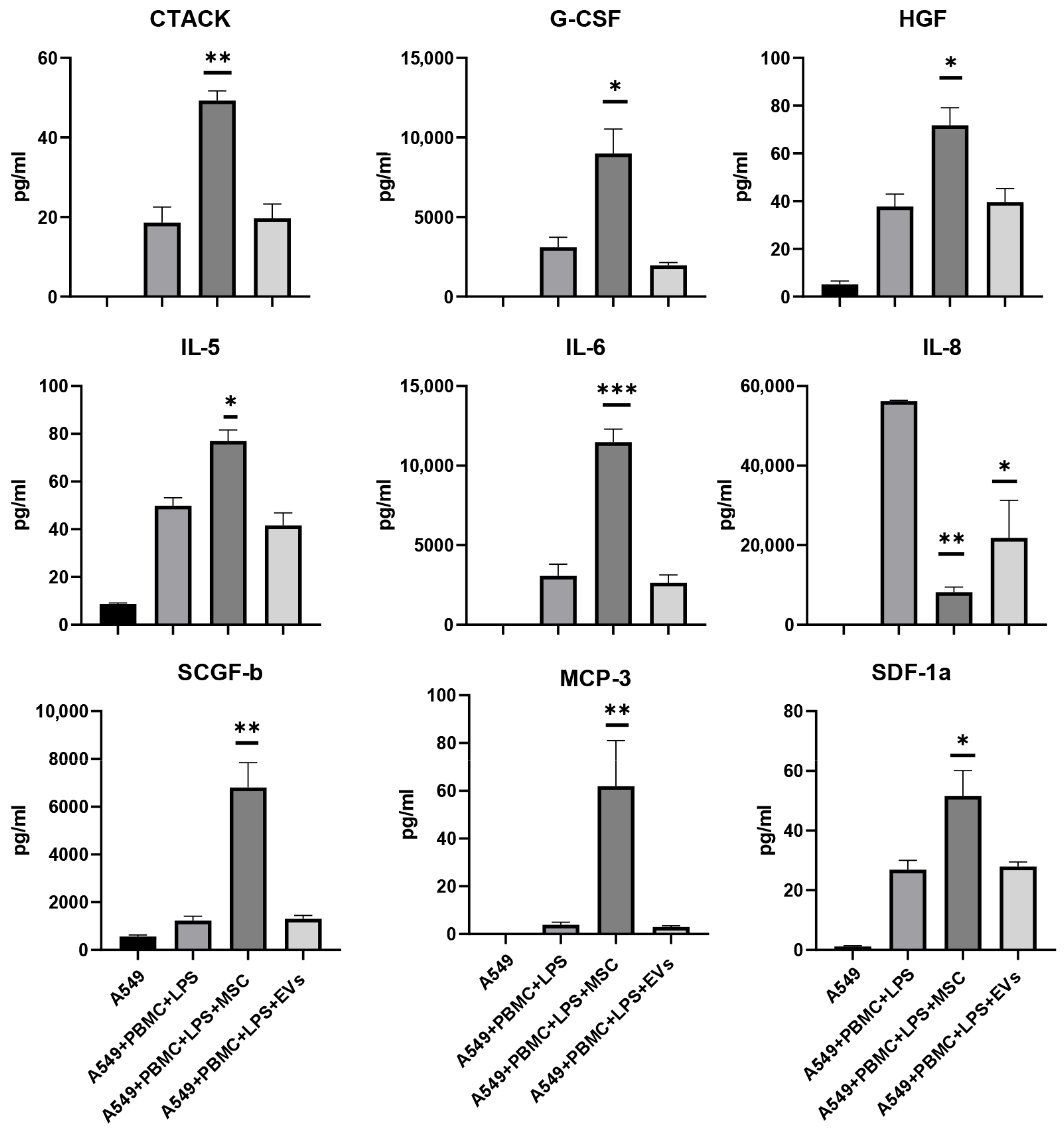
3.5. Influence of MSC and EVs on Cytokine/Chemokine Concentrations and Proliferation of A549 in Model System
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and Clinical Characteristics of 99 Cases of 2019 Novel Coronavirus Pneumonia in Wuhan, China: A Descriptive Study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- CDC Coronavirus Disease 2019 (COVID-19). Available online: https://www.cdc.gov/coronavirus/2019-ncov/index.html (accessed on 14 July 2022).
- Wu, Z.; McGoogan, J.M. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases from the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Fan, Y.; Lai, Y.; Han, T.; Li, Z.; Zhou, P.; Pan, P.; Wang, W.; Hu, D.; Liu, X.; et al. Coronavirus Infections and Immune Responses. J. Med. Virol. 2020, 92, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Cruz, F.F.; Rocco, P.R.M. The Potential of Mesenchymal Stem Cell Therapy for Chronic Lung Disease. Expert Rev. Respir. Med. 2020, 14, 31–39. [Google Scholar] [CrossRef]
- Subbarao, K.; McAuliffe, J.; Vogel, L.; Fahle, G.; Fischer, S.; Tatti, K.; Packard, M.; Shieh, W.-J.; Zaki, S.; Murphy, B. Prior Infection and Passive Transfer of Neutralizing Antibody Prevent Replication of Severe Acute Respiratory Syndrome Coronavirus in the Respiratory Tract of Mice. J. Virol. 2004, 78, 3572–3577. [Google Scholar] [CrossRef]
- Karikalan, B.; Darnal, H.K. Immune Status of COVID-19 Patients with Reference to SARS and MERS. J. Pure Appl. Microbiol. 2020, 14, 817–821. [Google Scholar] [CrossRef]
- Roberts, A.; Deming, D.; Paddock, C.D.; Cheng, A.; Yount, B.; Vogel, L.; Herman, B.D.; Sheahan, T.; Heise, M.; Genrich, G.L.; et al. A Mouse-Adapted SARS-Coronavirus Causes Disease and Mortality in BALB/c Mice. PLoS Pathog. 2007, 3, e5. [Google Scholar] [CrossRef]
- Astashkina, A.; Mann, B.; Grainger, D.W. A Critical Evaluation of in Vitro Cell Culture Models for High-Throughput Drug Screening and Toxicity. Pharmacol. Ther. 2012, 134, 82–106. [Google Scholar] [CrossRef]
- Miyake, K. Innate Recognition of Lipopolysaccharide by Toll-like Receptor 4-MD-2. Trends Microbiol. 2004, 12, 186–192. [Google Scholar] [CrossRef]
- Türker Şener, L.; Albenïz, G.; Dïnç, B.; Albenïz, I. ICELLigence Real-time Cell Analysis System for Examining the Cytotoxicity of Drugs to Cancer Cell Lines (Review). Exp. Ther. Med. 2017, 14, 1866–1870. [Google Scholar] [CrossRef]
- Vardakas, P.; Skaperda, Z.; Tekos, F.; Kouretas, D. ROS and COVID. Antioxidants 2022, 11, 339. [Google Scholar] [CrossRef]
- Hossein-Khannazer, N.; Shokoohian, B.; Shpichka, A.; Aghdaei, H.A.; Timashev, P.; Vosough, M. An Update to "Novel Therapeutic Approaches for Treatment of COVID-19". J. Mol. Med. 2021, 99, 303–310. [Google Scholar] [CrossRef]
- Hossein-khannazer, N.; Shokoohian, B.; Shpichka, A.; Aghdaei, H.A.; Timashev, P.; Vosough, M. Novel Therapeutic Approaches for Treatment of COVID-19. J. Mol. Med. 2020, 98, 789–803. [Google Scholar] [CrossRef]
- Agarwal, A.; Rochwerg, B.; Lamontagne, F.; Siemieniuk, R.A.; Agoritsas, T.; Askie, L.; Lytvyn, L.; Leo, Y.-S.; Macdonald, H.; Zeng, L.; et al. A Living WHO Guideline on Drugs for Covid-19. BMJ 2020, 370, m3379. [Google Scholar] [CrossRef]
- The RECOVERY Collaborative Group Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021, 384, 693–704. [CrossRef]
- Yang, G.; Zhang, S.; Wang, Y.; Li, L.; Li, Y.; Yuan, D.; Luo, F.; Zhao, J.; Song, X.; Zhao, Y. Aptamer Blocking S-TLR4 Interaction Selectively Inhibits SARS-CoV-2 Induced Inflammation. Signal Transduct. Target Ther. 2022, 7, 120. [Google Scholar] [CrossRef]
- Kaushik, D.; Bhandari, R.; Kuhad, A. TLR4 as a Therapeutic Target for Respiratory and Neurological Complications of SARS-CoV-2. Expert Opin. Ther. Targets 2021, 25, 491–508. [Google Scholar] [CrossRef]
- Manik, M.; Singh, R.K. Role of Toll-like Receptors in Modulation of Cytokine Storm Signaling in SARS-CoV-2-induced COVID-19. J. Med. Virol. 2022, 94, 869–877. [Google Scholar] [CrossRef]
- Chen, X.; Shan, Y.; Wen, Y.; Sun, J.; Du, H. Mesenchymal Stem Cell Therapy in Severe COVID-19: A Retrospective Study of Short-Term Treatment Efficacy and Side Effects. J. Infect. 2020, 81, 647–679. [Google Scholar] [CrossRef]
- Leng, Z.; Zhu, R.; Hou, W.; Feng, Y.; Yang, Y.; Han, Q.; Shan, G.; Meng, F.; Du, D.; Wang, S.; et al. Transplantation of ACE2- Mesenchymal Stem Cells Improves the Outcome of Patients with COVID-19 Pneumonia. Aging Dis. 2020, 11, 216–228. [Google Scholar] [CrossRef]
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In Vitro Cultivation of Human Tumors: Establishment of Cell Lines Derived from a Series of Solid Tumors. J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef]
- Giaever, I.; Keese, C.R. Monitoring Fibroblast Behavior in Tissue Culture with an Applied Electric Field. Proc. Natl. Acad. Sci. USA 1984, 81, 3761–3764. [Google Scholar] [CrossRef]
- Silachev, D.N.; Goryunov, K.V.; Shpilyuk, M.A.; Beznoschenko, O.S.; Morozova, N.Y.; Kraevaya, E.E.; Popkov, V.A.; Pevzner, I.B.; Zorova, L.D.; Evtushenko, E.A.; et al. Effect of MSCs and MSC-Derived Extracellular Vesicles on Human Blood Coagulation. Cells 2019, 8, E258. [Google Scholar] [CrossRef]
- Smith, B.T. Cell Line A549: A Model System for the Study of Alveolar Type II Cell Function. Am. Rev. Respir. Dis. 1977, 115, 285–293. [Google Scholar] [CrossRef]
- Li, X.C. A New Type of Programmed Inflammatory Cell Death: PANoptosis. Am. J. Transplant. 2021, 21, 3507. [Google Scholar] [CrossRef]
- Lee, S.; Karki, R.; Wang, Y.; Nguyen, L.N.; Kalathur, R.C.; Kanneganti, T.-D. AIM2 Forms a Complex with Pyrin and ZBP1 to Drive PANoptosis and Host Defence. Nature 2021, 597, 415–419. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, W.; Shi, X.; Ding, J.; Liu, W.; He, H.; Wang, K.; Shao, F. Chemotherapy Drugs Induce Pyroptosis through Caspase-3 Cleavage of a Gasdermin. Nature 2017, 547, 99–103. [Google Scholar] [CrossRef]
- Bhatia, M.; Moochhala, S. Role of Inflammatory Mediators in the Pathophysiology of Acute Respiratory Distress Syndrome. J. Pathol. 2004, 202, 145–156. [Google Scholar] [CrossRef]
- Immune Status of COVID-19 Patients with Different Disease Severity. Available online: https://aig-journal.ru/articles/Sostoyanie-immunnoi-sistemy-u-pacientov-s-razlichnoi-stepenu-tyajesti-COVID-19.html (accessed on 28 July 2022).
- Drummen, G.P.C.; Gadella, B.M.; Post, J.A.; Brouwers, J.F. Mass Spectrometric Characterization of the Oxidation of the Fluorescent Lipid Peroxidation Reporter Molecule C11-BODIPY (581/591). Free Radic. Biol. Med. 2004, 36, 1635–1644. [Google Scholar] [CrossRef]
- Li, H.; Yan, B.; Gao, R.; Ren, J.; Yang, J. Effectiveness of Corticosteroids to Treat Severe COVID-19: A Systematic Review and Meta-Analysis of Prospective Studies. Int. Immunopharmacol. 2021, 100, 108121. [Google Scholar] [CrossRef]
- Tomazini, B.M.; Maia, I.S.; Cavalcanti, A.B.; Berwanger, O.; Rosa, R.G.; Veiga, V.C.; Avezum, A.; Lopes, R.D.; Bueno, F.R.; Silva, M.V.A.O.; et al. Effect of Dexamethasone on Days Alive and Ventilator-Free in Patients with Moderate or Severe Acute Respiratory Distress Syndrome and COVID-19: The CoDEX Randomized Clinical Trial. JAMA 2020, 324, 1307–1316. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Feng, Z.; Wang, F.-S. Mesenchymal Stem Cell Treatment for COVID-19. eBioMedicine 2022, 77, 103920. [Google Scholar] [CrossRef] [PubMed]
- Mason, R.J. Pathogenesis of COVID-19 from a Cell Biology Perspective. Eur. Respir. J. 2020, 55, 2000607. [Google Scholar] [CrossRef] [PubMed]
- Nova, Z.; Skovierova, H.; Calkovska, A. Alveolar-Capillary Membrane-Related Pulmonary Cells as a Target in Endotoxin-Induced Acute Lung Injury. Int. J. Mol. Sci. 2019, 20, 831. [Google Scholar] [CrossRef]
- Calkovska, A.; Kolomaznik, M.; Calkovsky, V. Alveolar Type II Cells and Pulmonary Surfactant in COVID-19 Era. Physiol. Res. 2021, 70, S195–S208. [Google Scholar] [CrossRef] [PubMed]
- Foster, K.A.; Oster, C.G.; Mayer, M.M.; Avery, M.L.; Audus, K.L. Characterization of the A549 Cell Line as a Type II Pulmonary Epithelial Cell Model for Drug Metabolism. Exp. Cell Res. 1998, 243, 359–366. [Google Scholar] [CrossRef]
- Cooper, J.R.; Abdullatif, M.B.; Burnett, E.C.; Kempsell, K.E.; Conforti, F.; Tolley, H.; Collins, J.E.; Davies, D.E. Long Term Culture of the A549 Cancer Cell Line Promotes Multilamellar Body Formation and Differentiation towards an Alveolar Type II Pneumocyte Phenotype. PLoS ONE 2016, 11, e0164438. [Google Scholar] [CrossRef]
- Pontelli, M.C.; Castro, Í.A.; Martins, R.B.; La Serra, L.; Veras, F.P.; Nascimento, D.C.; Silva, C.M.; Cardoso, R.S.; Rosales, R.; Gomes, R.; et al. SARS-CoV-2 Productively Infects Primary Human Immune System Cells in Vitro and in COVID-19 Patients. J. Mol. Cell Biol. 2022, 14, mjac021. [Google Scholar] [CrossRef]
- Kazmierski, J.; Friedmann, K.; Postmus, D.; Emanuel, J.; Fischer, C.; Jansen, J.; Richter, A.; Bosquillon de Jarcy, L.; Schüler, C.; Sohn, M.; et al. Nonproductive Exposure of PBMCs to SARS-CoV-2 Induces Cell-Intrinsic Innate Immune Responses. Mol. Syst. Biol. 2022, 18, e10961. [Google Scholar] [CrossRef]
- Pelaia, C.; Tinello, C.; Vatrella, A.; De Sarro, G.; Pelaia, G. Lung under Attack by COVID-19-Induced Cytokine Storm: Pathogenic Mechanisms and Therapeutic Implications. Ther. Adv. Respir. Dis. 2020, 14, 1753466620933508. [Google Scholar] [CrossRef]
- Gong, J.; Dong, H.; Xia, Q.-S.; Huang, Z.-Y.; Wang, D.-K.; Zhao, Y.; Liu, W.-H.; Tu, S.-H.; Zhang, M.-M.; Wang, Q.; et al. Correlation Analysis between Disease Severity and Inflammation-Related Parameters in Patients with COVID-19: A Retrospective Study. BMC Infect. Dis. 2020, 20, 963. [Google Scholar] [CrossRef]
- Chan, L.; Karimi, N.; Morovati, S.; Alizadeh, K.; Kakish, J.E.; Vanderkamp, S.; Fazel, F.; Napoleoni, C.; Alizadeh, K.; Mehrani, Y.; et al. The Roles of Neutrophils in Cytokine Storms. Viruses 2021, 13, 2318. [Google Scholar] [CrossRef]
- Akbari, H.; Tabrizi, R.; Lankarani, K.B.; Aria, H.; Vakili, S.; Asadian, F.; Noroozi, S.; Keshavarz, P.; Faramarz, S. The Role of Cytokine Profile and Lymphocyte Subsets in the Severity of Coronavirus Disease 2019 (COVID-19): A Systematic Review and Meta-Analysis. Life Sci. 2020, 258, 118167. [Google Scholar] [CrossRef]
- Liu, Y.; Tan, W.; Chen, H.; Zhu, Y.; Wan, L.; Jiang, K.; Guo, Y.; Tang, K.; Xie, C.; Yi, H.; et al. Dynamic Changes in Lymphocyte Subsets and Parallel Cytokine Levels in Patients with Severe and Critical COVID-19. BMC Infect. Dis. 2021, 21, 79. [Google Scholar] [CrossRef]
- Pu, D.; Wang, W. Toll-like Receptor 4 Agonist, Lipopolysaccharide, Increases the Expression Levels of Cytokines and Chemokines in Human Peripheral Blood Mononuclear Cells. Exp. Ther. Med. 2014, 8, 1914–1918. [Google Scholar] [CrossRef]
- Fajgenbaum, D.C.; June, C.H. Cytokine Storm. N. Engl. J. Med. 2020, 383, 2255–2273. [Google Scholar] [CrossRef]
- Teijaro, J.R. Cytokine Storms in Infectious Diseases. Semin. Immunopathol. 2017, 39, 501–503. [Google Scholar] [CrossRef]
- Liu, T.; Feng, M.; Wen, Z.; He, Y.; Lin, W.; Zhang, M. Comparison of the Characteristics of Cytokine Storm and Immune Response Induced by SARS-CoV, MERS-CoV, and SARS-CoV-2 Infections. J. Inflamm. Res. 2021, 14, 5475–5487. [Google Scholar] [CrossRef]
- Chimenti, L.; Morales-Quinteros, L.; Puig, F.; Camprubi-Rimblas, M.; Guillamat-Prats, R.; Gómez, M.N.; Tijero, J.; Blanch, L.; Matute-Bello, G.; Artigas, A. Comparison of Direct and Indirect Models of Early Induced Acute Lung Injury. Intensive Care Med. Exp. 2020, 8, 62. [Google Scholar] [CrossRef]
- Al-Ani, B.; ShamsEldeen, A.M.; Kamar, S.S.; Haidara, M.A.; Al-Hashem, F.; Alshahrani, M.Y.; Al-Hakami, A.M.; Kader, D.H.A.; Maarouf, A. Lipopolysaccharide Induces Acute Lung Injury and Alveolar Haemorrhage in Association with the Cytokine Storm, Coagulopathy and AT1R/JAK/STAT Augmentation in a Rat Model That Mimics Moderate and Severe Covid-19 Pathology. Clin. Exp. Pharmacol. Physiol. 2022, 49, 483–491. [Google Scholar] [CrossRef]
- Wang, F.; Xu, L.; Dong, G.; Zhu, M.; Liu, L.; Wang, B. PIM2 Deletion Alleviates Lipopolysaccharide (LPS)-Induced Respiratory Distress Syndrome (ARDS) by Suppressing NLRP3 Inflammasome. Biochem. Biophys. Res. Commun. 2020, 533, 1419–1426. [Google Scholar] [CrossRef]
- Yang, X.; Ma, L. Post-Treatment with Propofol Inhibits Inflammatory Response in LPS-Induced Alveolar Type II Epithelial Cells. Exp. Ther. Med. 2022, 23, 249. [Google Scholar] [CrossRef]
- Kaspi, H.; Semo, J.; Abramov, N.; Dekel, C.; Lindborg, S.; Kern, R.; Lebovits, C.; Aricha, R. MSC-NTF (NurOwn®) Exosomes: A Novel Therapeutic Modality in the Mouse LPS-Induced ARDS Model. Stem Cell Res. Ther. 2021, 12, 72. [Google Scholar] [CrossRef]
- Hasanvand, A. COVID-19 and the Role of Cytokines in This Disease. Inflammopharmacology 2022, 30, 789–798. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.-H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An Inflammatory Cytokine Signature Predicts COVID-19 Severity and Survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]
- Chi, Y.; Ge, Y.; Wu, B.; Zhang, W.; Wu, T.; Wen, T.; Liu, J.; Guo, X.; Huang, C.; Jiao, Y.; et al. Serum Cytokine and Chemokine Profile in Relation to the Severity of Coronavirus Disease 2019 in China. J. Infect. Dis. 2020, 222, 746–754. [Google Scholar] [CrossRef]
- Nigar, S.; Shah, S.T.; Setu, M.A.A.; Dip, S.D.; Ibnat, H.; Islam, M.T.; Akter, S.; Jahid, I.K.; Hossain, M.A. Relative Expression of Proinflammatory Molecules in COVID-19 Patients Who Manifested Disease Severities. J. Med. Virol. 2021, 93, 5805–5815. [Google Scholar] [CrossRef]
- Khalil, B.A.; Elemam, N.M.; Maghazachi, A.A. Chemokines and Chemokine Receptors during COVID-19 Infection. Comput. Struct. Biotechnol. J. 2021, 19, 976–988. [Google Scholar] [CrossRef]
- Giamarellos-Bourboulis, E.J.; Netea, M.G.; Rovina, N.; Akinosoglou, K.; Antoniadou, A.; Antonakos, N.; Damoraki, G.; Gkavogianni, T.; Adami, M.-E.; Katsaounou, P.; et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe 2020, 27, 992–1000.e3. [Google Scholar] [CrossRef]
- Stefanowicz-Hajduk, J.; Ochocka, J.R. Real-Time Cell Analysis System in Cytotoxicity Applications: Usefulness and Comparison with Tetrazolium Salt Assays. Toxicol. Rep. 2020, 7, 335–344. [Google Scholar] [CrossRef]
- Karki, R.; Sharma, B.R.; Tuladhar, S.; Williams, E.P.; Zalduondo, L.; Samir, P.; Zheng, M.; Sundaram, B.; Banoth, B.; Malireddi, R.K.S.; et al. Synergism of TNF-α and IFN-γ Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell 2021, 184, 149–168.e17. [Google Scholar] [CrossRef] [PubMed]
- Christgen, S.; Zheng, M.; Kesavardhana, S.; Karki, R.; Malireddi, R.K.S.; Banoth, B.; Place, D.E.; Briard, B.; Sharma, B.R.; Tuladhar, S.; et al. Identification of the PANoptosome: A Molecular Platform Triggering Pyroptosis, Apoptosis, and Necroptosis (PANoptosis). Front. Cell Infect. Microbiol. 2020, 10, 237. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kanneganti, T.-D. From Pyroptosis, Apoptosis and Necroptosis to PANoptosis: A Mechanistic Compendium of Programmed Cell Death Pathways. Comput. Struct. Biotechnol. J. 2021, 19, 4641–4657. [Google Scholar] [CrossRef] [PubMed]
- Ryter, S.W.; Kim, H.P.; Hoetzel, A.; Park, J.W.; Nakahira, K.; Wang, X.; Choi, A.M.K. Mechanisms of Cell Death in Oxidative Stress. Antioxid. Redox Signal 2007, 9, 49–89. [Google Scholar] [CrossRef]
- Cecchini, R.; Cecchini, A.L. SARS-CoV-2 Infection Pathogenesis Is Related to Oxidative Stress as a Response to Aggression. Med. Hypotheses 2020, 143, 110102. [Google Scholar] [CrossRef]
- Zinovkin, R.A.; Grebenchikov, O.A. Transcription Factor Nrf2 as a Potential Therapeutic Target for Prevention of Cytokine Storm in COVID-19 Patients. Biochemistry 2020, 85, 833–837. [Google Scholar] [CrossRef]
- Muhammad, Y.; Kani, Y.A.; Iliya, S.; Muhammad, J.B.; Binji, A.; El-Fulaty Ahmad, A.; Kabir, M.B.; Umar Bindawa, K.; Ahmed, A. Deficiency of Antioxidants and Increased Oxidative Stress in COVID-19 Patients: A Cross-Sectional Comparative Study in Jigawa, Northwestern Nigeria. SAGE Open Med. 2021, 9, 2050312121991246. [Google Scholar] [CrossRef]
- Plotnikov, E.Y.; Morosanova, M.A.; Pevzner, I.B.; Zorova, L.D.; Manskikh, V.N.; Pulkova, N.V.; Galkina, S.I.; Skulachev, V.P.; Zorov, D.B. Protective Effect of Mitochondria-Targeted Antioxidants in an Acute Bacterial Infection. Proc. Natl. Acad. Sci. USA 2013, 110, E3100–E3108. [Google Scholar] [CrossRef]
- Rani, S.; Ryan, A.E.; Griffin, M.D.; Ritter, T. Mesenchymal Stem Cell-Derived Extracellular Vesicles: Toward Cell-Free Therapeutic Applications. Mol. Ther. 2015, 23, 812–823. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, J.; Feng, B.; Lin, F.; Zhou, J.; Liu, J.; Shi, X.; Lu, X.; Pan, Q.; Yu, J.; et al. Immunosuppressive Effect of Mesenchymal Stem Cells on Lung and Gut CD8+ T Cells in Lipopolysaccharide-Induced Acute Lung Injury in Mice. Cell Prolif. 2021, 54, e13028. [Google Scholar] [CrossRef]
- Lee, J.W.; Fang, X.; Gupta, N.; Serikov, V.; Matthay, M.A. Allogeneic Human Mesenchymal Stem Cells for Treatment of E. Coli Endotoxin-Induced Acute Lung Injury in the Ex Vivo Perfused Human Lung. Proc. Natl. Acad. Sci. USA 2009, 106, 16357–16362. [Google Scholar] [CrossRef]
- Panganiban, R.A.M.; Day, R.M. Hepatocyte Growth Factor in Lung Repair and Pulmonary Fibrosis. Acta Pharmacol. Sin. 2011, 32, 12–20. [Google Scholar] [CrossRef]
- Zhao, F.-Y.; Cheng, T.-Y.; Yang, L.; Huang, Y.-H.; Li, C.; Han, J.-Z.; Li, X.-H.; Fang, L.-J.; Feng, D.-D.; Tang, Y.-T.; et al. G-CSF Inhibits Pulmonary Fibrosis by Promoting BMSC Homing to the Lungs via SDF-1/CXCR4 Chemotaxis. Sci. Rep. 2020, 10, 10515. [Google Scholar] [CrossRef]
- Lee, B.-C.; Kang, I.; Yu, K.-R. Therapeutic Features and Updated Clinical Trials of Mesenchymal Stem Cell (MSC)-Derived Exosomes. J. Clin. Med. 2021, 10, 711. [Google Scholar] [CrossRef]
- Kurte, M.; Vega-Letter, A.M.; Luz-Crawford, P.; Djouad, F.; Noël, D.; Khoury, M.; Carrión, F. Time-Dependent LPS Exposure Commands MSC Immunoplasticity through TLR4 Activation Leading to Opposite Therapeutic Outcome in EAE. Stem Cell Res. Ther. 2020, 11, 416. [Google Scholar] [CrossRef]
- Hou, Y.S.; Liu, L.Y.; Chai, J.K.; Yu, Y.H.; Duan, H.J.; Hu, Q.; Yin, H.N.; Wang, Y.H.; Zhuang, S.B.; Fan, J.; et al. Lipopolysaccharide Pretreatment Inhibits LPS-Induced Human Umbilical Cord Mesenchymal Stem Cell Apoptosis via Upregulating the Expression of Cellular FLICE-Inhibitory Protein. Mol. Med. Rep. 2015, 12, 2521–2528. [Google Scholar] [CrossRef][Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shevtsova, Y.A.; Goryunov, K.V.; Babenko, V.A.; Pevzner, I.B.; Vtorushina, V.V.; Inviyaeva, E.V.; Krechetova, L.V.; Zorova, L.D.; Plotnikov, E.Y.; Zorov, D.B.; et al. Development of an In Vitro Model of SARS-CoV-Induced Acute Lung Injury for Studying New Therapeutic Approaches. Antioxidants 2022, 11, 1910. https://doi.org/10.3390/antiox11101910
Shevtsova YA, Goryunov KV, Babenko VA, Pevzner IB, Vtorushina VV, Inviyaeva EV, Krechetova LV, Zorova LD, Plotnikov EY, Zorov DB, et al. Development of an In Vitro Model of SARS-CoV-Induced Acute Lung Injury for Studying New Therapeutic Approaches. Antioxidants. 2022; 11(10):1910. https://doi.org/10.3390/antiox11101910
Chicago/Turabian StyleShevtsova, Yulia A., Kirill V. Goryunov, Valentina A. Babenko, Irina B. Pevzner, Valentina V. Vtorushina, Evgeniya V. Inviyaeva, Lyubov V. Krechetova, Ljubava D. Zorova, Egor Y. Plotnikov, Dmitry B. Zorov, and et al. 2022. "Development of an In Vitro Model of SARS-CoV-Induced Acute Lung Injury for Studying New Therapeutic Approaches" Antioxidants 11, no. 10: 1910. https://doi.org/10.3390/antiox11101910
APA StyleShevtsova, Y. A., Goryunov, K. V., Babenko, V. A., Pevzner, I. B., Vtorushina, V. V., Inviyaeva, E. V., Krechetova, L. V., Zorova, L. D., Plotnikov, E. Y., Zorov, D. B., Sukhikh, G. T., & Silachev, D. N. (2022). Development of an In Vitro Model of SARS-CoV-Induced Acute Lung Injury for Studying New Therapeutic Approaches. Antioxidants, 11(10), 1910. https://doi.org/10.3390/antiox11101910








