Current Knowledge on Mechanisms Preventing Photosynthesis Redox Imbalance in Plants
Abstract
1. Introduction
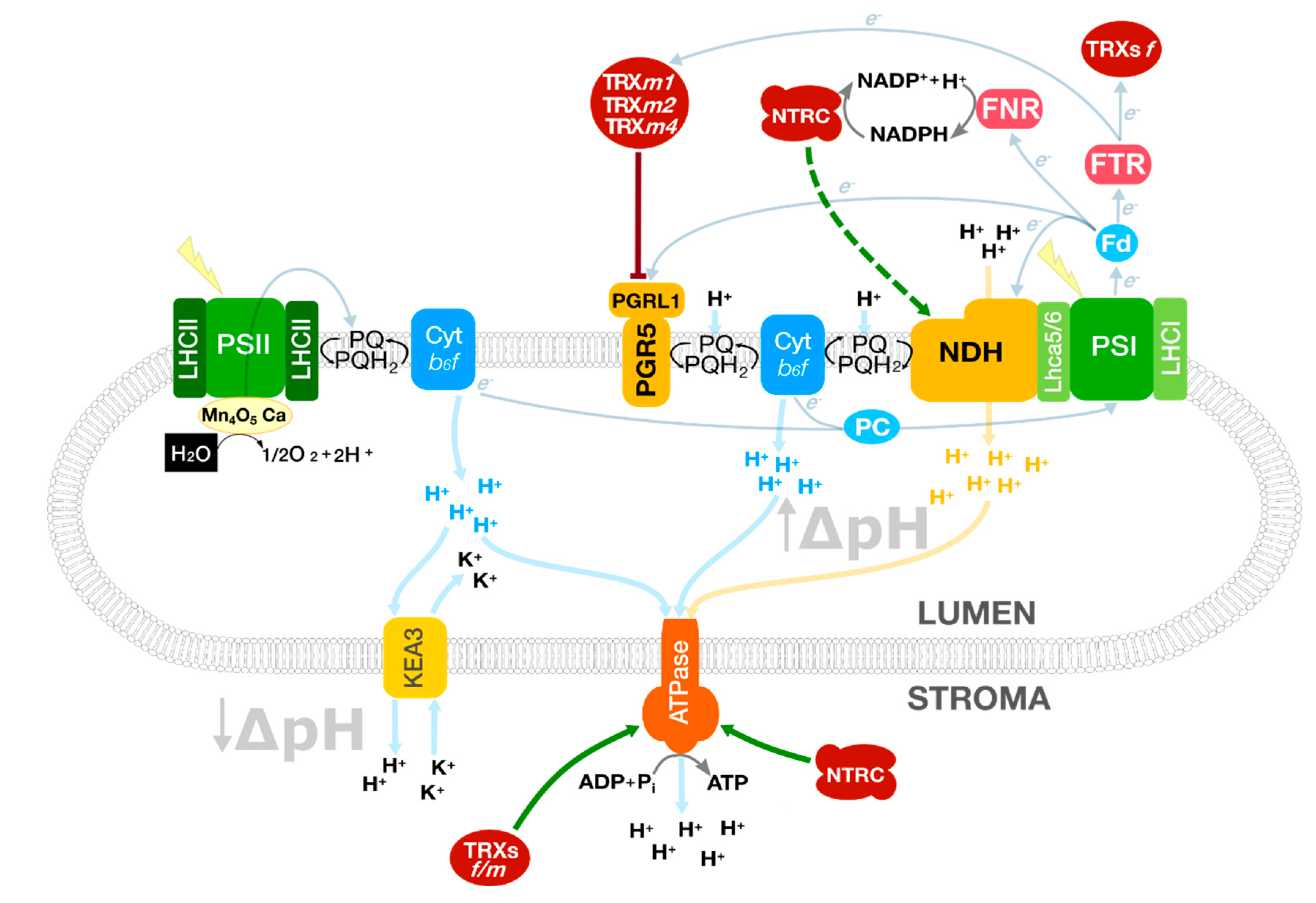
2. Balancing Photosynthesis through Cyclic Electron Flow
2.1. The NDH Complex
2.2. The PGR5/PGRL1 Complex
2.3. Ferredoxins: Active Players Balancing Linear and Cyclic Electron Flows?
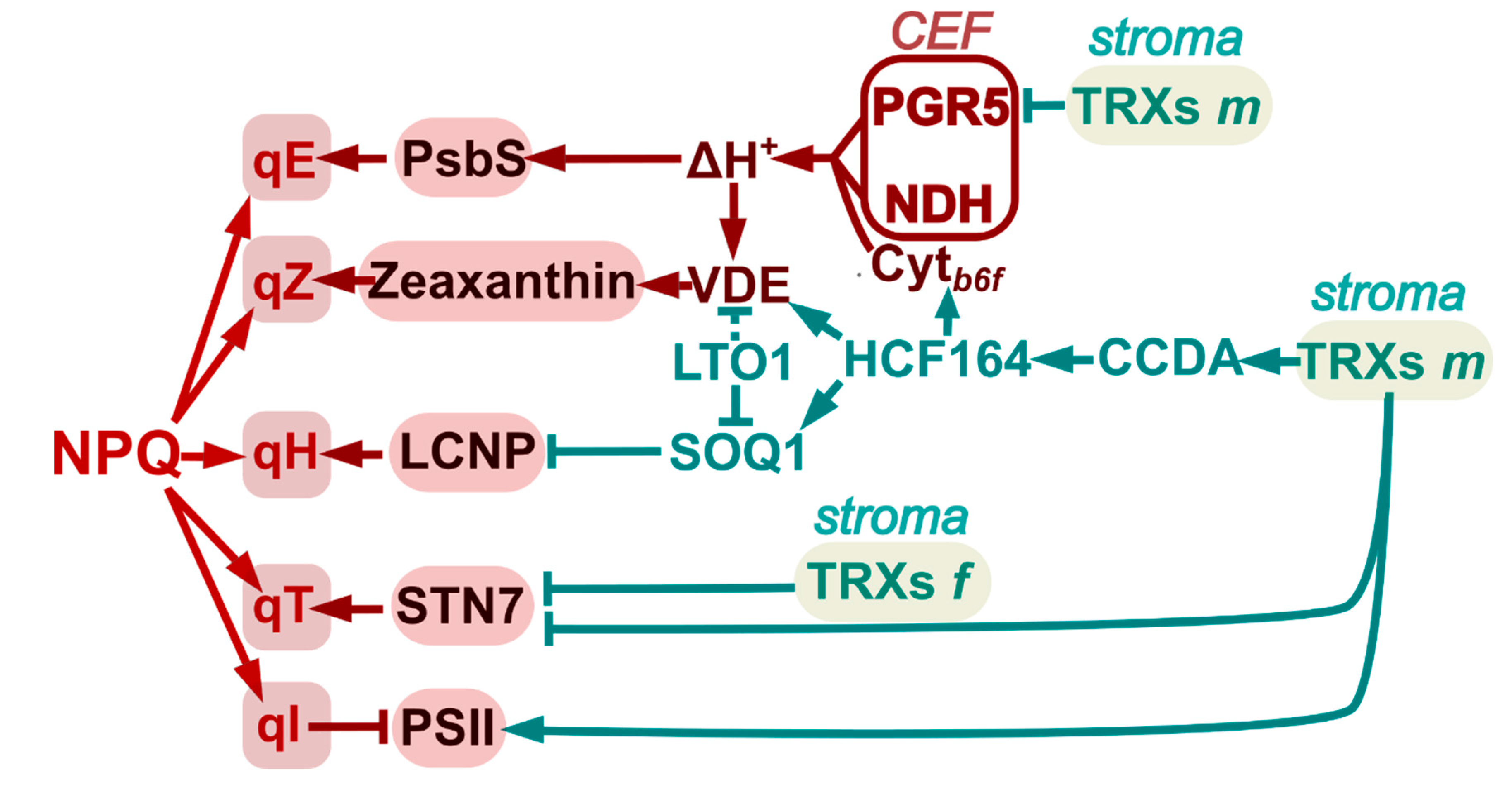
3. Redox Regulation of Non-Photochemical Quenching
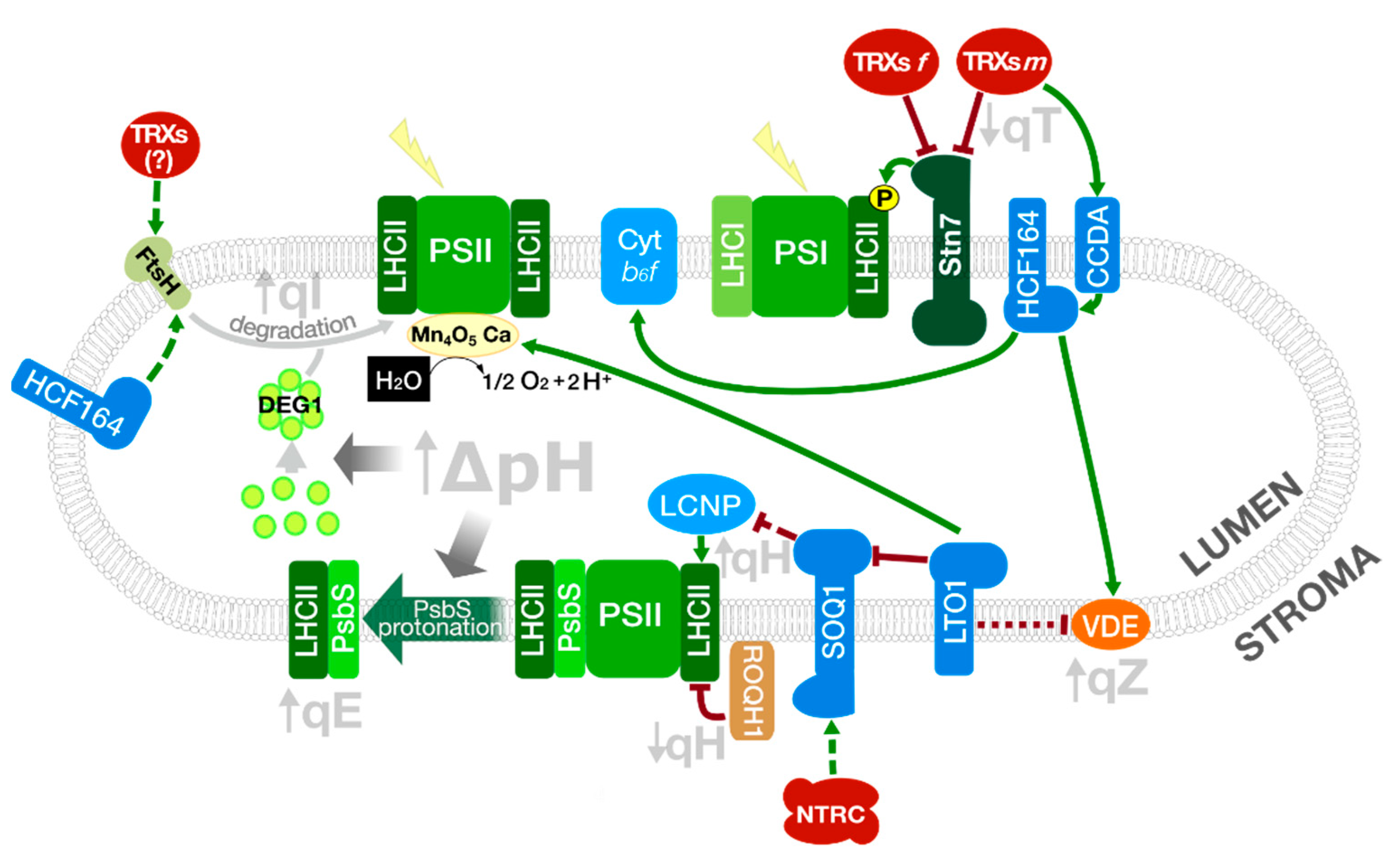
3.1. Photo-Protective Quenching in LHCII: Is Lipocalin Subjected to Redox Regulation?
3.2. Redistribution of Excitation Energy between the PSs: Role of Redox Regulation of LHCII Kinase and Cyt b6f Assembly in State Transitions
3.3. Photodamage and Repair of PSII: Redox Regulation of D1 Degradation and PsbO Stability
4. Plastid Terminal Oxidase: A Multitasking Redox Security Valve
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Govindjee, D.S.; Björn, L.O. Evolution of the Z-scheme of photosynthesis: A perspective. Photosynth. Res. 2017, 133, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Baracaldo, P.; Cardona, T. On the origin of oxygenic photosynthesis and cyanobacteria. New Phytol. 2020, 225, 1440–1446. [Google Scholar] [CrossRef] [PubMed]
- Grossman, A.R.; Bhaya, D.; Apt, K.E.; Kehoe, D.M. Light-harvesting complexes in oxygenic photosynthesis: Diversity, control, and evolution. Annu. Rev. Genet. 1995, 29, 231–288. [Google Scholar] [CrossRef] [PubMed]
- Schmid, V.H. Light-harvesting complexes of vascular plants. Cell Mol. Life Sci. 2008, 65, 3619–3639. [Google Scholar] [CrossRef]
- Munekage, Y.; Hashimoto, M.; Miyake, C.; Tomizawa, K.; Endo, T.; Tasaka, M.; Shikanai, T. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 2004, 429, 579–582. [Google Scholar] [CrossRef]
- Nawrocki, W.J.; Bailleul, B.; Picot, D.; Cardol, P.; Rappaport, F.; Wollman, F.A.; Joliot, P. The mechanism of cyclic electron flow. Biochim. Biophys. Acta Bioenerg. 2019, 1860, 433–438. [Google Scholar] [CrossRef]
- Blank, C.E.; Sánchez-Baracaldo, P. Timing of morphological and ecological innovations in the cyanobacteria--a key to understanding the rise in atmospheric oxygen. Geobiology 2010, 8, 1–23. [Google Scholar] [CrossRef]
- Lyons, T.W.; Reinhard, C.T.; Planavsky, N.J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 2014, 506, 307–315. [Google Scholar] [CrossRef]
- Hodgskiss, M.S.W.; Crockford, P.W.; Peng, Y.; Wing, B.A.; Horner, T.J. A productivity collapse to end earth’s great oxidation. Proc. Natl. Acad. Sci. USA 2019, 116, 17207–17212. [Google Scholar] [CrossRef]
- Schmidt, R.; Schippers, J.H. ROS-mediated redox signalling during cell differentiation in plants. Biochim. Biophys. Acta 2015, 1850, 1497–1508. [Google Scholar] [CrossRef]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, B.C.; Oelmüller, R. Reactive oxygen species generation and signalling in plants. Plant Signal Behav. 2012, 7, 1621–1633. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Allakhverdiev, S.I.; Murata, N. A new paradigm for the action of reactive oxygen species in the photoinhibition of photosystem II. Biochim. Biophys. Acta 2006, 1757, 742–749. [Google Scholar] [CrossRef]
- Mehler, A.H. Studies on reactions of illuminated chloroplasts. I. Mechanism of the reduction of oxygen and other Hill reagents. Arch. Biochem. Biophys. 1951, 33, 65–77. [Google Scholar] [CrossRef]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Wrzaczek, M.; Brosché, M.; Kangasjärvi, J. ROS signalling loops-production, perception, regulation. Curr. Opin. Plant Biol. 2013, 16, 575–582. [Google Scholar] [CrossRef]
- Werdan, K.; Heldt, H.W.; Milovancev, M. The role of pH in the regulation of carbon fixation in the chloroplast stroma. Studies on CO2 fixation in the light and dark. Biochim. Biophys. Acta 1975, 396, 276–292. [Google Scholar] [CrossRef]
- Motohashi, K.; Hisabori, T. CcdA is a thylakoid membrane protein required for the transfer of reducing equivalents from stroma to thylakoid lumen in the higher plant chloroplast. Antioxid. Redox Signal 2010, 13, 1169–1176. [Google Scholar] [CrossRef]
- Miki, H.; Funato, Y. Regulation of intracellular signalling through cysteine oxidation by reactive oxygen species. J. Biochem. 2012, 151, 255–261. [Google Scholar] [CrossRef]
- Rehder, D.S.; Borges, C.R. Cysteine sulfenic acid as an intermediate in disulfide bond formation and nonenzymatic protein folding. Biochemistry 2010, 49, 7748–7755. [Google Scholar] [CrossRef] [PubMed]
- López-Calcagno, P.E.; Howard, T.P.; Raines, C.A. The CP12 protein family: A thioredoxin-mediated metabolic switch? Front. Plant Sci. 2014, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Gütle, D.D.; Roret, T.; Hecker, A.; Reski, R.; Jacquot, J.P. Dithiol disulphide exchange in redox regulation of chloroplast enzymes in response to evolutionary and structural constraints. Plant Sci. 2017, 255, 1–11. [Google Scholar] [CrossRef]
- Serrato, A.J.; Romero-Puertas, M.C.; Lázaro-Payo, A.; Sahrawy, M. Regulation by S-nitrosylation of the Calvin-Benson cycle fructose-1,6-bisphosphatase in Pisum sativum. Redox Biol. 2018, 14, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, J.; Liu, B.; Feng, D.; Da, Q.; Wang, P.; Shu, S.; Su, J.; Zhang, Y.; Wang, J.; et al. Evidence for a role of chloroplastic m-type thioredoxins in the biogenesis of photosystem II in Arabidopsis. Plant Physiol. 2013, 163, 1710–1728. [Google Scholar] [CrossRef]
- Yoshida, K.; Hisabori, T. Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc. Natl. Acad. Sci. USA 2016, 113, 3967–3976. [Google Scholar] [CrossRef]
- Cejudo, F.J.; González, M.-C.; Pérez Ruiz, J.M. Redox regulation of chloroplast metabolism. Plant Physiol. 2021, 186, 9–21. [Google Scholar] [CrossRef]
- Serrato, A.J.; Rojas-González, J.A.; Torres-Romero, D.; Vargas, P.; Mérida, A.; Sahrawy, M. Thioredoxins m are major players in the multifaceted light-adaptive response in Arabidopsis thaliana. Plant J. 2021, 108, 120–133. [Google Scholar] [CrossRef]
- Holmgren, A.; Björnstedt, M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995, 252, 199–208. [Google Scholar] [CrossRef]
- Schürmann, P.; Buchanan, B.B. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid. Redox Signal 2008, 10, 1235–1274. [Google Scholar] [CrossRef]
- Serrato, A.J.; Fernández-Trijueque, J.; Barajas-López, J.D.; Chueca, A.; Sahrawy, M. Plastid thioredoxins: A one-for-all redox-signalling system in plants. Front. Plant Sci. 2013, 4, 463. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P.; Thormählen, I.; Daloso, D.M.; Fernie, A.R. The unprecedented versatility of the plant thioredoxin system. Trends Plant Sci. 2017, 22, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Okegawa, Y.; Motohashi, K. Chloroplastic thioredoxin m functions as a major regulator of Calvin cycle enzymes during photosynthesis in vivo. Plant J. 2015, 84, 900–913. [Google Scholar] [CrossRef] [PubMed]
- Nikkanen, L.; Rintamäki, E. Thioredoxin-dependent regulatory networks in chloroplasts under fluctuating light conditions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130224. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Hara, S.; Hisabori, T. Thioredoxin selectivity for thiol-based redox regulation of target proteins in chloroplasts. J. Biol. Chem. 2015, 290, 14278–14288. [Google Scholar] [CrossRef]
- Mallén-Ponce, M.J.; Huertas, M.J.; Sánchez-Riego, A.M.; Florencio, F.J. Depletion of m-type thioredoxin impairs photosynthesis, carbon fixation, and oxidative stress in cyanobacteria. Plant Physiol. 2021, 187, 1325–1340. [Google Scholar] [CrossRef]
- Serrato, A.J.; Pérez-Ruiz, J.M.; Spínola, M.C.; Cejudo, F.J. A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 43821–43827. [Google Scholar] [CrossRef]
- Pérez-Ruiz, J.M.; Spínola, M.C.; Kirchsteiger, K.; Moreno, J.; Sahrawy, M.; Cejudo, F.J. Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 2006, 18, 2356–2368. [Google Scholar] [CrossRef]
- Pérez-Ruiz, J.M.; Naranjo, B.; Ojeda, V.; Guinea, M.; Cejudo, F.J. NTRC-dependent redox balance of 2-Cys peroxiredoxins is needed for optimal function of the photosynthetic apparatus. Proc. Natl. Acad. Sci. USA 2017, 114, 12069–12074. [Google Scholar] [CrossRef]
- González, M.; Delgado-Requerey, V.; Ferrández, J.; Serna, A.; Cejudo, F.J. Insights into the function of NADPH thioredoxin reductase C (NTRC) based on identification of NTRC-interacting proteins in vivo. J. Exp. Bot. 2019, 70, 5787–5798. [Google Scholar] [CrossRef]
- Gurrieri, L.; Fermani, S.; Zaffagnini, M.; Sparla, F.; Trost, P. Calvin-Benson cycle regulation is getting complex. Trends Plant Sci. 2021, 26, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Lennartz, K.; Plücken, H.; Seidler, A.; Westhoff, P.; Bechtold, N.; Meierhoff, K. HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b(6)f complex in Arabidopsis. Plant Cell 2001, 13, 2539–2551. [Google Scholar] [CrossRef]
- Page, M.L.; Hamel, P.P.; Gabilly, S.T.; Zegzouti, H.; Perea, J.V.; Alonso, J.M.; Ecker, J.R.; Theg, S.M.; Christensen, S.K.; Merchant, S. A homolog of prokaryotic thiol disulfide transporter CcdA is required for the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J. Biol. Chem. 2004, 279, 32474–32482. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, K.; Hisabori, T. HCF164 receives reducing equivalents froms stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J. Biol. Chem. 2006, 281, 35039–35047. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, U.; Correa Galvis, V.; Kunz, H.H.; Strand, D.D. The regulation of the chloroplast proton motive force plays a key role for photosynthesis in fluctuating light. Curr. Opin. Plant Biol. 2017, 37, 56–62. [Google Scholar] [CrossRef]
- Yamori, W.; Shikanai, T. Physiological functions of cyclic electron transport around photosystem I in sustaining photosynthesis and plant growth. Annu. Rev. Plant Biol. 2016, 67, 81–106. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, L.R.; Froehlich, J.E.; Cruz, J.A.; Savage, L.J.; Kramer, D.M. Multi-level regulation of the chloroplast ATP synthase: The chloroplast NADPH thioredoxin reductase C (NTRC) is required for redox modulation specifically under low irradiance. Plant J. 2016, 87, 654–663. [Google Scholar] [CrossRef]
- Sekiguchi, T.; Yoshida, K.; Okegawa, Y.; Motohashi, K.; Wakabayashi, K.I.; Hisabori, T. Chloroplast ATP synthase is reduced by both f-type and m-type thioredoxins. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148261. [Google Scholar] [CrossRef]
- Uflewski, M.; Mielke, S.; Galvis, V.C.; von Bismarck, T.; Chen, X.; Tietz, E.; Ruß, J.; Luzarowski, M.; Sokolowska, E.; Skirycz, A.; et al. Functional characterization of proton antiport regulation in the thylakoid membrane. Plant Physiol. 2021. [Google Scholar] [CrossRef]
- Wang, C.; Yamamoto, H.; Narumiya, F.; Munekage, Y.N.; Finazzi, G.; Szabo, I.; Shikanai, T. Fine-tuned regulation of the K+/H+ antiporter KEA3 is required to optimize photosynthesis during induction. Plant J. 2017, 89, 540–553. [Google Scholar] [CrossRef]
- Shikanai, T. Chloroplast NDH: A different enzyme with a structure similar to that of respiratory NADH dehydrogenase. Biochim Biophys. Acta 2016, 1857, 1015–1022. [Google Scholar] [CrossRef]
- Peng, L.; Shikanai, T. Supercomplex formation with photosystem I is required for the stabilization of the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Physiol. 2011, 155, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, S.D.; Ibrahim, I.M.; Aryal, U.K.; Puthiyaveetil, S. Stoichiometry of protein complexes in plant photosynthetic membranes. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148141. [Google Scholar] [CrossRef]
- Nikkanen, L.; Toivola, J.; Trotta, A.; Guinea Diaz, M.; Tikkanen, M.; Aro, E.M.; Rintamäki, E. Regulation of cyclic electron flow by chloroplast NADPH-dependent thioredoxin system. Plant Direct 2018, 2, e00093. [Google Scholar] [CrossRef] [PubMed]
- Courteille, A.; Vesa, S.; Sanz-Barrio, R.; Cazalé, A.C.; Becuwe-Linka, N.; Farran, I.; Havaux, M.; Rey, P.; Rumeau, D. Thioredoxin m4 controls photosynthetic alternative electron pathways in Arabidopsis. Plant Physiol. 2013, 161, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Casano, L.M.; Martín, M.; Sabater, B. Hydrogen peroxide mediates the induction of chloroplastic Ndh complex under photooxidative stress in barley. Plant Physiol. 2001, 125, 1450–1458. [Google Scholar] [CrossRef]
- Queval, G.; Foyer, C.H. Redox regulation of photosynthetic gene expression. Phil. Trans. R Soc. B 2012, 367, 3475–3485. [Google Scholar] [CrossRef]
- Lascano, H.R.; Casano, L.M.; Martín, M.; Sabater, B. The activity of the chloroplastic Ndh complex is regulated by phosphorylation of the NDH-F subunit. Plant Physiol. 2003, 132, 256–262. [Google Scholar] [CrossRef]
- Lin, C.S.; Chen, J.; Chiu, C.C.; Hsiao, H.; Yang, C.J.; Jin, X.H.; Leebens-Mack, J.; de Pamphilis, C.W.; Huang, Y.T.; Yang, L.H.; et al. Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 2017, 90, 994–1006. [Google Scholar] [CrossRef]
- Nashilevitz, S.; Melamed-Bessudo, C.; Izkovich, Y.; Rogachev, I.; Osorio, S.; Itkin, M.; Adato, A.; Pankratov, I.; Hirschberg, J.; Fernie, A.R.; et al. An orange ripening mutant links plastid NAD(P)H dehydrogenase complex activity to central and specialized metabolism during tomato fruit maturation. Plant Cell 2010, 22, 1977. [Google Scholar] [CrossRef]
- DalCorso, G.; Pesaresi, P.; Masiero, S.; Aseeva, E.; Schünemann, D.; Finazzi, G.; Joliot, P.; Barbato, R.; Leister, D. A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 2008, 132, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Rantala, M.; Rantala, S.; Aro, E.-M. Composition, phosphorylation and dynamic organization of photosynthetic protein complexes in plant thylakoid membrane. Photoch. Photobiol. Sci. 2020, 19, 604. [Google Scholar] [CrossRef] [PubMed]
- Okegawa, Y.; Motohashi, K. M-Type Thioredoxins regulate the PGR5/PGRL1-dependent pathway by forming a disulfide-linked complex with PGRL1. Plant Cell 2020, 32, 3866–3883. [Google Scholar] [CrossRef] [PubMed]
- Hertle, A.P.; Blunder, T.; Wunder, T.; Pesaresi, P.; Pribil, M.; Armbruster, U.; Leister, D.L. PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol. Cell 2013, 49, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Okegawa, Y.; Tsuda, N.; Sakamoto, W.; Motohashi, K. Maintaining the chloroplast redox balance through the PGR5-dependent pathway and the Trx system is required for light-dependent activation of photosynthetic reactions. Plant Cell Physiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Dann, M.; Leister, D. Evidence that cyanobacterial Sll1217 functions analogously to PGRL1 in enhancing PGR5-dependent cyclic electron flow. Nat. Commun. 2019, 10, 5299. [Google Scholar] [CrossRef]
- Deák, Z.; Sass, L.; Kiss, É.; Vass, I. Characterization of wave phenomena in the relaxation of flash-induced chlorophyll fluorescence yield in cyanobacteria. Biochim. Biophys. Acta 2014, 1837, 1522–1532. [Google Scholar] [CrossRef]
- Peltier, G.; Aro, E.-M.; Shinakai, T. NDH-1 and NDH-2 plastoquinone reductase in oxygenic photosynthesis. Annu. Rev. Plant Biol. 2016, 67, 55–80. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, Q.S.; Zhao, W.; Liu, Z.; Ma, M.Y.; Zhong, M.Y.; Wang, M.X.; Xu, B. The highly efficient NDH-dependent photosystem I cyclic electron flow pathway in the marine angiosperm Zostera marina. Photosynth. Res. 2020, 144, 49–62. [Google Scholar] [CrossRef]
- Hanke, G.; Mulo, P. Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ. 2013, 36, 1071–1084. [Google Scholar] [CrossRef]
- Voss, I.; Goss, T.; Murozuka, E.; Altmann, B.; McLean, K.J.; Rigby, S.E.; Munro, A.W.; Scheibe, R.; Hase, T.; Hanke, G.T. FdC1, a novel ferredoxin protein capable of alternative electron partitioning, increases in conditions of acceptor limitation at photosystem I. J. Biol. Chem. 2011, 286, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Chen, S.; Voon, C.P.; Wong, K.B.; Tikkanen, M.; Lim, B.L. FdC1 and leaf-type ferredoxins channel electrons from Photosystem I to different downstream electron acceptors. Front. Plant Sci. 2018, 9, 410. [Google Scholar] [CrossRef] [PubMed]
- Hanke, G.T.; Kimata-Ariga, Y.; Taniguchi, I.; Hase, T. A post genomic characterization of Arabidopsis ferredoxins. Plant Physiol. 2004, 134, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Kato, H.; Shinzaki, Y.; Horiguchi, S.; Shikanai, T.; Hase, T.; Endo, T.; Nishioka, M.; Makino, A.; Tomizawa, K.; et al. Ferredoxin limits cyclic electron flow around PSI (CEF-PSI) in higher plants--stimulation of CEF-PSI enhances non-photochemical quenching of Chl fluorescence in transplastomic tobacco. Plant Cell Physiol. 2006, 47, 1355–1371. [Google Scholar] [CrossRef]
- Hanke, G.T.; Hase, T. Variable photosynthetic roles of two leaf-type ferredoxins in Arabidopsis, as revealed by RNA interference. Photochem. Photobiol. 2008, 84, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Lehtimäki, N.; Lintala, M.; Allahverdiyeva, Y.; Aro, E.M.; Mulo, P. Drought stress-induced upregulation of components involved in ferredoxin-dependent cyclic electron transfer. J. Plant Physiol. 2010, 167, 1018–1022. [Google Scholar] [CrossRef]
- Blanco, N.E.; Ceccoli, R.D.; Vía, M.V.; Voss, I.; Segretin, M.E.; Bravo-Almonacid, F.F.; Melzer, M.; Hajirezaei, M.R.; Scheibe, R.; Hanke, G.T. Expression of the minor isoform pea ferredoxin in tobacco alters photosynthetic electron partitioning and enhances cyclic electron flow. Plant Physiol. 2013, 161, 866–879. [Google Scholar] [CrossRef]
- Zhao, J.; Qiu, Z.; Ruan, B.; Kang, S.; He, L.; Zhang, S.; Dong, G.; Hu, J.; Zeng, D.; Zhang, G.; et al. Functional inactivation of putative photosynthetic electron acceptor ferredoxin C2 (FdC2) induces delayed heading date and decreased photosynthetic rate in rice. PLoS ONE 2015, 10, e0143361. [Google Scholar] [CrossRef]
- Sétif, P.; Fischer, N.; Lagoutte, B.; Bottin, H.; Rochaix, J.D. The ferredoxin docking site of photosystem I. Biochim. Biophys. Acta 2002, 1555, 204–209. [Google Scholar] [CrossRef][Green Version]
- Müller, P.; Li, X.P.; Niyogi, K.K. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001, 125, 1558–1566. [Google Scholar] [CrossRef]
- Pinnola, A.; Bassi, R. Molecular mechanisms involved in plant photoprotection. Biochem. Soc. Trans. 2018, 46, 467–482. [Google Scholar] [CrossRef] [PubMed]
- Malnoë, A.; Schultink, A.; Shahrasbi, S.; Rumeau, D.; Havaux, M.; Niyogi, K.K. The plastid lipocalin LCNP is required for sustained photoprotective energy dissipation in Arabidopsis. Plant Cell 2018, 30, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Nakano, H.; Yamamoto, H.; Shikanai, T. Contribution of NDH-dependent cyclic electron transport around photosystem I to the generation of proton motive force in the weak mutant allele of pgr5. Biochim. Biophys. Acta Bioenerg. 2019, 1860, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Hallin, E.I.; Guo, K.; Åkerlund, H.E. Violaxanthin de-epoxidase disulphides and their role in activity and thermal stability. Photosynth. Res. 2015, 124, 191–198. [Google Scholar] [CrossRef]
- Simionato, D.; Basso, S.; Zaffagnini, M.; Lana, T.; Marzotto, F.; Trost, P.; Morosinotto, T. Protein redox regulation in the thylakoid lumen: The importance of disulfide bonds for violaxanthin de-epoxidase. FEBS Lett. 2015, 589, 919–923. [Google Scholar] [CrossRef]
- Lu, Y.; Du, J.-J.; Yu, Z.-B.; Peng, J.-J.; Xu, J.-N.; Wang, X.-Y. Identification of potential targets for thylakoid oxidoreductase AtVKOR/LTO1 in chloroplasts. Protein Pept. Lett. 2015, 22, 219–225. [Google Scholar] [CrossRef]
- Da, Q.; Sun, T.; Wang, M.; Jin, J.; Li, M.; Feng, D.; Wang, J.; Wang, H.-B.; Liu, B. M-type thioredoxins are involved in the xanthophyll cycle and proton motive force to alter NPQ under low-light conditions in Arabidopsis. Plant Cell Rep. 2018, 37, 279–291. [Google Scholar] [CrossRef]
- Naranjo, B.; Mignée, C.; Krieger-Liszkay, A.; Hornero-Méndez, D.; Gallardo-Guerrero, L.; Cejudo, F.J.; Lindahl, M. The chloroplast NADPH thioredoxin reductase C, NTRC, controls non-photochemical quenching of light energy and photosynthetic electron transport in Arabidopsis. Plant Cell Environ. 2016, 39, 804–822. [Google Scholar] [CrossRef]
- Brooks, M.D.; Sylak-Glassman, E.J.; Fleming, G.R.; Niyogi, K.K. A thioredoxin-like/β-propeller protein maintains the efficiency of light harvesting in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, E2733–E2740. [Google Scholar] [CrossRef]
- Bru, P.; Nanda, S.; Malnoë, A. A genetic screen to identify new molecular players involved in photoprotection qH in Arabidopsis thaliana. Plants 2020, 9, 1565. [Google Scholar] [CrossRef]
- Nikkanen, L.; Guinea Díaz, M.; Toivola, J.; Tiwari, A.; Rintamäki, E. Multilevel regulation of non-photochemical quenching and state transitions by chloroplast NADPH-dependent thioredoxin reductase. Physiol. Plant 2019, 166, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Amstutz, C.L.; Fristedt, R.; Schultink, A.; Merchant, S.S.; Niyogi, K.K.; Malnoë, A. An atypical short chain dehydrogenase/reductase functions in the relaxation of sustained energy dissipation in the antenna of photosystem II in Arabidopsis. Nat. Plants 2020, 6, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, B.B.; Balmer, Y. Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 2005, 56, 187–220. [Google Scholar] [CrossRef] [PubMed]
- Finazzi, G.; Barbagallo, R.P.; Bergo, E.; Barbato, R.; Forti, G. Photoinhibition of Chlamydomonas reinhardtii in State 1 and State 2: Damages to the photosynthetic apparatus under linear and cyclic electron flow. J. Biol. Chem. 2001, 276, 22251–22257. [Google Scholar] [CrossRef]
- Lemeille, S.; Willig, A.; Depége-Fargeix, N.; Delessert, C.; Bassi, R.; Rochaix, J.-D. Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol. 2009, 7, e1000045. [Google Scholar] [CrossRef]
- Gabilly, S.T.; Dreyfuss, B.W.; Karamoko, M.; Corvest, V.; Kropat, J.; Page, M.D.; Merchant, S.S.; Hamel, P.P. CCS5, a thioredoxin-like protein involved in the assembly of plastid c-type cytochromes. J. Biol. Chem. 2010, 285, 29738–29749. [Google Scholar] [CrossRef]
- Shapiguzov, A.; Chai, X.; Fucile, G.; Longoni, P.; Zhang, L.; Rochaix, J.D. Activation of the Stt7/STN7 kinase through dynamic interactions with the cytochrome b6 f complex. Plant Physiol. 2016, 171, 82–92. [Google Scholar] [CrossRef]
- Wunder, T.; Liu, Q.; Aseeva, E.; Bonardi, V.; Leister, D.; Pribil, M. Control of STN7 transcript abundance and transient STN7 dimerisation are involved in the regulation of STN7 activity. Planta 2013, 237, 541–558. [Google Scholar] [CrossRef]
- Rintamäki, E.; Martinsuo, P.; Pursiheimo, S.; Aro, E.-M. Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc. Natl. Acad. Sci. USA 2000, 97, 11644–11649. [Google Scholar] [CrossRef]
- Thormälen, I.; Zupok, A.; Rescher, J.; Leger, J.; Weissenberger, S.; Groysman, J.; Orwat, A.; Chatel-Innocenti, G.; Issakidis-Bourguet, E.; Armbruster, U.; et al. Thioredoxins play a crucial role in dynamic acclimation of photosynthesis in fluctuating light. Mol. Plant 2017, 10, 168–182. [Google Scholar] [CrossRef]
- Ancín, M.; Fernández-San Millán, A.; Larraya, L.; Morales, F.; Veramendi, J.; Aranjuelo, I.; Farran, I. Overexpression of thioredoxin m in tobacco chloroplasts inhibits the protein kinase STN7 and alters photosynthetic performance. J. Exp. Bot. 2019, 70, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Pesaresi, P.; Hertle, A.; Pribil, M.; Kleine, T.; Wagner, R.; Strissel, H.; Ihnatowicz, A.; Bonardi, V.; Scharfenber, M.; Schneider, A.; et al. Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 2009, 21, 2402–2423. [Google Scholar] [CrossRef] [PubMed]
- Pesaresi, P.; Pribil, M.; Wunder, T.; Leister, D. Dynamics of reversible protein phosphorylation in thylakoids of flowering plants: The roles of STN7, STN8 and TAP38. Biochim. Biophys. Acta 2011, 1807, 887–896. [Google Scholar] [CrossRef]
- Allen, J.F.; Santabarbara, S.; Allen, C.A.; Puthiyaveetil, S. Discrete redox signalling pathways regulate photosynthetic light-harvesting and chloroplast gene transcription. PLoS ONE 2011, 6, e26372. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Gama, F.; Molina-Navarro, M.M.; Gualberto, J.M.; Claxton, R.; Naik, S.G.; Huynh, B.H.; Herrero, E.; Jacquot, J.P.; Johnson, M.K.; et al. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe–2S] clusters. EMBO J. 2008, 27, 1122–1133. [Google Scholar] [CrossRef] [PubMed]
- Talib, E.A.; Outten, C.E. Iron-sulfur cluster biogenesis, trafficking, and signalling: Roles for CGFS glutaredoxins and BolA proteins. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118847. [Google Scholar] [CrossRef] [PubMed]
- Järvi, S.; Suorsa, M.; Aro, E.-M. Photosystem II repair in plant chloroplasts-regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta 2015, 1847, 900–909. [Google Scholar] [CrossRef]
- Lima-Melo, Y.; Gollan, P.J.; Tikkanen, M.; Silveira, J.A.; Aro, E.-M. Consequences of photosystem-I damage and repair on photosynthesis and carbon use in Arabidopsis thaliana. Plant J. 2019, 97, 1061–1072. [Google Scholar] [CrossRef]
- Allahverdiyeva, Y.; Suorsa, M.; Rossi, F.; Pavesi, A.; Kater, M.M.; Antonacci, A.; Tadini, L.; Pribil, M.; Schneider, A.; Wanner, G.; et al. Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J. 2013, 75, 671–684. [Google Scholar] [CrossRef]
- Sun, X.; Fu, T.; Chen, N.; Guo, J.; Ma, J.; Zou, M.; Lu, C.; Zhang, L. The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol. 2010, 152, 1263–1273. [Google Scholar] [CrossRef]
- Kley, J.; Schmidt, B.; Boyanov, B.; Stolt-Bergner, P.C.; Kirk, R.; Ehrmann, M.; Knopf, R.R.; Naveh, L.; Adam, Z.; Clausen, T. Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure. Nat. Struct. Mol. Biol. 2011, 18, 728–731. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Ozawa, S.-I.; Takahashi, Y.; Sakamoto, W. D1 fragmentation in photosystem II repair caused by photo-damage of a two-step model. Photosynth. Res. 2015, 126, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Aro, E.M.; Suorsa, A.; Rokka, A.; Allahverdiyeva, Y.; Paakkarinen, V.; Saleem, A.; Battchikova, N.; Rintamäki, E. Dynamics of photosystem II: A proteomic approach to thylakoid protein complexes. J. Exp. Bot. 2005, 56, 347–356. [Google Scholar] [CrossRef]
- Lindahl, M.; Tabak, S.; Cseke, L.; Pichersky, E.; Andersson, B.; Adam, Z. Identification, characterization and cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J. Biol. Chem. 1996, 271, 29329–29334. [Google Scholar] [CrossRef] [PubMed]
- Zaltsman, A.; Ori, N.; Adam, Z. Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopis. Plant Cell 2005, 17, 2782–2790. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Sakamoto, W. FtsH protease in the thylakoid membrane: Physiological functions and the regulation of protease activity. Front. Plant Sci. 2018, 9, 855. [Google Scholar] [CrossRef]
- Nishimura, K.; Kato, Y.; Sakamoto, W. Chloroplast proteases: Updates on proteolysis within and across suborganellar compartments. Plant Physiol. 2016, 171, 2280–2293. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, M.; Kieselbach, T. Disulphide proteomes and interactions with thioredoxin on the track towards understanding redox regulation in chloroplasts and cyanobacteria. J. Proteom. 2009, 72, 416–438. [Google Scholar] [CrossRef]
- Hall, M.; Mata-Cabana, A.; Akerlund, H.E.; Florencio, F.J.; Schröder, W.P.; Lindahl, M.; Kieselbach, T. Thioredoxin targets of the plant chloroplast lumen and their implications for plastid function. Proteomics 2010, 10, 987–1001. [Google Scholar] [CrossRef]
- Karamoko, M.; Cline, S.; Redding, K.; Ruiz, N.; Hamel, P.P. Lumen thiol oxidoreductase 1, a disulphide bond-forming catalyst, is required for the assembly of photosystem II in Arabidopsis. Plant Cell 2011, 23, 4462–4475. [Google Scholar] [CrossRef]
- Ferreira, K.N.; Iverson, T.M.; Maghlaoui, K.; Barber, S.; Iwata, S. Arquitecture of the photosynthetic oxygen-evolving center. Science 2004, 303, 1831–1838. [Google Scholar] [CrossRef] [PubMed]
- Roberts, I.N.; Lam, X.T.; Miranda, H.; Kieselbach, T.; Funk, C. Degradation of PsbO by the Deg protease HhoA is thioredoxin dependent. PLoS ONE 2012, 7, e45713. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Lee, J.; Kim, Y.; Bae, D.; Kang, K.Y.; Yoon, S.C.; Lim, D. Defining the plant disulfide proteome. Electrophoresis 2004, 25, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Marchand, C.; Le Marechal, P.; Meyer, Y.; Decottignies, P. Comparative proteomic approaches for the isolation of proteins interacting with thioredoxin. Proteomics 2006, 6, 6528–6537. [Google Scholar] [CrossRef]
- Wyman, A.J.; Yocum, C.F. Structure and activity of the Photosystem II manganese-stabilizing protein: Role of the conserved disulfide bond. Photosynth. Res. 2005, 85, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, J.; Shutova, T.; Melnik, B.; Chernyshov, S.; Marchenkov, V.; Semisotnov, G.; Klimov, V.; Samuelsson, G. Importance of a single disulfide bond for the PsbO protein of photosystem II: Protein structure stability and soluble overexpression in Escherichia coli. Photosynth. Res. 2008, 98, 391–403. [Google Scholar] [CrossRef]
- Butenko, Y.; Lin, A.; Naveh, L.; Kupervaser, M.; Levin, Y.; Reich, Z.; Adam, Z. Differential roles of the thylakoid luminal Deg protease homologs in chloroplast proteostasis. Plant Physiol. 2018, 178, 1065–1080. [Google Scholar] [CrossRef]
- Havaux, M. Plastoquinone in and beyond photosynthesis. Trends Plant Sci. 2020, 25, 1252–1265. [Google Scholar] [CrossRef]
- Ksas, B.; Légeret, B.; Ferretti, U.; Chevalier, A.; Pospíšil, P.; Alric, J.; Havaux, M. The plastoquinone pool outside the thylakoid membrane serves in plant photoprotection as a reservoir of singlet oxygen scavengers. Plant Cell Environ. 2018, 41, 2277–2287. [Google Scholar] [CrossRef]
- Ksas, B.; Becuwe, N.; Chevalier, A.; Havaux, M. Plant tolerance to excess light energy and photooxidative damage relies on plastoquinone biosynthesis. Sci. Rep. 2015, 5, 10919. [Google Scholar] [CrossRef]
- Carol, P.; Stevenson, D.; Bisanz, C.; Breitenbach, J.; Sandmann, G.; Mache, R.; Coupland, G.; Kuntz, M. Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 1999, 11, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wright, D.A.; Wetzel, C.; Voytas, D.F.; Rodermel, S. The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 1999, 11, 43–55. [Google Scholar] [CrossRef]
- Kambakam, S.; Bhattacharjee, U.; Petrich, J.; Rodermel, S. PTOX mediates novel pathways of electron transport in etioplasts of Arabidopsis. Mol. Plant 2016, 9, 1240–1259. [Google Scholar] [CrossRef] [PubMed]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fu, A. The plastid terminal oxidase is a key factor balancing the redox state of thylakoid membrane. Enzymes 2016, 40, 143–171. [Google Scholar] [CrossRef]
- Thiers, K.L.L.; da Silva, J.H.M.; Sartori, G.R.; Dos Santos, C.P.; Saraiva, K.; Roque, A.; Arnholdt-Schmitt, B.; Costa, J.H. Polymorphisms in plastoquinol oxidase (PTOX) from Arabidopsis accessions indicate SNP-induced structural variants associated with altitude and rainfall. J. Bioenerg. Biomembr. 2019, 51, 151–164. [Google Scholar] [CrossRef]
- Feilke, K.; Ajlani, G.; Krieger-Liszkay, A. Overexpression of plastid terminal oxidase in Synechocystis sp. PCC 6803 alters cellular redox state. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160379. [Google Scholar] [CrossRef]
- Borisova-Mubarakshina, M.M.; Vetoshkina, D.V.; Ivanov, B.N. Antioxidant and signalling functions of the plastoquinone pool in higher plants. Physiol. Plant 2019, 166, 181–198. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, M.-C.; Cejudo, F.J.; Sahrawy, M.; Serrato, A.J. Current Knowledge on Mechanisms Preventing Photosynthesis Redox Imbalance in Plants. Antioxidants 2021, 10, 1789. https://doi.org/10.3390/antiox10111789
González M-C, Cejudo FJ, Sahrawy M, Serrato AJ. Current Knowledge on Mechanisms Preventing Photosynthesis Redox Imbalance in Plants. Antioxidants. 2021; 10(11):1789. https://doi.org/10.3390/antiox10111789
Chicago/Turabian StyleGonzález, María-Cruz, Francisco Javier Cejudo, Mariam Sahrawy, and Antonio Jesús Serrato. 2021. "Current Knowledge on Mechanisms Preventing Photosynthesis Redox Imbalance in Plants" Antioxidants 10, no. 11: 1789. https://doi.org/10.3390/antiox10111789
APA StyleGonzález, M.-C., Cejudo, F. J., Sahrawy, M., & Serrato, A. J. (2021). Current Knowledge on Mechanisms Preventing Photosynthesis Redox Imbalance in Plants. Antioxidants, 10(11), 1789. https://doi.org/10.3390/antiox10111789





